« Prev Next »
How does the heart beat out its regular rhythm for decades? How does a muscle cell contract, allowing us to move and breathe? How do our eyes process the light and color before them? In each case, specialized, excitable cells perform these activities by changing their electrical properties. These changes are regulated by the variety of ion channels found in the plasma membrane. What are ion channels, what kinds of ion channels are found in cells, how do they work, and how do they change the electrical properties of cells? These questions fascinate scientists and have been the subject of much research. Although the basics of how a cell becomes excitable have been understood for more than half a century, crucial details of how ion channels work are just now being discovered.
Ionic Theory of Action Potential Propagation
What Is an Ion Channel?
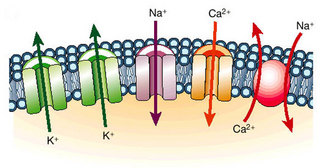
How Do Ion Channels Work?
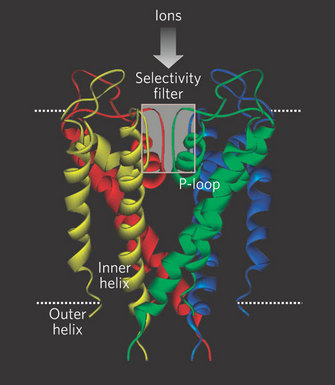
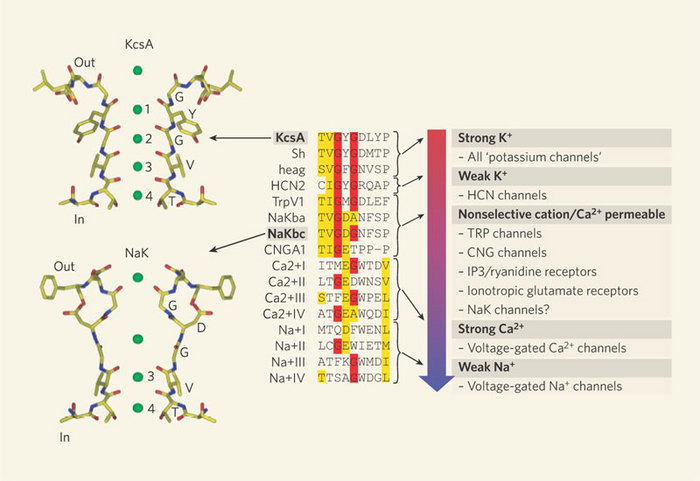
However, in 1998, Rod Mackinnon and colleagues succeeded in what many people thought was an impossible experiment: they solved the X-ray crystal structure of a K+ channel (Doyle et al. 1998; Figure 2). They used a K+ channel from the bacterium Streptomyces lividans (named KcsA) since it offered several experimental advantages for X-ray crystallography (smaller size, fewer hydrophobic domains, and it could be expressed in bacteria in large quantities). Furthermore, KcsA appeared to conduct K+ ions just like mammalian K+ channels, and it could even be blocked by some of the same toxins used to study mammalian channels. There were many important discoveries that came from this work, but with respect to selectivity, Mackinnon and colleagues made an important and fundamental discovery about how the K+ channel structure is arranged to select K+ ions over others, specifically Na+. The channel does this by permitting carbonyl oxygens — carbon molecules with a double bond to oxygen — on the polypeptide chain to strip off the water molecules surrounding the K+ ions. These oxygens function much like a "cage" to confine the K+ ions (Figure 3). However, this structure cannot strip away the water molecules surrounding Na+ ions. Therefore, the effective radius of the Na+ ion — which includes the water molecules — is much larger than that for K+, which effectively excludes Na+ from the channel. This important work with the Kcsa channel was followed by others, as it also revealed the mechanism of high rate of ion flux through the channel. However, it did not reveal the mechanism of voltage-sensitive gating, and this question remains controversial. For his important work on the structure of ion channels, Rod Mackinnon shared the Nobel Prize in Chemistry in 2003. Since this first crystal structure of an ion channel, the structures of a few other channels have also been solved. Each has revealed different mechanisms permitting the flux of ions across membranes. Given the tremendous diversity of channels, and their importance in normal cellular function, the quest to solve more ion channel structures remains one of the most exciting and active areas of current research.
Channelopathies
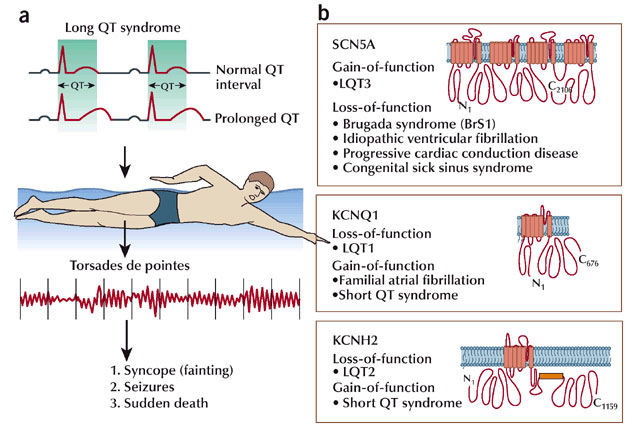
With the advances in molecular genetics described above, analyses of how defects in ion channel genes are associated with many human diseases has become possible. Indeed, epilepsies and cardiac dysfunction are just two examples of many illnesses stemming from mutations in ion channels. These are collectively referred to as channelopathies. For example, mutations in heart Na+ and K+ channels can lead to a cardiac channelopathy called Long QT syndrome (Ackerman 2004). People with these mutations are particularly susceptible to cardiac arrhythmias (Figure 4). With ever increasing ability to sequence whole genomes and perform genetic sequencing of patients, physicians and scientists now work together to discover how ion channel mutations lead to disease. Often, based on mutations identified in patients, scientists generate models that can recapitulate the effects of the mutation in a controlled environment. These models include genetically modified mice, cell culture models, and even tissues and cells collected from the patients themselves.
Summary
Ion channels are responsible for the transmembrane flux of ions that lead to the generation of action potentials. There is a stunning array of different types of channels that can be activated by different stimuli. Biophysical studies have begun to reveal the fundamental mechanisms responsible for the selectivity of a channel for one ion over another. And, both physicians and scientists are learning much about the role of ion channels in normal physiology from the discovery of human mutations. Nevertheless, the functions of many ion channels remain unknown, and their structure-function relationships are still undefined. Despite being very small structures, ion channels have large functions; they control the beating of a heart, the perception of sound or sight, or storage of a memory. Ultimately, these biological processes depend not on a single ion channel, but on all the ion channels in a cell and tissue network functioning in a coordinated manner. How this happens will continue to capture our imagination and attention for decades to come.
References and Recommended Reading
Ackerman, M. J. Cardiac channelopathies: it's in the genes. Nature Medicine 10, 463–464 (2004). doi:10.1038/nm0504-463.
Doyle, D. A., Morais Cabral, J. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998). doi: 10.1126/science.280.5360.69.
Hille, B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates, 2001).
Hodgkin, A. L. & Huxley, A. F. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. Journal of Physiology 116, 449–472 (1952a).
Hodgkin, A. L. & Huxley, A. F. The components of membrane conductance in the giant axon of Loligo. Journal of Physiology 116, 473–496 (1952b).
Hodgkin, A. L. & Huxley, A. F. The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. Journal of Physiology 116, 497–506 (1952c).
Hodgkin, A. L. & Huxley, A. F. A quantitative description of membrane current and its application to conduction and excitation in nerve. Journal of Physiology 117, 500–544 (1952d).
Hodgkin, A. L. & Huxley, A. F. Propagation of electrical signals along giant nerve fibers. Proceedings of the Royal Society B: Biological Science 140, 177–183 (1952e).
Hodgkin, A. L. & Huxley, A. F. Movement of sodium and potassium ions during nervous activity. Cold Spring Harbor Symposia on Quantitative Biology 17, 43–52 (1952f).
Hodgkin, A. L., Huxley, A. F. et al. Measurement of current-voltage relations in the membrane of the giant axon of Loligo. Journal of Physiology 116, 424–48 (1952).
Lai, H. C. & Jan, L. Y. The distribution and targeting of neuronal voltage-gated ion channels. Nature Reviews Neuroscience 7, 548–62 (2006). doi:10.1038/nrn1938.



 Figure 1: Ion channels function as pores to permit the flux of ions down their electrochemical potential gradient.
Figure 1: Ion channels function as pores to permit the flux of ions down their electrochemical potential gradient.