« Prev Next »
DNA is wound around clusters of eight histone proteins (H1–H8), and together, histones and DNA make up chromatin. Histones not only keep DNA organized, but they are also known to help regulate expression of genes. Specifically, modifications to histone proteins, such as methylation and acetylation, are thought to help keep genes active or silent, thus comprising a kind of code to be read by transcriptional regulators. The chromatin code is only just beginning to be cracked, however, and many questions remain. For example, do certain histone modifications always affect gene expression in the same way, or are there exceptions to the rules? To begin to address these exciting questions, researchers must first extend their study of histone modifications using a combination of chromatin immunoprecipitation (ChIP) and quantitative polymerase chain reaction (PCR). Ideally, any correlations that are noted between histone modification and gene expression will help reveal precisely how histones regulate expression.
The Histone Code
Current research in the area of histones and expression centers around what is known as the histone code. First proposed by researcher David Allis, this concept involves the notion that histone modifications can modulate the accessibility of DNA to transcription factors. For example, histone methylation might block DNA's access to transcription factors, while histone acetylation might change electrostatic interactions within the chromatin to open up DNA and allow transcription. These principles were especially well demonstrated by examining patterns of histone modification during blood cell development in chickens (Litt et al., 2001). Focusing on patterns of histone methylation and acetylation within the β-globin gene region, the investigators involved in this study detected changes that suggested interesting lessons for gene regulation.
The Chicken Study: A Closer Look
To examine changes in histone modification during blood cell development in chickens, the investigators started with whole chromatin from chicken blood cells of different ages or differentiation stages. They next broke up the DNA from each cell type and isolated histones that bore specific modifications, using antibodies and beads to pull down the modified histone of interest and its associated DNA, a technique known as chromatin immunoprecipitation, or ChIP. Next, the investigators queried the identity of the pulled-down DNA by PCR, using primer pairs to amplify short sequences spanning the entire β-globin gene region. The DNA that was pulled down by ChIP was robustly detected by PCR, while the DNA that was not pulled down was detected at significantly lower levels. Together, ChIP and quantitative PCR thus provided information about the pattern of histone modification, both across the β-globin gene region and over the entire course of blood cell development.
The investigators used this information to ask whether histone modification patterns exhibited a correlation with β-globin gene expression during blood cell development, which would provide evidence for the existence of a histone code of gene regulation (Figure 1). Of the three blood cell stages examined (HD24, 6C2, and 10-day erythrocyte cells), β-globin gene expression was only detected within 10-day erythrocytes. Interestingly, patterns of several types of histone modification, including acetylated (Ac) lysines 9 and 14 (K9, K14) and methylated (Me) lysine 4 (K4), revealed similar patterns along the β-globin gene in 10-day erythrocytes (Figure 1C). Importantly, a different pattern was observed in cells in which β-globin was not expressed, including HD24, 6C2, and brain cells (Figure 1A, B, D).
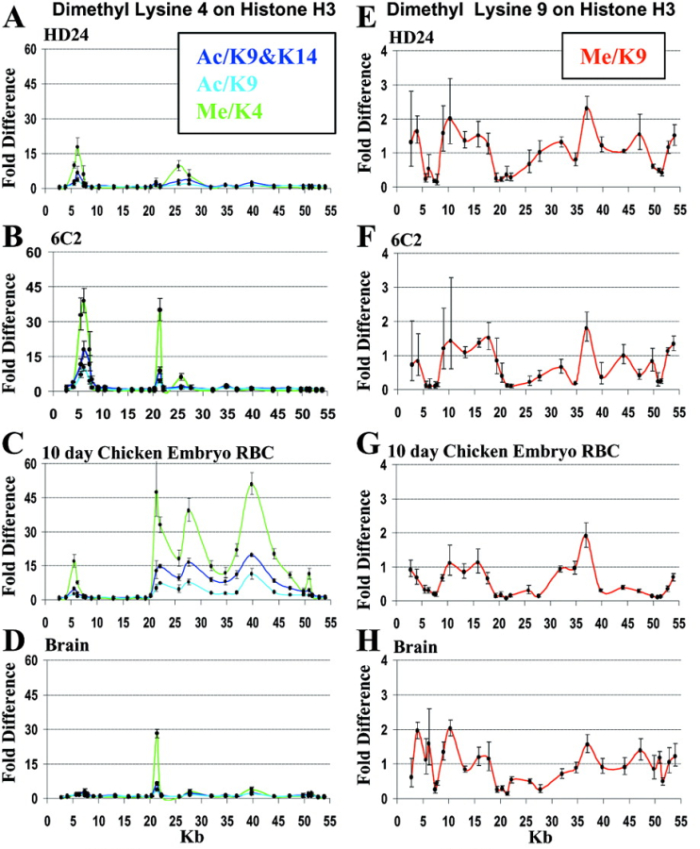
Interestingly, these "transcription-friendly" histone patterns were mutually exclusive with a different histone modification (Me/K9; Figure 1E–H). Moreover, regions with heightened concentrations of Me/K9 modifications were known to be transcriptionally silent regions. These observations strongly suggested that two major classes of histone modification might act in opposition to help specify a gene regulatory code for the β-globin gene. That is, transcriptional activation was associated with methylation at K4, which paralleled acetylation of K9 and K14, while transcriptionally inactive chromatin was associated with K9 methylation.
Summary
While this study supports the existence of gene regulation by a histone code, many more exciting questions remain. For example, how does the code end up there in the first place? This and other questions are under hot pursuit by researchers of the histone code.
References and Recommended Reading
Litt, M. D., et al. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293, 2453–2455 (2001)