« Prev Next »
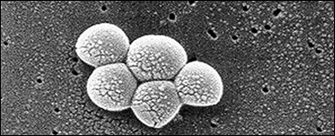
Mutation Rates and Bacterial Growth
Even if only a single S. aureus cell were to make its way into your wound, it would take only 10 generations for that single cell to grow into a colony of more than 1,000 (210 = 1,024), and just 10 more generations for it to erupt into a colony of more than 1 million (220 = 1,048,576). For a bacterium that divides about every half hour (which is how quickly S. aureus can grow in optimal conditions), that is a lot of bacteria in less than 12 hours. S. aureus has about 2.8 million nucleotide base pairs in its genome. At a rate of, say, 10-10 mutations per nucleotide base, that amounts to nearly 300 mutations in that population of bacteria within 10 hours!
To better understand the impact of this situation, think of it this way: With a genome size of 2.8 × 106 and a mutation rate of 1 mutation per 1010 base pairs, it would take a single bacterium 30 hours to grow into a population in which every single base pair in the genome will have mutated not once, but 30 times! Thus, any individual mutation that could theoretically occur in the bacteria will have occurred somewhere in that population—in just over a day.
Mutations, Antibiotic Resistance, and Staph Infections
Now, say that a few days after your initial infection with S. aureus, you decide to go to the local health center to have your wound examined. Maybe your finger is not healing as quickly as you had expected. Maybe its red color is a bit worrisome. Maybe the wound is starting to ooze a bit. Maybe you vaguely recall hearing or reading something about some kind of bacterial infection that is popping up on college campuses across the country and landing some students in the hospital. Concerned that your wound might be infected, the physician at the health center decides to prescribe an antibiotic.
Under a best-case scenario, the prescribed antibiotic would kill all of the replicating S. aureus cells in your body, mutant or otherwise, and your wound would quickly heal. After all, the potency of antibiotic treatment is why, when penicillin entered medical care in the 1940s, it was deemed a "miracle drug." Penicillin and other antibiotics have saved countless lives for more than half a century. Under a different scenario, however, any one of those mutations could give your S. aureus infection the ability to resist the particular drug you are being treated with. Luckily, in the real world, usually more than one mutation is required to generate drug resistance, and bacteria cannot double quite so quickly inside a person with a functioning immune system. But the problem still remains: The rapid division of bacterial cells causes them to evolve resistance to most treatments rather quickly.
Thus, although you are on antibiotics and you are otherwise healthy, a total of 600 mutations have accumulated by the time you go to bed that night. Any one of those mutations could give your staph infection the capacity to continue replicating, even in the presence of the antibiotic. All it takes is a single mutated S. aureus—one that, through one of a number of innovative biochemical means, does not die in the presence of whatever antibiotic the physician decided to prescribe—to render that antibiotic useless (at least for this particular infection). Moreover, when that mutant cell replicates, it will pass on its resistant phenotype to its daughter cells, and they to theirs. Thus, a rapidly growing proportion of the replicating bacteria still present in your body will be drug resistant. This is because the drug will kill only those cells that do not have the newly evolved drug-resistance capacity. Thus, the entire bacterial population will eventually become resistant to the prescribed antibiotic. When that happens, your infection will be said to be antibiotic resistant, and your physician will have to prescribe a different drug to combat it. MRSA: The Spread of Drug Resistance
In fact, there is a good chance that the staph infection you picked up from that contaminated doorknob is already antibiotic resistant. Most staph infections in humans are caused by methicillin-resistant Staphylococcus aureus, or MRSA, a drug-resistant phenotype that has been circulating for more than 45 years, almost as long as methicillin has been on the market. According to the U.S. Centers for Disease Control (CDC), in 2004, 63% of all reported staph infections in the United States were caused by MRSA (CDC, 2007). That figure represents a remarkable 300% increase in just 10 years' time. (In 1995, about 22% of all reported staph infections were MRSA, compared with only 2% in 1974.) The irony is that methicillin, a chemically modified version of penicillin, was developed in the 1950s as an alternative treatment for the growing proportion of staph infections already resistant to penicillin. At that time, about 60% of all staph infections were resistant to penicillin.
Needless to say, physicians no longer prescribe traditional antibiotics for methicillin-resistant staph infections (Micet, 2007). Instead, they usually administer "last-resort" intravenous vancomycin, although a growing number of doctors are now prescribing other newer antibiotics. Even with these options, scientists estimate that about 19,000 people in the United States die every year from MRSA (Klevens et al., 2007)—that's more than the number of U.S. residents and citizens that die from HIV/AIDS (about 17,000 every year). Of course, not all staph infections are deadly. In fact, about 30% to 40% of us have both methicillin-resistant and non-methicillin-resistant S. aureus living on the surface of our skin yet suffer no symptoms at all. Most deaths from S. aureus occur when what is normally "just" a skin infection enters the bloodstream and becomes invasive, affecting a person's internal anatomy. Moreover, most MRSA deaths occur in the hospital among patients being treated for other reasons and whose immune systems are too weak to fight off the infection, even when vancomycin is administered. In fact, MRSA used to occur only in hospitals. Also, as little as twenty years ago, MRSA did not spread via contaminated doorknobs (except in hospitals). The first so-called "community-acquired" MRSA infection—an infection occurring in a person neither hospitalized nor having had any recent contact with someone who was hospitalized—wasn't reported until the early 1990s. Since then, a growing number of MRSA cases and deaths have occurred outside of hospitals.
The Tip of the Antibiotic-Resistance Iceberg
As worrisome as MRSA is, it is just the tip of the iceberg, so to speak. In fact, there are a number of far more threatening drug-resistant bacteria in existence, such as Pseudomonas aeruginosa. P. aeruginosa poses a greater threat because it has certain biological features that make it more readily resistant to antibiotics than MRSA. For example, P. aeruginosa has a highly impermeable outer membrane, whereas MRSA does not. This outer membrane makes it more difficult for antibiotic chemical compounds to actually get inside the bacterial cell so that they can inflict damage. Moreover, once the antibiotic compounds are inside it, P. aeruginosa has what are known as efflux pumps, which can very quickly pump foreign compounds like antibiotics back out of the cell before they have a chance to do damage. MRSA does not have efflux pumps. Because of these biological features, P. aeruginosa infections either quickly evolve multidrug resistance or are drug-resistant from the start. Unlike with MRSA, however, the likelihood of picking up a P. aeruginosa infection from a doorknob in a school building is practically nil. P. aeruginosa infections occur mostly among hospital patients—at least for now.
In both the hospital and the community, antibiotic resistance has emerged as a major public health problem. In fact, some scientists consider it the most important public health problem of the twenty-first century. The problem exists not just because bacterial mutation rates lead to a rapid accumulation of mutations (including drug-resistant mutations), but also because of the selective pressures that antibiotics impose. If a drug-resistant phenotype were to evolve and there were no antibiotic present, then that phenotype would fare no better than any other bacterial phenotype. In other words, it wouldn't flourish, and it might even die out. It is only when antibiotics are used that drug-resistant phenotypes have a selective advantage and survive.
Of course, not all mutations confer resistance, and most probably have nothing at all to do with resistance. That said, bacterial populations with especially high mutation rates (so-called "hypermutable" strains) often have higher antibiotic resistance rates. For example, in a study of cystic fibrosis (CF) patients infected with P. aeruginosa (which is a major cause of sickness and death among CF patients), where more than a third of all CF patients had hypermutable P. aeruginosa infections, the hypermutable populations had higher resistance rates than isolates with "normal" mutation rates (Oliver et al., 2000). Plus, there are other ways that bacteria evolve resistance, in addition to spontaneous nucleotide base mutations. For instance, bacteria can acquire resistance genes through conjugation (i.e., from plasmid DNA) and from recombination with other bacterial DNA following transformation.
The emergence and spread of antibiotic resistance has become such an important public health problem that many federal and state public health agencies now distribute educational posters to encourage "good hygiene" practices—practices that prevent the spread of antibiotic-resistant bacteria from one person to another and help keep those mutant S. aureus bacteria off doorknobs. Maybe you have seen one of these posters in a school hallway, in a locker room, or in a public bathroom, imploring you to wash your hands with soap and water to help prevent disease.
To make MRSA matters even worse, in 2002, health care workers reported the first cases of vancomycin-resistant MRSA. In other words, even "last-resort" vancomycin doesn't always work. Therein lies the crisis: People are dying from "simple" bacterial infections, all because of a very low mutation rate.
Beyond Staphylococcus aureus
Bacterial resistance to antibiotics and other drugs is inevitable, and MRSA is just one example of why scientists must be so concerned about mutation rates. For instance, the inevitability of mutation and the development of resistance is one of the reasons why tuberculosis, thought to be essentially eradicated in the 1940s, is again on the rise in many countries. Multidrug-resistant strains of microorganisms will continue to be a major world health concern, keeping many scientists across disciplines occupied with the development and testing of novel therapies.
References and Recommended Reading
Klevens, R. M., et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Journal of the American Medical Association 298, 1763–1771 (2007)
Micet, S. T. Alternatives to vancomycin for the treatment of methicillin-resistant Staphyloccus aureus infections. Clinical Infectious Diseases 45, S184–S190 (2007)
Oliver, A., et al. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288, 1251–1254 (2000) doi:10.1126/science.288.5469.1251
U.S. Centers for Disease Control (CDC). MRSA: Methicillin-resistant Staphylococcus aureus in health care settings. (2007)