« Prev Next »
From Gene to Genome
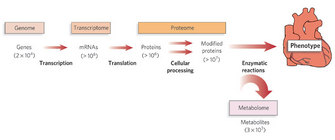
Over the last two decades, we have seen a shift of focus in the biomolecular sciences, from the desire to understand how a single gene functions, to understanding how all genes and gene products of a cell work together. This new paradigm for understanding interrelationships between networks of biological processes is termed systems biology, and when applied to the cell biology, it is primarily about the network of genes, mRNA and proteins. This approach to scientific analysis rests on a new way of looking at processes: understanding them within a much broader context than ever before. Consequently, this approach rests on recent technological progress that allows data to be analyzed in much larger amounts. What is the concept of systems biology of the cell — what are its roots, what are its aims, and (in a minor way) what are the technological assets it depends on? (Figure 1).
Molecular Biology Is Maturing
This approach has led to a staggering amount of data regarding individual genes. Scientists have been able to ask, and answer, hitherto unanswerable questions. On which chromosome are particular genes situated? What is their function? How and by which factors are they regulated? At the beginning of the twenty-first century, it is about time to make a new transition. Instead of focusing on the individual genes and their effects, biologists and biochemists have created an arsenal of techniques and methodologies to attack a question that is simple in itself, but which requires a lot of complex information to be answered: how is the combination of all genes within a cell able to govern all the reactions that go on in a cell? This question stems from an insight, characterized by one scientist, that "multi-scale dynamic complex systems formed by interacting macromolecules and metabolites, cells, organs, and organisms underlie most biological processes" (Vidal 2009). Yet, this is the goal of biology at the molecular level — to understand the collaborative functioning of the elements as being part of a system that forms a cell, a tissue, even an organism. Indeed, nothing in biology acts alone: everything acts in conjunction, opposition, and synergy with other elements. Genes regulate each other's activity, gene products join forces or inhibit each other, cells communicate, metabolites move from tissue to tissue, and everything is interconnected. Systems biology aims to understand this complexity.
Defining Systems Biology of the Cell
What does a systems biologist do? Systems biologists want to study all genes, expressed as messenger RNAs, and acting through proteins and metabolites, which play important roles in a specific cell or tissue, at a specific moment. In general, their efforts fall into four categories (defined by Kirschner 2005): quantitative measurement, creating mathematical models based on these quantitative data, reconstruction of how a cell reacts under different conditions, and the development of theories that will explain the large variation in different species in the way they react and respond to these conditions.
This goal has given rise to several new offshoots of the scientific technical subspecialties. First, there is the development of high-throughput analytical technology to be able to collect the vast array of data that systems biology requires. Fast and reliable methods have been developed to assess the different levels in a cell (the DNA or the genome, the RNA or the transcriptome, the protein pool or the proteome, and the metabolite pool or the metabolome). A few of the most important ones are discussed below. Second, there is the management of these vast sets of data, the field of bioinformatics (Rodrigo et al. 2008). Indeed, one of the most pressing needs emerging from this type of focus is an efficient way to store the ever growing collections of A's, T's, G's and C's (in databases such as the NCBI Genbank, or the protein-centered SWISS-PROT) which also allows for fast retrieval of data. Indeed, the amount of data that has come pouring in has grown exponentially (Boguski 1998). Apart from that, a number of applications had to be developed to facilitate a thorough analysis of these data. For example, there are analysis tools used to translate DNA to RNA and to protein sequences, identify sites for restriction enzymes to cut, and identify potential protein modifications. (See the collection at European Bioinformatics Institute, National Center for Biotechnolog Information, or Swiss Institute of Bioinformatics.) Other tools enable searches for homologous but not completely similar sequences, such as the widely known Basic Local Alignment Search Tool or BLAST (Altschul et al. 1990).
Moreover, there is the marriage between biology and mathematics. Using sets of differential equations, scientists try to summarize the biological data they amassed into working, robust mathematical models (Klein & Hölzel 2006, Westerhoff et al. 2009). In this respect, systems biology stems from earlier attempts to draw up a number of simple physical and mathematical models on what is called "biological self-organization." This approach is motivated by questions such as, how do biological structures come into existence, based on the properties of their constituent parts, and given their unique thermodynamic situation (Prigogyne 1967)? Models like those covered aspects of cell development, such as morphogenesis (Turing 1952, Ciliberto et al. 2003), microtubule formation (Pollard 2003), leaf phyllotaxis (Smith et al. 2006) or separate metabolic pathways (Polle 2001). In the future, they are bound to describe the behavior of the complete cell. Additionally, these models will allow for finely-tuned targeted manipulation of a cell's metabolism, and thus for more efficiency in genetic engineering (Goryanin et al. 1999, Patil et al. 2004).
On the Horizon
Molecular biology is currently at the center of a firestorm of new questions supported by new technologies. Out of this has emerged new methodologies like genomic sequencing and whole genome comparisons, which enable us to change the way we study living cells (Figure 2). These developments are not merely new steps along the beaten track, but herald a new way of thinking about and in biology (Westerhoff & Palsson 2004). After all, until now, biological complexity could only be understood through the study of the individual parts. In molecular biology and biochemistry, the questions have always been more of a qualitative nature, such as, have we cloned the relevant gene or not? Is it present and active? In which cells? Systems biology is bound to take away that burden, to become much more quantitative, and to offer a more integrated perspective on the inner workings of a cell, without having to resort to vagueness. This makes systems biology a difficult yet exact type of science. For many biologists (and in particular molecular biologists), equations were something that belonged in physics labs, and advanced mathematics and in depth statistics were a less prevalent and optional tool, with the exception of field ecologists, for whom multivariate statistics are typically present from the start. They will have to learn to cope with equations that will at least parallel those of advanced physics in their complexity. However, the rewards are enormous, and their magnitude is probably only beginning to surface.
References and Recommended Reading
Altschul, S. F., Gish, W. et al. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410 (1990).
Boguski, M. S. Bioinformatics - a new era. Trends in Biochemical Sciences, Supplement, Trends Guide to Bioinformatics, 1–3 (1998).
Ciliberto, A., Novak, B. et al. Mathematical model of the morphogenesis checkpoint in budding yeast. The Journal of Cell Biology 163, 61243–61254. doi: 10.1083/jcb.200306139.
Goryanin, I., Hodgman T. C. et al. Mathematical simulation and analysis of cellular metabolism and regulation. Bioinformatics 15, 749–758 (1999).
Kirschner, M. W. The meaning of systems biology. Cell 121, 503–504 (2005). doi: 10.1016/j.cell.2005.05.005
Klein, C. A. & Hölzel, D. Systemic cancer progression and tumor dormancy: mathematical models meet single cell genomics. Cell Cycle 5, 1788–1798 (2006).
Maxam, A. M. & Gilbert, W. A new method for sequencing DNA. Proceedings of the National Academy of Sciences, USA 74, 560–564 (1977).
Mullis, K. & Faloona, F. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods in Enzymology 155, 335–350 (1987).
Mullis, K., Faloona, F. et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symposia on Quantitative Biology 51, 263–273 (1986).
Pollard, T. D. The cytoskeleton, cellular motility and the reductionist agenda. Nature 422, 741–745 (2003).
Polle, A. Dissecting the superoxide dismutase-ascorbate-glutathione pathway in chloroplasts by metabolic modelling. Computer simulations as a step towards flux analysis. Plant Physiology 126, 445–462 (2001).
Prigogyne, I. Introduction to Thermodynamics of Irreversible Processes. 3rd ed. New York, NY: Interscience, 1967.
Rodrigo, A., Bertels, F. et al. The perils of plenty: what are we going to do with all these genes? Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences 363, 3893–3902 (2008). doi: 10.1098/rstb.2008.0173
Sanger, F., Nicklen, S. et al. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences, USA 74, 5463–5467 (1977).
Smith, R. S., Guyomarc'h, S. et al. A plausible model of phyllotaxis. Proceedings of the National Academy of Sciences, USA 103, 1301–1306 (2006). doi: 10.1073/pnas.0510457103
Turing, A. M. The chemical basis of morphogenesis. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences 237, 37–72 (1952).
Vidal, M. A unifying view of 21st century systems biology. FEBS Letters 583, 3891–3894 (2009). doi: 10.1016/j.febslet.2009.11.024
Watson, J. D. & Crick, F. H. C. Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Westerhoff, H. V. & Palsson, B. O. The evolution of molecular biology into systems biology. Nature Biotechnology 22, 1249–1252 (2004). doi: 10.1038/nbt1020.
Westerhoff, H. V., Winder, C. et al. Systems Biology: the elements and principles of Life. FEBS Letters 583, 3882–3890 (2009). doi: 10.1016/j.febslet.2009.11.018



 Figure 1: Systems biology allows us to think broadly.
Figure 1: Systems biology allows us to think broadly.