« Prev Next »
Long before the human genome was sequenced, scientists were able to map the location of genes to specific human chromosomes. In order to do this, researchers did not need to know the DNA sequence of each gene they were mapping. However, they did need to be able to track gene transmission in family pedigrees based on distinct phenotypes associated with different alleles of the same gene.
Recall that if two genes are located close to each other on the same chromosome, it is less likely that a recombination event will occur between them during gamete formation; indeed, it is highly likely that the genes are linked, so their two alleles will cosegregate. Thus, to map genes to specific chromosomes, scientists needed to identify other phenotypes associated with previously mapped genes that cosegregated with alleles of the gene they were trying to map. Cosegregation meant that the two genes were neighbors on the same chromosome. Next, scientists could use data on recombination frequencies to determine the distance between the two genes. This ability to map genes to specific human chromosomes and to define chromosomal gene neighborhoods provided an invaluable platform for the launch of the Human Genome Project and for mapping disease genes.
An Uncoiler Element Marks the Spot: Mapping a Human Gene to an Autosome for the First Time
Perhaps the most significant advance in mapping human autosomes occurred in 1968, when Donahue et al. published the first report describing the assignment of a gene to a human autosome. More specifically, Donahue and colleagues demonstrated that the Duffy blood group locus (called Fy), the key component of blood group heterozygosity, mapped to human chromosome 1.
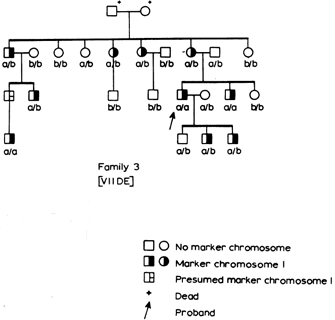
Fast forward to the mid-1960s, when Roger Donahue was a Ph.D. candidate in human genetics at Johns Hopkins University, and he was learning how to prepare metaphase chromosome spreads using his own white blood cells. (This was just prior to the development of chromosome banding techniques in 1971.) When preparing one of these spreads, Donahue noted that one of his two copies of chromosome 1 had what he described as an "uncoiler (Un) element" located next to the centromere; this region of the chromosome was loose and extended (Figures 1 and 2). Donahue then asked his family members to donate their blood samples, and he noted that the Un element was inherited in a dominant manner, although it was not associated with any deleterious phenotypes. Moreover, he predicted that the Un alteration was due to a change in the coiling structure of the DNA in that region (Donahue et al., 1968). At that time, the uncoiler phenotype of chromosome 1 was estimated to occur in 3 out of every 1,000 people.
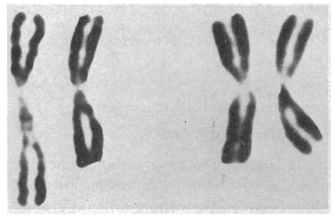
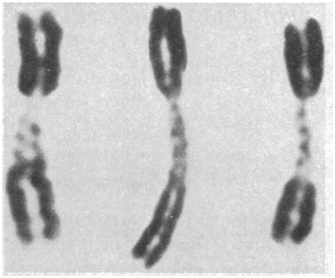
As part of his analysis, Donahue also examined the status of many different blood-cell markers among his family members, including ABO blood type status, Rh status, Duffy blood group type status, and more. He was interested in determining whether any of the blood-cell markers cosegregated with the Un element. Despite a small pedigree and relatively few blood-cell markers, Donahue found that the Un element nearly always segregated with the Fya allele within his family, as shown in the pedigree in Figure 3. In this pedigree, the first set of parents must have had the genotypes Fya/Fyb and Fyb/Fyb; after all, all of their offspring were either Fya/Fyb or Fyb/Fyb. In addition, of these offspring, three out of four Fya/Fyb individuals had the Un element, whereas none of the Fyb/Fyb individuals had this element, suggesting that the Un element cosegregates with the Fya allele. Indeed, in most cases, offspring who inherit the Fya allele from an Fya/Fyb parent with the Un element also have the Un element, and offspring who inherit the Fyb allele from an Fya/Fyb parent with the Un element never show the Un element.
Donahue combined data from the analysis of his family together with data from three much smaller families, two of which carried the Un allele and one of which had a chromosomal inversion in the same region of chromosome 1, to carry out a linkage analysis. Complete linkage is associated with a recombination frequency of 0%, whereas no linkage is associated with a recombination frequency of 50% (independent assortment) between the Un and Fy loci. Based on his calculations, Donahue estimated that the Un and Fy loci were separated by a genetic distance of 2.5 map units on chromosome 1 (Donahue et al., 1968).
Studies conducted prior to Donahue's research had reported that a congenital cataract locus, called Cae, was tightly linked to the Duffy blood group locus. However, the chromosomal location of the Cae locus was not known. Based on the linkage between Un and Fy and between Fy and Cae, Donahue predicted that the Cae locus was also present on chromosome 1 (Donahue et al., 1968). However, the relative order of the three genes could not be determined with the data in hand. Nonetheless, Donahue's keen insight coupled with his fortuitous discovery provided new vision to human geneticists by demonstrating that physical changes in chromosome structure could be combined with pedigree analyses to map genes to specific human chromosomes.
Pregenome Success Stories
In addition to identifying linkage between altered chromosome structure and a given gene, researchers also recognized that there are sites in our chromosomes that can vary from one person to the next. These sites are referred to as polymorphisms, and scientists were able to develop specific DNA probes that recognize and bind to these polymorphic DNA regions. For example, in some cases, the altered DNA sequence leads to a change in a restriction enzyme site, and a characteristic banding pattern occurs when a DNA probe is hybridized to the chromosomal DNA in a Southern blot analysis; this altered pattern of bands is referred to as a restriction fragment length polymorphism (RFLP). RFLP analyses can be used in combination with large pedigrees to determine RFLP patterns that are linked to various diseases, such as Huntington's disease. Those DNA probes that show a disease-associated RFLP can in turn be used to probe human-mouse somatic cell hybrids to determine the chromosomal location of the disease-associated gene. Indeed, by 1991, the list of human disease-associated genes identified using RFLP-based approaches was impressively long.
The same DNA probes used for RFLP analyses can also be used in a technique called fluorescence in situ hybridization. In this case, the probe is labeled with a fluorescent tag and incubated with a metaphase chromosome spread. The DNA probe will hybridize to its corresponding chromosomal region, and the fluorescent signal can be detected and mapped.
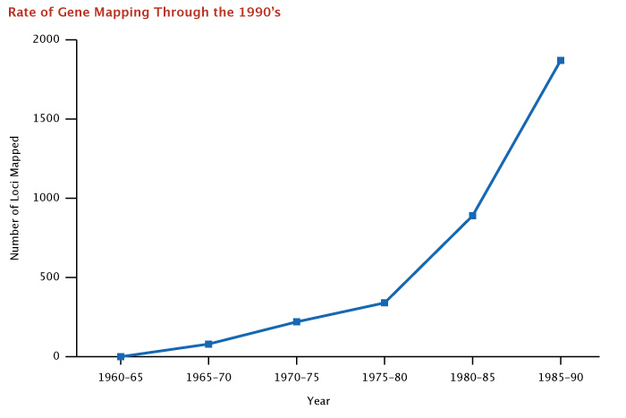
By using chromosomal banding patterns to identify chromosomal deletions or translocations, large pedigrees to facilitate linkage analyses, polymorphic regions of our chromosomal DNA that differ from one person to the next, and human-mouse somatic cell hybrids, researchers were able to rapidly establish links between specific genes and their corresponding chromosomes. Figure 4 (McKusick, 1991) shows the rapid increase in gene mapping that occurred following Donahue's initial mapping of the Duffy blood group locus to chromosome 1 in 1968.
A Launch Pad for the Human Genome Project
By mapping human genes to their respective chromosomal locations, researchers set the stage for the Human Genome Project. The discovery of recombinant DNA technology a few years earlier provided the tool necessary for such a large-scale project. The Human Genome Project led to the development of several important resources, including a public genome database (GDB) for depositing sequence information. Indeed, a database called Online Mendelian Inheritance in Man (OMIM), which is the online version of Victor McKusick's catalog "Mendelian Inheritance in Men" first published in 1966, annotates the allelic variants associated with every human gene. As the Human Genome Project proceeded, these databases and others grew exponentially. The ability to physically map gene sequences to specific chromosome regions provided the Human Genome Project with a critical starting point as researchers proceeded to fill in the tremendous gaps in the DNA sequence between these early pioneer genes.
References and Recommended Reading
McKusick, V. A. Current trends in mapping human genes. Federation of American Societies for Experimental Biology Journal 5, 12–20 (1991) (link to article)