« Prev Next »
Inside the nucleus of a mammalian cell preparing for cell division, the chromatin strands are long, thin, fragile, and twisted helices that curve, loop, and tangle with each other — like noodles in a bowl of soup. Given this analogy, how can the cell separate the noodles into two identical portions, without breaking any? In eukaryotic cells, this problem occurs during every mitotic phase a cell enters. To resolve the mess, DNA must be condensed into discrete, compact form: chromosomes.
In the eukaryotic genome, DNA associates with proteins to form densely packed chromatin, a highly coiled and compact structure. The two types of chromatin, heterochromatin and euchromatin, are functionally and structurally distinct regions of the genome. Heterochromatin is densely packed and inaccessible to transcription factors so it is rendered transcriptionally silent (Richards and Elgin 2002). Euchromatin, on the other hand, is less condensed, more accessible, and therefore transcriptionally active (Hennig 1999). What follows is a discussion of heterochromatin, a transcriptionally inactive form that is vital to the stability of chromosomes throughout the cell cycle, and how studies in fission yeast have revealed key molecular drivers of heterochromatin formation.
What Is Heterochromatin?
Researchers previously viewed heterochromatin as a "genetic junkyard" with little biological function (Pardue and Hennig 1990). Intriguingly, mutations affecting heterochromatin formation at centromeres ultimately lead to defects in chromosome segregation, thus resulting in genome instability. Indeed, scientists now recognize the significant role that heterochromatin plays in maintaining the structural and functional integrity of specific chromosomal regions, such as centromeres and telomeres. Preserving such chromosomal integrity prevents tumor development. How do we define heterochromatin, biochemically? Distinct posttranslational modifications on histones, including methylation of histone H3 at lysine 9, are characteristics of transcriptionally inactive heterochromatin.
Centromeres and Heterochromatin
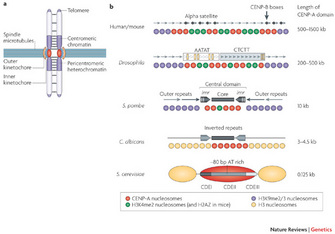
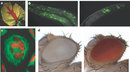
Centromeres perform essential functions for accurate chromosome segregation in two ways. One essential function is that centromeres attract cohesin, a protein that "glues" sister chromatids together in place. Another function is that a centromere serves as the physical locus (location) on a chromosome where kinetochores assemble to form the attachment site of spindle microtubules (Figure 1a). In metazoa, clusters of repetitive DNA are packaged into heterochromatin at the centromere region (Figure 1a, b). Typically, heterochromatin forms at pericentromeric (near the cellular centromere) and telomere regions that comprise repetitive DNA sequences (Bühler and Gasser 2009).
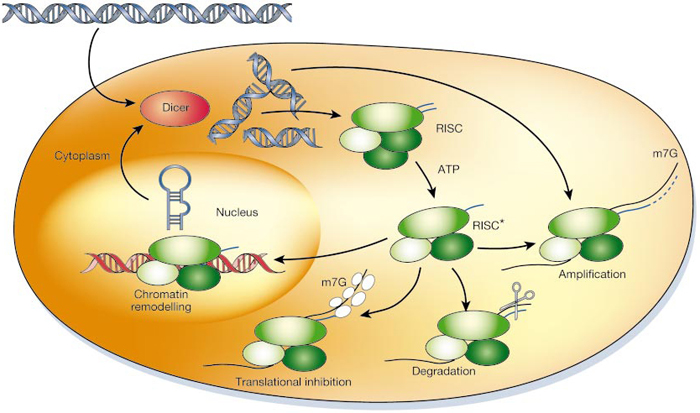
At centromeres, heterochromatin formation is directed by RNA interference (RNAi) a naturally occurring process in the nucleus of eukaryotic cells that silences gene expression (Figure 2). RNAi during heterochromatin formation is an important step in the assembly of chromosome segregation machinery at mitosis. A favored laboratory model organism, the fission yeast Schizosaccharomyces pombe, is a unicellular eukaryote suitable for genetic analysis of this complex process, as its centromere structure and RNAi machinery resemble those in metazoa. Since the 1950s, experiments in fission yeast have enlightened our understanding of cell cycle events, due to the organism's genetic tractability. Fission yeast have an entirely sequenced genome, and substantial genetic structural homology to complex eukaryotes (Wood et al. 2002; Mitchison 1957). What follows is an exploration of the potential involvement of two protein kinases in the epigenetic regulation of centromeric chromatin in fission yeast.
Why is S. pombe a Good Model Organism for the Study of RNAi in Heterochromatin?
S. pombe is a unicellular fungus with a rod-like shape which divides symmetrically. Remarkably, as a eukaryote with a small genome size of 4,979 protein-encoding genes, the structure of centromeres in fission yeast resembles that in metazoa. Within this yeast cell's chromosomes, the centromeres are also similar to those of metazoa, with large, repetitive DNA and complex structures that span 35–110 kb (kilobases). Also, unlike many eukaryotes, S. pombe does not possess redundant genes encoding key proteins required for RNAi pathways (Weaver 2008). Consequently, it is a good system to test the function of a single molecule in the RNAi signaling pathway, because a single gene perturbation is likely to have a measureable effect. In this way, the experiments are straightforward and the results are easier to interpret than in other eukaryotes carrying genes with redundant functions in RNAi processes.
RNAi and Heterochromatin Pre-mRNA Splicing Factors
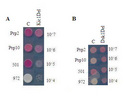
But how are these splicing factors regulated? Previous studies identified two proteins, Dsk1 (dis1-suppressing protein kinase) and Kic1 (kinase in the CLK/STY family) which may interact with a splicing factor involved in the RNAi-directed silencing. These protein kinases phosphorylate and regulate the cellular localization of some splicing factors such as serine/arginine (SR) proteins present in all metazoa (Tang et al. 2003; Tang et al. 2000; Tang et al. 1998). Covalent modification such as phosphorylation by protein kinases is a fundamental regulatory mechanism in all living organisms, and a reverse phosphorylation/dephosphorylation cascade is a major form of signal transduction in a cell. Do Dsk1 or Kic1 protein kinases influence RNAi-directed heterochromatin formation at centromeres in fission yeast? To answer this question, we first need to understand a genetic assay routinely used in fission yeast experiments.
A Clever Genetic Assay for Factors Involved in RNAi-Directed Silencing
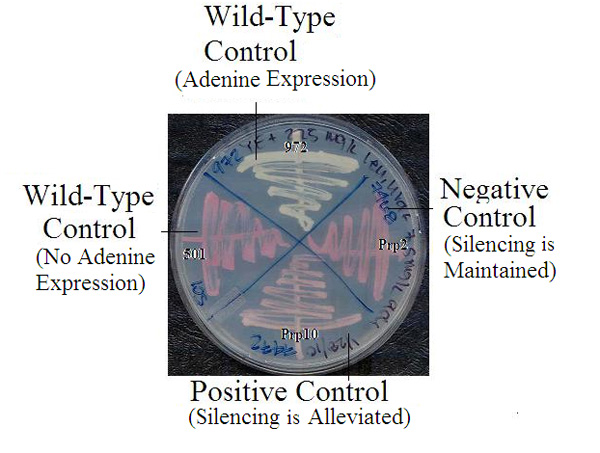
Robin Allshire's laboratory developed a robust and clever genetic assay to identify proteins involved in the heterochromatin formation at S. pombe centromeres. In this experimental system, they inserted a marker gene called ade6+ into a pericentromeric territory near the central domain, within outer repeats (Figure 1b). This region of the fission yeast genome typically exhibits heterochromatin organization in wild-type cells. Any genes from this region should not be expressed if heterochromatin formation is maintained.
The following is an example of how this powerful technique was recently used by an undergraduate researcher. To investigate whether the two protein kinases are involved in heterochromatin formation at centromeres, Nicole Duggan and her advisor, Irene Tang, employed a reverse "destructive" genetic logic which consists of three steps: mutating a gene (destructive), studying the resulting phenotype, and inferring relevant gene functions in the RNAi-directed transcriptionally silencing. They created the fission yeast strains that harbor the ade6+ reporter centromere, but have either dsk1+ or kic1+ deleted. The rationale was that the silencing state of marker ade6+ gene indicated the maintenance of the heterochromatin structure, whereas its expression reflected the alleviation of the silent state.
How did these researchers monitor the expression of ade6+ inserted in the centromeric region? There is a simple assay system for assessing genetic activity in fission yeast. If the reporter gene remains silent, the cells are defective in synthesizing adenine. When grown in media without sufficient supply of adenine, red-colored yeast colonies appear, due to accumulation of red intermediates in the metabolic pathway. Normally, yeast colonies would be white-colored if the gene was active. So a color comparison can show at a single glance whether or not the gene in question is active in the yeast colonies. Interestingly, partial alleviation of the silencing state (meaning partial expression) can result in intermediately-colored pink colonies (Figure 4). Duggan and Tang used this system to monitor gene silencing.
Are Dsk1 and Kic1 involved?
Summary
Transcriptionally inactive heterochromatin plays a vital role in sustaining stable structure of specialized chromosomal regions with repetitive DNA, such as centromeres and telomeres. Loss of integrity in these chromosomal areas can lead to detrimental effects and drive cancer development. Fission yeast centromere structure and RNAi-directed heterochromatin formation are highly analogous to those in metazoa, and thus provide a valuable model for dissecting the complex molecular mechanism. A robust genetic screen used to identify proteins in the RNAi pathway revealed that the protein kinase Kic1 may influence RNAi-directed heterochromatin formation and transcriptional gene silencing in fission yeast.
References and Recommended Reading
Allshire, R. C. & Karpen, G. H., Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nature Reviews Genetics 9, 923–937 (2008).
Bayne, E. H. et al. Splicing Factors Facilitate RNAi-Directed Silencing in Fission Yeast. Science. 322, 602–606. (2008).
Bühler, M. & Gasser, S. M. Silent chromatin at the middle and ends: lessons from yeasts. EMBO J. 28, 2149–2161. (2009).
Ekwall, K., Cranston, G. & Allshire, R. C. Fission Yeast Mutants That Alleviate Transcriptional Silencing in Centromeric Flanking Repeats and Disrupt Chromosome Segregation. Genetics. 153, 1153–1169. (1999).
Hannon, G. J. RNA interference. Nature. 418, 244–251. (2002).
Hannon, G. J. & Rossi, J. J, Unlocking the potential of the human genome with RNA interference. Nature. 341, 371–378. (2004).
Hennig, W. Heterochromatin. Chromosoma. 108, 1–9. (1999).
Ishii, K. et al. Heterochromatin Integrity Affects Chromosome Reorganization After Centromere Dysfunction. Science. 321, 1088–1091. (2008).
Lippman, Z.& Mattienssen, R. The role of RNA interference in heterochromatic silencing. Nature. 431, 364–370. (2004)
Mitchison, J. M. The growth of single cells. Schizosaccharomyces pombe. Expl. Cell Res. 13, 244–262. (1957).
Pardue, M. L. & Hennig, W. Heterochromatin: junk or collectors items? Chromosoma. 100, 3–7. (1990).
Richards, E. J. & Elgin, S. C. R. Epigenetic Codes for Heterochromatin Formation and Silencing: Rounding up the Usual Suspects. Cell. 108, 489–500. (2002).
Tang, Z. et al. The Kic1 kinase of Schizosaccharomyces pombe is a CLK/STY orthologue that regulates cell-cell separation. Experimental Cell Research. 283, 101–115. (2003).
Tang, Z. et al. Biochemical and Genetic Conservation of Fission Yeast Dsk1 and Human SR Protein-Specific Kinase 1. Molecular and Cellular Biology. 20, 816–824. (2000).
Tang, A., Yanagida, M., & Lin, R-J. Fission Yeast Mitotic Regulator Dsk1 Is an SR Protein-specific Kinase. Journal of Biological Chemistry. 273, 5963–5969. (1998).
Weaver, R.F. Molecular Biology, Fourth Edition. Boston, MA: McGraw Hill Press (2008).
White, S. A. & Allshire, R.C., Current Topics in Microbiology and Immunology, Chapter 8: RNAi-Mediated Chromatin Silencing in Fission Yeast. Springer Berlin Heidelberg (2008).
Wood, V. et al. The genome sequence of Schizosaccharomyces pombe. Nature. 415, 871–880. (2002).



 Figure 1: Centromere structure and organization.
Figure 1: Centromere structure and organization.