« Prev Next »
Bacteria are typically exposed to an ever-changing environment in which nutrient availability may increase or decrease radically. Bacteria respond to such variations in their environment by altering their gene expression pattern; thus, they express different enzymes depending on the carbon sources and other nutrients available to them. It would be wasteful to synthesize, for example, lactose-metabolizing enzymes in the absence of lactose. However, when lactose is the only available carbon source, bacteria must quickly induce lactose-metabolizing enzymes, or else they will die. In bacteria, this sort of genetic regulation is mediated at the level of transcription.
Bacterial Operons Are Coregulated Gene Clusters
Bacterial genes are organized into operons, or clusters of coregulated genes. In addition to being physically close in the genome, these genes are regulated such that they are all turned on or off together. Grouping related genes under a common control mechanism allows bacteria to rapidly adapt to changes in the environment.
The best-studied examples of operons are from the bacterium Escherichia coli (E. coli), and they involve the enzymes of lactose metabolism and tryptophan biosynthesis. Because the lactose (lac) operon shares many features with other operons, its organization and regulation are described in detail below.
The lac Operon
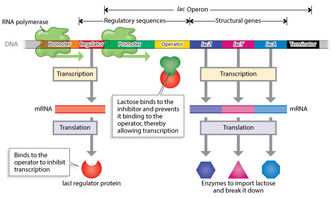
The organization of genes into an operon allows for simultaneous expression of all the genes that are located in cis (i.e., on the same contiguous piece of DNA) in the operon. Several features contribute to this characteristic of operons (Figure 1). First, all of the operon's genes are downstream of a single promoter. This promoter serves as a recognition site for the transcriptional machinery of the RNA polymerase complex. Second, all genes in an operon actually become part of a single messenger RNA molecule, which is subsequently translated into individual protein gene products.
Several other regulatory sequences also ensure coordinated regulation of the lac operon. These include the operator and the terminator. The operator is a special DNA sequence located between the promoter sequence and the structural genes that enables repression of the entire lac operon, following binding by the inhibitor (lac i) protein. Expression of the lac operon is, in fact, regulated by the presence of lactose itself. The ability to turn lactose-metabolizing genes on or off as a group therefore provides an efficient way to quickly adapt to environmental changes. The terminator, on the other hand, instructs the transcriptional machinery to terminate transcription. As such, the promoter serves as a transcriptional start site, the terminator serves as a stop site, and the operator helps determine whether transcription will occur.
Because of the common control mechanism for all of the genes in the lac operon, mutations in this operon can have multiple, or pleiotropic, effects. For example, mutations affecting the promoter can prevent all of the operon's genes from being expressed, because RNA polymerase will be unable to bind and commence transcription. Other mutations may affect expression of only some of the genes in the operon. For instance, nonsense mutations in the lac z gene encoding beta-galactosidase might interfere with expression of downstream permease and transacetylase genes by causing the ribosome to fall off prior to their translation. This kind of mutation is said to have a "polar" effect on the pathway in that it affects downstream genes but not upstream genes.
Thus, although bacteria may be considered simpler organisms than humans, it is clear that bacterial gene regulation is extremely efficient and that the bacterial genome is highly organized. Bacteria appear to be perfectly adapted to a variety of environments, and they are ready to respond to whatever environmental changes they encounter by employing elegant and complex regulatory mechanisms.
References and Recommended Reading
Jacob, F., & Monod, J. On the regulation of gene activity. Cold Spring Harbor Symposia on Quantitative Biology 26, 193–211 (1962)
Lawrence, J. G. Shared strategies in gene organization among prokaryotes and eukaryotes. Cell 110, 407–413 (2002)



 Figure 1: The lac operon in E.coli.
Figure 1: The lac operon in E.coli.