« Prev Next »
How can only 25,000–30,000 protein-coding genes in humans produce the massive variety of proteins, cells, and tissues that exist in our bodies? Alternative splicing is one major mechanism that makes the most of the precursor messenger RNAs (pre-mRNAs) transcribed from these few genes by processing the pre-mRNA into a diverse array of mature mRNAs that encode distinct proteins. Signal transduction pathways and cell cycle regulation are directly tied with regulation of alternative splicing. Scientists estimate that 15–60 percent of human genetic diseases involve splicing errors, making understanding splicing mechanisms and regulation an important area of research.
What Is Alternative Splicing, and Why Is It Important?
When scientists analyzed the initial sequence of the human genome, they were surprised by the relatively small number of protein-coding genes in humans compared with less complex organisms, such as fruit flies. How can so few genes encode all the information found in human cells?
A critical finding regarding the prevalence of alternative splicing was that a majority of human genes produce a wide variety of messenger RNAs (mRNA) that in turn encode distinct proteins (Johnson et al. 2003). Scientists estimate that 15–60 percent of human genetic diseases involve splicing mutations, either through direct mutation of the splice-site signals or through disruption of other components of the splicing pathway (Wang & Cooper 2007). Therefore, understanding what information in pre-mRNAs determines alternative splicing and how cells regulate alternative splicing is of critical importance.
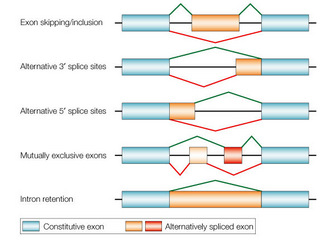
How does the splicing machinery distinguish between exons, which are part of the mature mRNA, and introns, which are removed from the pre-mRNA? Alternative splicing adds an extra layer of complexity, because regulatory sequences that sometimes designate an exon's inclusion into the mature mRNA dictate the exclusion of that exon under other conditions.
RNA splicing requires specific sequences in the pre-mRNA that mark where introns and exons are located. Occasionally, mutations in these sequences can lead to the production of mRNAs that are out of frame or unstable. Such mutations cause a problem with protein production despite the mutation being in noncoding sequence. The process of splicing is concurrent with mRNA transcription, with possible splicing regulatory roles for factors involved in chromatin structure, transcription, and other steps (Nilsen & Graveley 2010).
How Do Scientists Identify Alternatively Spliced mRNAs?
Scientists have estimated the prevalence of alternative splicing on a genome-wide scale using high-throughput approaches including microarrays, analysis of expressed sequence tags, and sequencing (Wang et al. 2008). In these experiments, scientists isolate RNA from cells and then convert it into complementary DNA, or cDNA, which they can tag with a fluorescent label during the reverse transcription process. They then analyze the cDNA through its ability to bind to small fragments of DNA tethered to a small chip, or microarray. If the chip includes DNA sequences that span exon-intron junctions or other sequences designating an alternatively spliced mRNA, then scientists can detect how common that alternatively spliced isoform is relative to other forms of the same pre-mRNA. In addition, they can also compare the splice isoforms among different tissues or developmental stages. Direct sequencing of the spliced mRNA is another technique researchers use to determine the frequency of alternatively spliced mRNA. Again, scientists isolate RNA from cells and then reverse transcribe it into the more stable cDNA for further analysis.
Recent predictions based on deep sequencing of cDNA fragments suggest that more than 90 percent of human pre-mRNA transcripts are alternatively spliced, many in a tissue-specific or developmental stage-specific fashion (Wang et al. 2008). How, though, do distinct cell types or cellular conditions regulate the production of multiple mRNAs from one primary RNA transcript? What signals are required in the pre-mRNA, and what splicing factors identify and splice the alternative exons? A recent study has attempted to answer this question using a computational approach by comparing features of RNAs derived from mouse tissue. The researchers' larger goal was to uncover a "splicing code" analogous to the "histone code" (Barash et al. 2010). Scientists are testing the applicability of the code deciphered based on mouse RNA data to other organisms, including humans. They are also investigating how variations in the splicing code contribute to disease.
Specific RNA Sequences Are Required for Regulation of Alternative Splicing
If we want to understand the splice code, then we need to analyze the existing information about constitutive and alternative-splicing sequences. Additional sequences are designated as splicing enhancers or silencers, and serine-arginine rich (SR) proteins bind these sequences to further regulate alternative splicing. Studying these diverse modes of mRNA splicing, which produces distinct isoforms, requires additional experiments. Microarray, transcriptome analysis, and other systems approaches provide information on the number and sequences of mRNA isoforms. For instance, how do we identify splicing enhancers and splicing silencers that are bound by SR proteins?
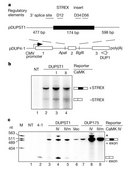
Xie and Black (2001) studied alternative splicing of potassium channel transcripts. These channels are essential during transmission of electrical signals in the brain, and the resulting alternatively spliced proteins have differing electrical properties. The pre-mRNA of the large gene SLO that encodes a potassium channel is too cumbersome to manipulate as a whole. Therefore, they designed minigenes with the relevant alternative exon, the so-called STREX exon (stress axis-related exon), but not the entire gene of interest (Figure 2).
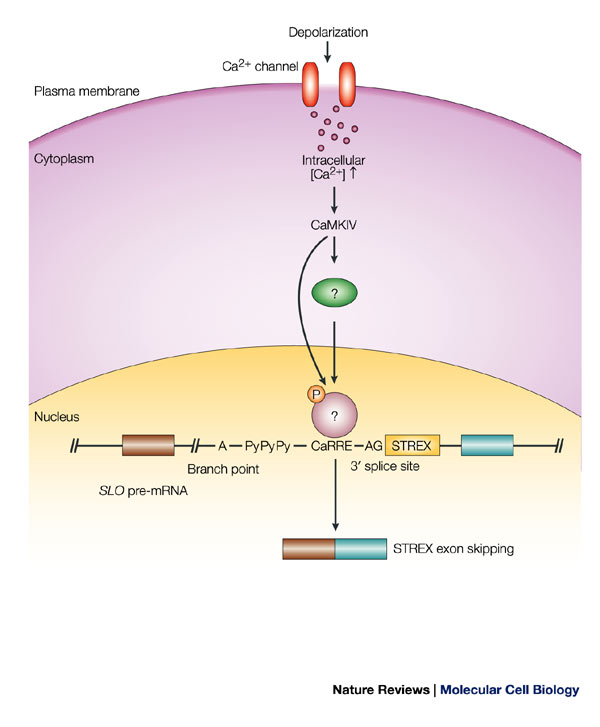
Alternatively spliced transcripts that include the STREX exon encode potassium channels that are more sensitive to Ca2+ than ones without it. Based on this observation, Xie and Black hypothesized that Ca2+/calmodulin-dependent protein kinases (CaMKs) play a role in repression of STREX exon splicing. Figure 2a illustrates the minigene they transfected into HEK cells, with or without plasmid expressing active forms of CaMKs. Following transfection, they isolated RNA and determined the amount of STREX exon inclusion using a radioactively labeled primer (Figure 2a; primer is called DUP1). If CaMKs act according to Xie and Black's hypothesis, the researchers would detect an increase in the amount of the smaller, -STREX, product compared with the larger, +STREX, band. Figure 2 shows the results of their assay. Lane M is a size marker they use for comparison with experimental products. As controls, they isolated RNA from untransfected cells, lane 1, which produced no product in this assay, and from cells transfected with a plasmid that did not contain the STREX exon (lane 2), which produced only the -STREX band, as expected. Lane 3 shows the splicing pattern of RNA from cells transfected with the +STREX plasmid, called DUPST1, and the majority of the product includes STREX. The data in Lane 4 confirm that CaMK IV does increase the amount of -STREX. This experimental sample has the STREX-containing plasmid along with CaMK IV, and now the STREX exon is not included as often.
What sequences in the alternative STREX exon account for the cells' response to CaMK IV? To investigate this question, they made a series of mutations in the STREX mini-gene and transfected these genes into cells with or without CaMK IV. They identified a purine-rich sequence that acts as a CaMK IV response element and have found that sequence in other genes expressed in the nervous system. Figure 3 summarizes scientists' understanding of the CaMK IV-mediated signaling pathway leading to skipping of the STREX exon (Shin & Manley 2004, Figure 5). Scientists have not yet identified the target(s) of the CaMK IV kinase and have not determined if the CaMK IV kinase phosphorylates SR proteins required for alternative splicing.
Cell Signaling Regulates mRNA Isoform Production
The RNA sequences required for splicing regulation are present in the pre-mRNA, the production of which is similar in different cell types. How, then, do cells determine what mature mRNA isoform to produce under different conditions? Cheng, Yaffe, and Sharp studied the regulation of the production of variants of CD44, a transmembrane glycoprotein, by cell signaling (Cheng, Yaffe, & Sharp 2006). They hypothesized that growth factor signaling influences the production of specific CD44 isoforms, which in turn act as signaling molecules, leading to a positive feedback loop (Cheng, Yaffe, & Sharp 2006). To test their hypothesis, they stimulated cells deprived of growth factors in the media with growth factor-rich media and then measured the production of different CD44 mRNA isoforms over time using RT-PCR. Addition of growth factors stimulated alternative splicing, as seen by the increase in band intensity in the variants relative to the standard and loading control. They examined the role of one of the variants in the positive feedback loop by knocking down its expression with small interfering RNA (siRNA) and found that expression of the alternative isoform is required for cell cycle progression (Cheng, Yaffe, & Sharp 2006). How does misregulation of the CD44 positive feedback loop contribute to uncontrolled cell growth? Scientists are actively pursuing these questions in their research.
Modification of Splicing Factors Influences Alternative Splicing
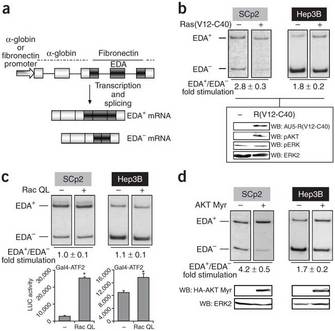
Because the pre-mRNA of fibronectin was large, Blaustein and colleagues developed its minigene, which contains three alternatively spliced regions. They transfected a plasmid containing the minigene with the extra domain A (EDA) exon into different cell types with or without a plasmid encoding the Ras protein, which stimulates EDA inclusion. Figure 4a illustrates the fibronectin minigene and the possible spliced products, EDA+ or EDA- mRNA (Blaustein et al. 2005, Figure 1a and 1b). After transfection, they isolated RNA and performed RT-PCR, determining the ratio of EDA+ to EDA- by measuring band intensities. The Ras(V12-C40) is a GTPase that activates a phosphatidylinositol 3-kinase (PI 3-kinase); the V12-C40 indicates that it is a double mutant; the researchers showed elsewhere that these mutations confer an "always on" state to the protein. Addition of Ras(V12-C40) led to an increase in EDA inclusion (EDA+), especially in the mammary epithelial cell line SCp2; Hep3B is a human hepatoma cell line. The panel below the RT-PCR data confirms that transfected cells expressed Ras(V12-C40) and that those cells also increased expression of phosphorylated AKT kinase, a kinase associated with activation of the PI 3-kinase signaling pathway. The other two panels in the bottom part of Figure 4 demonstrate that they added equal amounts of protein for their analysis. (Note that WB stands for western blot, indicating that they are analyzing protein rather than RNA in this part of the figure.)
Blaustein and colleagues concluded that activation of the PI 3-kinase signaling pathway by Ras(V12-C40) in turn activated AKT kinase, resulting in a change in the fibronectin splicing pattern to increased EDA exon inclusion. They had already identified sequences in the pre-mRNA in the EDA neighborhood necessary for regulated splicing, as well as the SR proteins that bind. Through additional experiments they fleshed out the cascade of events connecting AKT kinase to SR protein phosphorylation (Blaustein et al. 2005). Current areas of research include determining additional targets of Akt2 and other protein kinases, as well as finding how signaling networks interact to regulate alternative splicing and other aspects of gene expression.
A Splicing Regulator Inhibits Splicing During Mitosis
An important approach for demonstrating direct effects of specific splicing regulators on particular pre-mRNAs is to use a nuclear or cytoplasmic cell extract with radioactively labeled model pre-mRNA substrate. Shin and Manley performed this experiment in HeLA cell extracts supplemented with purified recombinant SR protein SRp38 (Sing & Manley 2002). SRp38 inhibited splicing of model substrates, but only if it was in the dephosphorylated state. They further characterized SRp38 splicing inhibition through preparation of mitotic cell extracts in comparison with the standard extract. The dephosphorylated form of SRp38 is present in the mitotic extracts, and splicing is inhibited. It is unclear why cells have an SR protein dedicated to splicing inhibition.
Although scientists have determined many factors contributing to regulation of alternative pre-mRNA splicing, their current challenge is to integrate results across multiple labs and research projects to reveal common threads or trends. For instance, they can compare characteristic sequences of the gene and corresponding pre-mRNA, as well as splicing factors, across multiple experiments. These metacomparisons will integrate large amounts of data, toward the ultimate goal of understanding alternative splicing at the systems level. Such analyses will likely uncover connections among features such as chromatin structure, transcription, and RNA processing across different cells and tissues. In addition, scientists are applying knowledge about alternative splicing and diseases to design therapies that restore appropriate regulation.
Summary
References and Recommended Reading
Barash, Y. et al. Deciphering the splicing code. Nature 465, 53–59 (2010) doi:10.1038/nature09000.
Blaustein, M. et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nature Structural and Molecular Biology 12, 1037–1044 (2005) doi:10.1038/nsmb1020.
Chen, M. & Manley, J. L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nature Reviews Molecular Cell Biology 10, 741–754 (2009) doi:10.1038/nrm2777.
Cheng, C., Yaffe, M. B., & Sharp, P. A. 2006. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes and Development 20, 1715–1720 (2006) doi:10.1101/gad.1430906.
Johnson, J. M. et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144 (2003) doi:10.1126/science.1090100.
Nilsen, T. W. & Graveley, B. R. Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010) doi:10.1038/nature08909.
Shin, C. & Manley, J. L. The SR protein SRp38 represses splicing in M phase cells. Cell 111, 407–417 (2002) doi:10.1016/S0092-8674(02)01038-3.
Shin, C. & Manley, J. L. Cell signalling and the control of pre-mRNA splicing. Nature Reviews Molecular Cell Biology 5, 727–738 (2004) doi:10.1038/nrm1467.
Stamm, S. Regulation of alternative splicing by reversible protein phosphorylation. Journal of Biological Chemistry 283, 1223–1227 (2008) doi:10.1074/jbc.R700034200.
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008) doi:10.1038/nature07509.
Wang, G-S. & Cooper, T. A. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nature Reviews Genetics 8, 749–761 (2007) doi:10.1038/nrg2164.
Xie, J. & Black, D. L. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature 410, 936–939 (2001) doi:10.1038/35073593.



 Figure 1: A schematic representation of alternative splicing
Figure 1: A schematic representation of alternative splicing