Abstract
Usutu (USUV), West Nile (WNV), and Zika virus (ZIKV) are neurotropic arthropod-borne viruses (arboviruses) that cause severe neurological disease in humans. However, USUV-associated neurological disease is rare, suggesting a block in entry to or infection of the brain. We determined the replication, cell tropism and neurovirulence of these arboviruses in human brain tissue using a well-characterized human fetal organotypic brain slice culture model. Furthermore, we assessed the efficacy of interferon-β and 2′C-methyl-cytidine, a synthetic nucleoside analogue, in restricting viral replication. All three arboviruses replicated within the brain slices, with WNV reaching the highest titers, and all primarily infected neuronal cells. USUV- and WNV-infected cells exhibited a shrunken morphology, not associated with detectable cell death. Pre-treatment with interferon-β inhibited replication of all arboviruses, while 2′C-methyl-cytidine reduced only USUV and ZIKV titers. Collectively, USUV can infect human brain tissue, showing similarities in tropism and neurovirulence as WNV and ZIKV. These data suggest that a blockade to infection of the human brain may not be the explanation for the low clinical incidence of USUV-associated neurological disease. However, USUV replicated more slowly and to lower titers than WNV, which could help to explain the reduced severity of neurological disease resulting from USUV infection.
Similar content being viewed by others
Introduction
Arthropod-borne viruses (arboviruses) are transmitted to humans by vector species, such as mosquitoes. The continued geographical expansion of these vector species, due to factors such as climate change, leads to increasing contact of naïve populations with newly emerging and re-emerging arboviruses1. Many arboviruses pose a significant threat to human health due to their ability to cause severe neurological disease in humans.
Usutu virus (USUV), West Nile virus (WNV) and Zika virus (ZIKV) are mosquito-borne arboviruses of the Flaviviridae family, genus Orthoflavivirus. The medical importance of ZIKV was highlighted following a rapid global expansion that led to large-scale outbreaks in Micronesia in 2007, Polynesia in 2013, and South America between 2015 and 20162. ZIKV infection during pregnancy can result in congenital Zika syndrome, characterized by severe neurological abnormalities in newborns3. However, neurological complications arising from infection with ZIKV have also been reported in adults4. WNV is responsible for recurrent outbreaks of West Nile neuroinvasive disease in many regions throughout the globe, and is increasingly emerging in Europe, leading to severe neurological complications, including encephalitis and paralysis5. USUV continues to emerge in Europe, with its presence identified in 14 European countries6 and in the United Kingdom7. Further, as the mosquito vectors, reservoir hosts and climatic conditions present in North America are similar to those in Europe, there may be a risk of USUV emergence in the Americas in the near future8. While USUV is closely related to and co-circulates with WNV in Europe, it has only been associated with sporadic cases of neurological disease in humans9,10, even though a higher seroprevalence in regions of USUV and WNV co-circulation has been identified11.
Despite the clinical significance of these arboviruses, the underlying cause of the differing severity and incidence of neurological disease resulting from infection with WNV and USUV is not yet understood. A critical factor impeding progress in this field is the lack of robust and relevant experimental models that faithfully recapitulate the in vivo crosstalk between neurons, immune cells and glia within the human brain. Conventional cell culture systems often lack the complexity of the human brain12,13, while animal models may not fully replicate human pathophysiology14,15.
The current study employed an innovative, well-characterized human fetal organotypic brain slice culture (hfOBSC) platform16, to address the existing research gap regarding the differential neurovirulence of emerging flaviviruses. This human brain model serves as a bridge to connect conventional in vitro and in vivo model systems, facilitating experimentation while preserving the microenvironment of human fetal brain tissue. The primary goal of this study was to model the early events of infection of the brain to determine whether USUV is able to infect human brain tissue to a similar extent as WNV. To achieve this, we investigated the cell tropism, virus spread, and neurovirulence exhibited by these closely related arboviruses within the hfOBSC platform. Identification of differences in these characteristics could aid in understanding the contrasting neurological outcomes resulting from infection with USUV versus WNV. We further compare these data with ZIKV, which has been previously preliminarily characterized in other human fetal brain models17,18. This methodological advancement and direct comparison address the urgent need for a more physiologically relevant CNS model16,19,20, promising valuable insights into the neurotropic characteristics of these arboviruses.
Results
Human fetal organotypic brain slice culture model facilitates studies on virus-host interactions in the human brain
To investigate the virus-host interactions of USUV, WNV and ZIKV within the human brain, we employed a recently published, innovative and well-characterized human fetal organotypic brain slice culture model16. Briefly, before initiating infections, we first confirmed previous characterization of the hfOBSC model by IHC for brain cell type specific markers (Fig. 1). The data showed robust staining for neuronal marker, MAP2, signifying the presence of neuronal cell types and for the astrocytic marker, GFAP, highlighting the abundance of astrocytes within the cultured brain slices. Additionally, we observed a scattered presence of the microglial marker, IBA1, and oligodendrocyte marker, OLIG2, indicating a diverse distribution of glial cells throughout the hfOBSC. As expected, considering the fetal source of the human brain tissue16,18,21, a substantial number of cells exhibited positive staining for the stem cell marker, SOX2. Indeed, we observed that some cells co-expressed cell markers, indicating a more progenitor, rather than mature, state22,23,24 (Fig. S1). These data showcase that culturing hfOBSCs successfully maintains the cellular composition of the main brain cell types observed in human fetal brain specimens of 17–20 weeks post-gestation25,26.
Characterisation of human fetal organotypic brain slice cultures. Immunohistochemistry staining of hfOBSC sections at 14 days post-culture establishment. Antibodies specific to SOX2 (stem cell marker), MAP2 (neuronal marker), OLIG2 (oligodendrocyte marker), GFAP (astrocyte marker) and IBA1 (microglial marker) were used to identify the main brain cell types present in sequential sections. Representative images of serial sections stained for the 4 markers of hfOBSC from 3 donors. Scale bars of large images and the zoomed inset represent 50 and 10 μm, respectively.
Human fetal organotypic brain slice culture model supports productive USUV, WNV and ZIKV infection
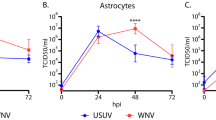
To assess the comparative susceptibility of human brain tissue to infection with USUV, WNV and ZIKV, we infected hfOBSC from 3 different donors and monitored viral replication over a 3-day period. The 72-h end-point was determined based upon the first experiment in which we observed titers of USUV and WNV to plateau after 48 hpi. USUV initially replicated to significantly higher titers than ZIKV (p = 0.0003), but showed similar titers by 48 hpi (Fig. 2A). WNV consistently showed higher titers than both USUV and ZIKV at all time points. At 72hpi, the distribution of infection throughout the brain slices was comparable between the three arboviruses (Fig. 2B and Supp videos S1-3). However, despite the differences in titers (Fig. 2A), infection percentages ranged between 2 and 4% and were not significantly different between the three viruses (Fig. 2C). Overall, these data show that the hfOBSC model is permissive and supports productive USUV, WNV and ZIKV infection, indicating that USUV is able to infect human brain tissue to a similar extent as ZIKV and that there is limited donor-to-donor variability.
Human fetal organotypic brain slice cultures support productive USUV, WNV and ZIKV infection. (A) Replication kinetics of USUV, WNV and ZIKV on hfOBSC cultures. Brain slices were inoculated with 106 TCID50 units of USUV, WNV, or ZIKV, then washed with PBS following the inoculation period. Titers of infectious virus in the culture supernatant were determined after 0, 24, 48, and 72 h post-infection by titration and assessment of cytopathic effect on Vero cells. Data shown are expressed as the mean ± SD of 3 independent experiments with 3–4 replicates per condition, per experiment. Error bars represent SD. ** p = 0.0031 (24hpi)/p = 0.0072 (72hpi). ***p = 0.0003. ****p < 0.0001; 2-way ANOVA. (B) 3D rendering showing the distribution of infection across representative brain slices from 3 donors. Renderings were made with z-stack confocal images of immunofluorescent stained whole, 3D tissue cleared hfOBSC infected with 106 TCID50 USUV, WNV or ZIKV. The hfOBSC slices were fixed in 4% paraformaldehyde at 72hpi. Flavivirus envelope protein and nuclei are shown in red and grey, respectively. Scale bars represent 400 μm. (C) Infection percentages of whole, 3D tissue-cleared brain slices quantified in their entirety using immunofluorescent staining for flavivirus envelope protein and nuclei. Data shown represents the mean ± SD of 2 independent experiments. Error bars represent SD. Ns, non-significant. One-way ANOVA.
USUV, WNV and ZIKV infect MAP2 positive cells in human fetal organotypic brain slices
As ZIKV has previously been described to infect post-mitotic committed neurons in ex vivo human fetal brain18, we determined whether USUV and WNV also preferentially infected neurons (MAP2-positive cells) in our hfOBSC model. For all three arboviruses, the majority of virus-infected cells co-stained for MAP2, thereby indicating a similar cell tropism (Fig. 3A,B). Notably, the size of infected cells differed between the viruses. Compared to ZIKV, USUV- and especially WNV-infected cells were smaller (Fig. 3C).
USUV, WNV and ZIKV infect MAP2-positive cells in the human fetal organotypic brain slice cultures: (A) Immunofluorescent staining of flavivirus envelope protein and the neuronal marker, MAP2, in hfOBSCs infected with 106 TCID50 USUV, ZIKV and WNV. The hfOBSCs were fixed with 4% paraformaldehyde at 72 h post-infection (72 hpi). Representative images of 3 donors. Scale bars represent 20 μm. (B) Quantification of the percentage of infected cells that costain for the neuronal marker MAP2. Full thickness, 40 × magnification z-stacks from 2 hfOBSC slices were quantified per condition. Data are presented as mean ± SD. (C) Quantification of the area of infected cell bodies in USUV-, WNV- and ZIKV-infected hfOBSCs at 72 hpi. Each data point represents one cell. Infected cells from 2 independent donors were quantified. **p = 0.012. ****p < 0.0001; one-way ANOVA.
USUV, WNV and ZIKV infection does not induce detectable cell death in human fetal organotypic brain slices
Neurovirulence can manifest through direct CPE within virally infected cells, or as a consequence of indirect immunopathology, both of which can lead to cell death27. Given that various programmed cell death pathways, such as apoptosis and necrosis, can alter cell size27,28, we aimed to investigate whether infection of the brain slices led to increased markers of cell-death, and whether this differed between the viruses. Cell death was determined by (1) LDH assay on conditioned culture media and (2) TUNEL staining on sections of virus- and mock-infected hfOBSC. However, surprisingly, none of the three arboviruses caused a significant increase in LDH release (Fig. S2A) or TUNEL staining (Fig. S2B) of virus- compared to mock-infected hfOBSC. These data indicated that, despite evident differences in the morphology of infected cells, USUV, WNV, and ZIKV infection did not induce a detectable increase in cell death in the hfOBSC model.
IFN-β inhibits replication of USUV, WNV and ZIKV in human fetal organotypic brain slices
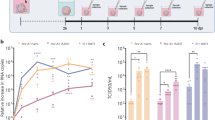
Type I IFNs have previously been used as a successful intervention in WNV-associated neurological disease29,30,31,32,33. However, this treatment is not always successful, especially in immunocompromised patients32,34,35, therefore development of more efficacious antivirals to treat flaviviral neurological disease is required. In anticipation of this, we determined the applicability of the hfOBSC as a preclinical model to test antiviral drugs. The hfOBSCs were pretreated for 24 h and throughout the infection course with predefined concentrations of human IFN-β or 2′C-methylcytidine (2CMC), a synthetic nucleoside analog with documented activity against WNV and Yellow Fever Virus36,37. We found that IFN-β treatment totally abolished replication of all three arboviruses, whereas 2CMC reduced the titers of ZIKV (p < 0.0001) and USUV (p = 0.0038) significantly but did not have a significant impact on the replication of WNV (Fig. 4).
Pre-treatment of human fetal brain slices with interferon-β prevents infection with USUV, WNV and ZIKV. Infectious viral titers at 48 h post-infection of hfOBSCs infected with 106 TCID50 USUV, WNV or ZIKV following over-night pre-treatment with the antiviral 2’C methyl-cytidine (25 μM) or interferon β (IFN-β; 50 ng/mL). Data are presented as mean ± SD for 3–4 replicates each of 3 donors per condition. Dotted line indicates limit of detection. **p = 0.0038. ****p < 0.0001; 2-way ANOVA.
Discussion
WNV is a neurotropic arbovirus that poses a significant threat to public health worldwide, leading to thousands of cases of neuroinvasive disease and hundreds of deaths every year, both in North America5, and increasingly in Europe38. In contrast, the closely related USUV has caused clinical disease in around 100 individuals, with diagnosable neurological disease reported in only 11 cases6, and there have been no reports of fatal infection11,39. The molecular mechanisms underlying this varying capacity to cause severe neurological disease in humans are still unknown, underscoring the unmet need for an appropriate experimental human brain model. Such a model requires the complexity of the in vivo crosstalk between relevant cell types in the CNS, that is absent in in vitro culture systems12,13, and the potential for translation to humans, which is lacking in commonly used animal models14,15.
Here, we employed a recently developed, innovative and well-characterized ex vivo model system of human, fetal brain tissue that provides a three-dimensional, physiologically relevant, human brain architecture16. While it does not fully recapitulate the cell composition and architecture of the adult brain, our model contains the main relevant cell types of the human brain, including microglia, to determine the cell tropism and pathogenesis of neurotropic viruses, such as herpes simplex virus13,31. With this ex vivo system, we aimed to investigate and compare the early events of infection to model the disease stage immediately following neuroinvasion of virus. We have shown that USUV is able to infect and replicate to a similar extent as ZIKV, while WNV replicates faster and to higher peak titers. Our observation that USUV, WNV and ZIKV can infect cells expressing MAP2, supports previous studies showing infection of neuronal cells by USUV, WNV and ZIKV18,40,41,42,43. The increased ability of WNV to infect and replicate within human neural tissue may contribute to the heightened severity of disease observed clinically, once the virus has gained access to brain tissue, but USUV still appears well able to replicate efficiently. This finding could suggest that the reduced incidence of USUV-associated neurological disease in humans may not be due to an inability to productively infect brain tissue, but may stem from an inability of this virus to gain access to the brain during natural infection. Future work should aim to determine the routes of neuroinvasion used by USUV, and how the lower titres observed with USUV may relate to the development and severity of neurological disease with this virus.
The size of infected cells was smaller in the USUV and WNV infected conditions, compared with ZIKV. Such variation could indicate virus-specific variation in tropism for neuronal subtypes of different sizes, or could result from induction of cell death pathways, such as apoptosis and necrosis, leading to cell shrinkage28,41. Yet, contrary to other studies18,40,41,42,43, we did not observe virus-induced cell death. However, previous work has shown comparable data to ours when investigating WNV infection of murine brain slice cultures, with minimal detection of WNV-induced apoptosis and LDH release at 3 dpi44,45, but increasing at later time-points45. Despite relatively high viral titers, the percentages of infected cells at 3 dpi were relatively low, which could explain the lack of detectable cell death resulting from infection of the hfOBSCs. Surprisingly, the infection percentage did not differ between viruses, despite the observed differences in titers. As we measure the presence of infectious virus in the supernatants of hfOBSCs, these data may indicate a variation in the amount of viral progeny released per infected cell, as has been shown for different strains of WNV46. This characteristic is difficult to determine in the complex hfOBSC system, but future work could employ more standardized, simplistic neuronal model systems to further delineate potential differences between USUV and WNV.
In WNV disease cases, neurological symptoms begin within 3 days after onset of viremia during which time there is an assumed invasion of virus into the CNS from the blood47, thereby indicating that within this timeframe, infection with WNV should lead to sufficient damage to the CNS to induce disease. One limitation of this study is that the hfOBSC model lacks blood-derived immune cells, which are important drivers of protection but also damage to brain tissue27,28,48. Indeed, pathological examination of fulminant cases of WNV has revealed extensive immune infiltration and immunopathology both in presence and absence of viral antigen49,50,51,52,53,54,55,56,57, indicating that damage to the CNS likely results from indirect immune mechanisms, in addition to potential direct effects induced by viral infection of neurons. Therefore, future work should investigate the direct and indirect mechanisms of cell death with addition of human immune cells over a longer timescale to model the clinically relevant environment that leads to WNV-associated pathology of the human brain, and thereby identify potential differences between USUV and WNV in their ability to induce such (immuno)pathology.
In addition to studying neuropathogenesis, we have shown that the hfOBSC model can also be used to assess the efficacy of intervention strategies. Within the human fetal brain tissue, IFN-β treatment was highly effective as it inhibited replication of all three arboviruses. 2CMC demonstrated a more nuanced antiviral effect, but still significantly reduced the titers of ZIKV and USUV. However, the impact of 2CMC treatment on WNV replication was not significant. This differential response to treatment may indicate variations in the interaction of the nucleoside analogue with the viral polymerases, which has been suggested to influence flaviviral pathogenesis58. These findings therefore prompt further exploration into the specific mechanisms influencing antiviral efficacy between viruses, within the complex microenvironment of human brain tissue. Our study further emphasises the importance of employing complex, human-relevant models to study therapeutic restriction of viral replication, as has been indicated with previous work on ZIKV59, and more recently with SARS-CoV-260.
In conclusion, we have shown that USUV can infect and replicate in ex vivo human brain tissue, showing no blockade to infection of the brain that could explain the reduced clinical incidence of USUV-related neurological disease compared to WNV. Future work should focus on investigating the routes of neuroinvasion to identify possible blockades preventing USUV from gaining access to the CNS. However, we found that USUV replicated more slowly and to lower titers than WNV, which could contribute to the reduced severity and fatality observed in cases of USUV-associated neurological disease compared with WNV.
In USUV- and WNV-infected brain slice cultures, the size of infected cells was smaller than ZIKV-infected cells. These findings could indicate tropism for different neuronal subtypes, or be as a result of morphological changes induced by infection with USUV and WNV, but not ZIKV, that could suggest an impact on the function of the infected cells. Finally, this study demonstrates the applicability of the hfOBSC model to study the pathogenesis of different neurotropic (arbo)viruses as well as to assess the efficacy of novel intervention strategies.
Methods
Preparation of human fetal organotypic brain slice cultures
Human fetal brain tissues from legally terminated second trimester pregnancies (17–20 weeks) were obtained by the Human Immune System (HIS)-Mouse Facility of Academic Medical Center (AMC; Amsterdam, The Netherlands), after written informed consent of the mothers for the use of tissues in research and with approval of the Medical Ethical Review Board of the AMC (MEC: 03/038) and Erasmus MC (MEC-2017-009). Abortions were not performed for medical indications and fetuses did not have any major anatomical deformities (checked by ultrasound prior to the procedure) or trisomy. The procedure was performed by in utero dissection and removal of fetal tissue specimens did not enable recovery of intact organs. Study procedures were performed according to the Declaration of Helsinki, and in compliance with relevant Dutch laws and institutional guidelines. The tissues obtained were anonymized and non-traceable to the donors. On request by the researchers, only gender and gestational age is provided. Upon removal of exterior blood vessels and meninges, brain tissue fragments (~ 0.5 × 0.5 cm) were cut into 350 µm-thick slices using a vibratome (Leica; type VT1200S) in artificial cerebrospinal fluid (aCSF) under constant oxygenation (95% O2, 5% CO2) as described previously61. Slices were transferred to 12 mm transwell plates with polyester membrane inserts (4 µm pore size; Corning) to recuperate the tissue in recovery medium composed of a 7:3 (v/v) mixture of Neurobasal media and advanced DMEM/F12 culturing medium (both Life Technologies) supplemented with 20% heat-inactivated fetal bovine serum (FBS) and antibiotics. Following 1 h (hr) incubation in a CO2 incubator at 37 °C, the recovery medium was replaced with optimized hfOBSC serum-free culture medium containing a 7:3 (v/v) mixture of Neurobasal media and advanced DMEM/F12 culturing medium with essential growth factors. To ensure fluidic flow in the transwell system, 750 µl medium was added to the basolateral compartment and 50 µl medium added to the apical compartment. The culture medium of the hfOBSCs was refreshed every 48 h. Detailed information on the development, characterization and culture conditions of the hfOBSC has been described recently16.
Virus strains and culturing
All viruses were grown and passaged on Vero cells (African green monkey kidney epithelial cells, ATCC CCL-81) at a multiplicity of infection of 0.01 for 5–6 days in Dulbecco’s modified Eagle’s medium (DMEM; Lonza) with 2% FBS (Sigma-Aldrich), 100 U/ml penicillin, 100 ug/ml streptomycin (Lonza) and 2 mM L-glutamine (Lonza). Supernatants were harvested at the indicated times post-infection, spun down at 4000 g for 10 min and aliquoted and frozen at − 80 °C. The virus strains used in this study were USUV (lineage Africa 3, GenBank accession MH891847.1), WNV (lineage 2, GenBank accession OP762595.1) and ZIKV (Suriname 2016, KU937936). The USUV and WNV strains used represent prevalent strains currently circulating in Europe62,63, thereby modelling the current risk situation in Europe. The ZIKV strain used represents the Asian lineage responsible for the large scale outbreaks of congenital disease in the Americas64. All virus stocks were sequenced and used at passage 3.
Flavivirus infection and antiviral treatment of human fetal organotypic brain slice cultures
Brain slice cultures were used for experiments after day 3 post-sample acquisition and culture establishment. All experiments were performed on tissues from 3 independent donors, which were obtained on different days, for which 3 to 4 hfOBSC cultures per condition were used in each experiment. In case of antiviral treatment, media from both the apical and basolateral compartments was removed and replaced with culture medium supplemented with 50 ng/ml recombinant human interferon beta (IFN-β; Peprotech), 25 µM 2′-C-Methylcytidine (2CMC), (which was kindly provided by Johan Neyts; Lab of Virology, Antiviral Drug & Vaccine Research, KU Leuven, Belgium) a nucleoside analogue which acts to inhibit viral RNA dependent RNA polymerases, or the respective vehicle consisting of PBS plus 0.025% DMSO. 2′C methylcytidine dosage and toxicity was determined on Vero cells. Vero cells were pretreated with doubling doses for 2 h prior to MOI 0.01 infection with USUV, then incubated for 5 days. The minimum effective dose that completely inhibited USUV-induced CPE was 12.5 µM. The minimum toxic dose in which 2′C methylcytidine alone killed uninfected Vero cells was 100 µM (data not shown). Drug treated hfOBSCs were incubated overnight at 37 °C prior to infection and treatment was maintained throughout the infection course. On the day of infection, all culture medium of the apical compartment was removed before addition of 106 TCID50 virus inoculum. As determination of the exact cell number in each hfOBSC culture used for infection was not possible, we standardized the inoculation dose to input of viral titer, rather than cell number. This relatively high inoculation dose was determined to maximize the chance of infection, and based on previous literature using ZIKV on ex vivo fetal brain3. The hfOBSCs were returned to the incubator at 37 °C for 1 h and subsequently washed 2–3 times with PBS to remove the inoculum, allowing for detection of virus release into the supernatant with time. The hfOBSC transwells were transferred to a clean culture plate and 750 µl of culture medium was added to the basolateral compartment and 50 µl added to the apical compartment. The complete volume of medium in both compartments was replaced every 24 h. The harvested supernatants were stored at − 80 °C for virus titration.
Virus titration
Tenfold serial dilutions of culture supernatants were inoculated onto a semiconfluent monolayer of Vero cells in a 96-well plate (2.3 × 104 cells/well) in 3 technical replicates. Cytopathic effect (CPE) was used as read out and determined at 6 days post-infection (6 dpi). Virus titers were calculated as the 50% tissue culture infective dose (TCID50) using the Spearman-Kärber method65. An initial 1:10 dilution of supernatant resulted in a detection limit of 31.6 TCID50/ml.
In situ analysis of brain slices
Formalin-fixed paraffin-embedded (FFPE) brain slices were serially sectioned at 4 µm thickness. Heat-induced antigen retrieval was performed using conventional citric acid buffer (pH 6.0). Consecutive sections from three different levels of the brain slices (i.e. apical, middle and basal level) were immunohistochemically stained (IHC) with the following primary antibodies: mouse anti-SOX2 (stem cell marker, R&D, 1:100), rabbit anti-Iba1 (microglia marker; Wako, 1:500), rabbit anti-GFAP (glial fibrillary acidic protein, astrocyte marker; Dako, 1:500), rabbit anti-Olig2 (oligodendrocyte transcription factor 2, oligodendrocyte marker; clone EPR2673, Abcam, 1:200) or guinea pig anti-MAP2 (microtubule-associated protein 2, neuron marker; Synaptic Systems, 1:300). Next, sections were washed and incubated with the appropriate secondary antibody including rabbit anti-mouse Ig biotinylated (Dako, 1:200), goat anti-rabbit Ig biotinylated (Dako, 1:200), or rabbit anti-guinea-pig IgG (H + L) horseradish peroxidase (HRP)-conjugated (Invitrogen, 1:200), respectively. The HRP-labeled streptavidin (Dako, 1:300) was applied to sections with biotinylated antibodies, followed by 3-amino-9-ethylcarbazole substrate. Sections were counterstained with hematoxylin, mounted with Kaiser’s glycerol, and scanned using the Hamamatsu NanoZoomer 2.0 HT (Hamamatsu).
3D tissue clearing and immunofluorescent staining
Whole brain slices were fixed in 4% paraformaldehyde (PFA) for a minimum of 24 h before undergoing tissue clearing and immunofluorescent (IF) staining with the 3D Cell Culture Clearing Kit (Abcam) as per the manufacturers protocol. Primary antibodies used were: mouse anti-flavivirus envelope protein (D1-4G2-4-15 hybridoma; ATCC, USA, 1:250), rabbit anti-MAP2 (Millipore, 1:200), guinea-pig anti-MAP2 (Synaptic Systems, 1:200) and rabbit anti-SOX2 (Abcam, 1:100). Next, sections were washed and incubated with the appropriate secondary antibody including donkey anti-mouse IgG AF488/555, donkey anti-rabbit AF488/555 and donkey anti-guinea-pig AF647 (Invitrogen). ToPro (DNA staining; Thermo Fisher Scientific, 1:1000) was used to stain cell nuclei. Images were obtained using a Zeiss LSM 700 laser scanning microscope.
Cryosectioning and immunofluorescent staining
The PFA-fixed whole brain slices were incubated in 30% sucrose at 4 °C for 24 h and subsequently placed in Optimal cutting temperature compound (Agar Scientific) before snap freezing on dry ice. Frozen tissue was then cut into 5 µm thick sections using a cryostat (Thermo Fisher Scientific, type HM525nx). Slides were permeabilized and blocked with 0.5% triton-X100 (Sigma) and 5% BSA (Aurion) before addition of the primary antibodies including mouse anti-flavivirus envelope protein (D1-4G2-4-15 hybridoma; ATCC, 1:250), guinea-pig anti-MAP2 (Synaptic Systems, 1:200) and rabbit anti-GFAP (Millipore, 1:200). Next, sections were washed and incubated with the appropriate secondary antibody including donkey anti-mouse IgG AF488/AF555 or donkey anti-mouse IgG AF488/AF555 (Invitrogen). Hoechst 33,342 (DNA staining; Invitrogen, 1:1000) was used to stain cell nuclei. Images were obtained using a Zeiss LSM 700 laser scanning microscope.
Terminal deoxynucleotidyl transferase mediated dUTP nick end labeling (TUNEL) assay
The TUNEL assay was performed using the ApopTag® Plus in situ apoptosis fluorescein S7111 detection kit (Sigma) according to the manufacturer’s protocol, following instructions for a combined IF staining. Sections were treated with TrueBlack® (Biotium) after antigen retrieval to decrease autofluorescence, followed by staining with the following primary antibodies: polyclonal rabbit anti-CC3 (cleaved caspase 3 protein; Cell Signaling Technology, 1:300 dilution), polyclonal rabbit anti-pMLKL (phosphorylation of mixed lineage kinase domain-like protein; Abcam, 1:250) and monoclonal mouse anti-GSDMD (gasdermin D protein; Abnova, 1:250). Next, sections were washed and incubated with the appropriate secondary antibody including donkey anti-rabbit AF555 (1:250) or goat anti-mouse IgG2a AF647 (1:250) (all from Invitrogen).
Lactate dehydrogenase assay (LDH)
The viability of the hfOBSC was determined by lactate dehydrogenase (LDH) assay. The LDH assays were performed on conditioned medium of hfOBSC cultures using the LDH-Cytoxicity Assay kit (Abcam) according to the manufacturer’s protocol. LDH levels of brain slices incubated in lysis buffer according to the manufacturer’s instructions were used as 100% cell death positive control.
Image processing
Images were subjected to processing and file-type conversion using ImageJ software (version 1.53t, National Institutes of Health, Bethesda, MD). Processed images were then rendered in 3D using Dragonfly software (Version 2021.1 for [Windows]; Comet Technologies Canada Inc., Montreal, Canada). This software is available at https://www.theobjects.com/dragonfly. Quantitative analysis of infected cell area and infection percentages of whole slices were done using the ‘Cell detection’ and batch-processing tools in QuPath 0.3.2 software66 based upon staining for viral envelope protein.
Statistical analysis
Quantitative data were analyzed and statistics was carried out using Prism 8.0.2 (GraphPad).
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
References
Rocklöv, J. & Dubrow, R. Climate change: An enduring challenge for vector-borne disease prevention and control. Nat. Immunol. 21, 479–483 (2020).
Chang, C., Ortiz, K., Ansari, A. & Gershwin, M. E. The Zika outbreak of the 21st century. J. Autoimmun. 68, 1 (2016).
Tahotná, A., Brucknerová, J. & Brucknerová, I. Zika virus infection from a newborn point of view. TORCH or TORZiCH?. Interdiscip. Toxicol. 11, 241 (2018).
Muñoz, L. S., Parra, B. & Pardo, C. A. Neurological implications of Zika virus infection in adults. J. Infect. Dis. 216, S897 (2017).
Historic Data (1999–2022) | West Nile Virus | CDC. at <https://www.cdc.gov/westnile/statsmaps/historic-data.html>
Surveillance, prevention and control of West Nile virus and Usutu virus infections in the EU/EEA. EFSA Supporting Publications 20, (2023).
Vilibic-Cavlek, T. et al. Epidemiology of usutu virus: The European scenario. Pathogens 9, 1–19 (2020).
Gill, C. M. et al. Usutu virus disease: A potential problem for North America?. J. Neurovirol. 26, 149–154 (2020).
Clé, M. et al. Neurocognitive impacts of arbovirus infections. J. Neuroinflammation 17, (2020).
Simonin, Y. et al. Human usutu virus infection with atypical neurologic presentation, Montpellier, France, 2016. Emerg. Infect. Dis. 24, 875 (2018).
Grottola, A. et al. Usutu virus infections in humans: A retrospective analysis in the municipality of Modena, Italy. Clin. Microbiol. Infect. 23, 33–37 (2017).
Jensen, C. & Teng, Y. Is it time to start transitioning from 2D to 3D cell culture?. Front. Mol. Biosci. 7, 513823 (2020).
Walczak, P. A., Perez-Esteban, P., Bassett, D. C. & Hill, E. J. Modelling the central nervous system: tissue engineering of the cellular microenvironment. Emerg. Top Life Sci. 5, 507–517 (2021).
Setia, H. & Muotri, A. R. Brain organoids as a model system for human neurodevelopment and disease. Semin. Cell Dev. Biol. 95, 93–97 (2019).
Jucker, M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat. Med. 16, 1210–1214 (2010).
Rashidi, A. S. et al. Herpes simplex virus infection induces necroptosis of neurons and astrocytes in human fetal organotypic brain slice cultures. J. Neuroinflammation 21, 1–16 (2024).
Bulstrode, H. et al. Myeloid cell interferon secretion restricts Zika flavivirus infection of developing and malignant human neural progenitor cells. Neuron 110, 3936-3951.e10 (2022).
Lin, M. Y. et al. Zika virus infects intermediate progenitor cells and post-mitotic committed neurons in human fetal brain tissues. Sci. Rep. 7, 1–8 (2017).
Croft, C. L., Futch, H. S., Moore, B. D. & Golde, T. E. Organotypic brain slice cultures to model neurodegenerative proteinopathies. Mol. Neurodegener. 14, 1–11 (2019).
Alaylioğlu, M., Dursun, E., Yilmazer, S. & Gezen Ak, D. A Bridge between in vitro and in vivo studies in neuroscience: Organotypic brain slice cultures. Arch. Neuropsychiatry 57, 333 (2020).
Kelley, K. W. & Pașca, S. P. Human brain organogenesis: Toward a cellular understanding of development and disease. Cell 185, 42–61 (2022).
Witusik, M. et al. Successful elimination of non-neural cells and unachievable elimination of glial cells by means of commonly used cell culture manipulations during differentiation of GFAP and SOX2 positive neural progenitors (NHA) to neuronal cells. BMC Biotechnol. 8, 1–12 (2008).
Dráberová, E. et al. Class III β-tubulin is constitutively coexpressed with glial fibrillary acidic protein and Nestin in midgestational human fetal astrocytes: Implications for phenotypic identity. J. Neuropathol. Exp. Neurol. 67, 341–354 (2008).
Zecevic, N. Specific characteristic of radial glia in the human fetal telencephalon. Glia 48, 27–35 (2004).
Bhaduri, A. et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142 (2020).
Eze, U. C., Bhaduri, A., Haeussler, M., Nowakowski, T. J. & Kriegstein, A. R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 24, 584–594 (2021).
Imre, G. Cell death signalling in virus infection. Cell Signal 76, 109772 (2020).
Fricker, M., Tolkovsky, A. M., Borutaite, V., Coleman, M. & Brown, G. C. Neuronal cell death. Physiol. Rev. 98, 813–880 (2018).
Kalil, A. C. et al. Use of interferon-α in patients with West Nile encephalitis: Report of 2 cases. Clin. Infect. Dis. 40, 764–766 (2005).
Lewis, M. & Amsden, J. R. Successful treatment of West Nile virus infection after approximately 3 weeks into the disease course. Pharmacother. J. Human Pharmacol. Drug Ther. 27, 455–458 (2007).
Kasule, S. N., Gupta, S., Patron, R. L., Grill, M. F. & Vikram, H. R. Neuroinvasive West Nile virus infection in solid organ transplant recipients. Transplant Infect. Dis. 25, e14004 (2023).
Winston, D. J. et al. Donor-derived west nile virus infection in solid organ transplant recipients: Report of four additional cases and review of clinical, diagnostic, and therapeutic features. Transplantation 97, 881 (2014).
Sayao, A.-L. et al. Calgary experience with West Nile virus neurological syndrome during the late summer of 2003. Can. J. Neurol. Sci. 31, 194–203 (2004).
Chan-Tack, K. M. & Forrest, G. Failure of interferon alpha-2b in a patient with West Nile virus meningoencephalitis and acute flaccid paralysis. Scand. J. Infect Dis. 37, 944–946 (2005).
Penn, R. G. et al. Persistent neuroinvasive West Nile virus infection in an Immunocompromised patient. Clin. Infect. Dis. 42, 680–683 (2006).
Benzaria, S. et al. 2′-C-Methyl branched pyrimidine ribonucleoside analogues: Potent inhibitors of RNA virus replication. Antivir. Chem. Chemother. 18, 225–242 (2007).
Julander, J. G. et al. Efficacy of 2′-C-methylcytidine against yellow fever virus in cell culture and in a hamster model. Antivir. Res. 86, 261 (2010).
Historical data by year - West Nile virus seasonal surveillance. at <https://www.ecdc.europa.eu/en/west-nile-fever/surveillance-and-disease-data/historical>
Agliani, G. et al. Pathological features of West Nile and Usutu virus natural infections in wild and domestic animals and in humans: A comparative review. One Health 16, 100525 (2023).
Peng, B. H. & Wang, T. West Nile virus induced cell death in the central nervous system. Pathogens 8, 215 (2019).
Pan, Y., Cheng, A., Wang, M., Yin, Z. & Jia, R. The dual regulation of apoptosis by flavivirus. Front. Microbiol. 12, 654494 (2021).
Salinas, S. et al. Deleterious effect of Usutu virus on human neural cells. PLoS Negl. Trop. Dis. 11, e0005913 (2017).
Yang, S. et al. Zika virus-induced neuronal apoptosis via increased mitochondrial fragmentation. Front. Microbiol. 11, 598203 (2020).
Vig, P. J. S. et al. Differential expression of genes related to innate immune responses in ex vivo spinal cord and cerebellar slice cultures infected with west Nile Virus. Brain Sci. 9, 1 (2018).
Clarke, P. et al. Death receptor-mediated apoptotic signaling is activated in the brain following infection with west nile virus in the absence of a peripheral immune response. J. Virol. 88, 1080–1089 (2014).
Lim, S. M. et al. Characterization of the mouse neuroinvasiveness of selected European strains of West nile virus. PLoS One 8, 74575 (2013).
Solomon, T., Ooi, M. H., Beasley, D. W. C. & Mallewa, M. West Nile encephalitis. BMJ: Br. Med. J. 326, 865 (2003).
Bai, F., Ashley Thompson, E., Vig, P. J. S. & Arturo Leis, A. Current understanding of West nile virus clinical manifestations, immune responses, neuroinvasion, and immunotherapeutic implications. Pathogens 8, 193 (2019).
Bosanko, C. M. et al. West Nile virus encephalitis involving the substantia nigra: Neuroimaging and pathologic findings with literature review. Arch. Neurol. 60, 1448–1452 (2003).
Kelley, T. W., Prayson, R. A., Ruiz, A. I., Isada, C. M. & Gordon, S. M. The neuropathology of West Nile virus meningoencephalitis: A report of two cases and review of the literature. Am. J. Clin. Pathol. 119, 749–753 (2003).
Armah, H. B. et al. Systemic distribution of West Nile virus infection: Postmortem immunohistochemical study of six cases. Brain Pathol. 17, 354–362 (2007).
Guarner, J. et al. Clinicopathologic study and laboratory diagnosis of 23 cases with West Nile virus encephalomyelitis. Hum. Pathol. 35, 983–990 (2004).
Goldsmith, C. et al. Fatal West nile virus encephalitis in a renal transplant recipient. Am. J. Clin. Pathol. 121, 26–31 (2004).
Schafernak, K. T. & Bigio, E. H. West Nile virus encephalomyelitis with polio-like paralysis & nigral degeneration. Can. J. Neurol. Sci. 33, 407–410 (2006).
Reddy, P. et al. West Nile virus encephalitis causing fatal CNS toxicity after hematopoietic stem cell transplantation. Bone Marrow Transplant 33, 109–112 (2004).
Penn, R. G. et al. Persistent neuroinvasive west nile virus infection in an immunocompromised patient. Clin. Antivir. Dis. 42, 680–683 (2006).
Omalu, B. I., Shakir, A. A., Wang, G., Lipkin, W. I. & Wiley, C. A. Fatal fulminant pan-meningo-polioencephalitis due to west nile virus. Brain Pathol. 13, 465–472 (2003).
Gaibani, P., Cavrini, F., Gould, E. A., Rossini, G. & Pierro, A. Comparative genomic and phylogenetic analysis of the first usutu virus isolate from a human patient presenting with neurological symptoms. PLoS One 8, 64761 (2013).
Mumtaz, N. et al. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antivir. Res. 146, 161–163 (2017).
Toussi, S. S., Hammond, J. L., Gerstenberger, B. S. & Anderson, A. S. Therapeutics for COVID-19. Nat. Microbiol. 8, 771–786 (2023).
Schäfer, C. B., Gao, Z., De Zeeuw, C. I. & Hoebeek, F. E. Temporal dynamics of the cerebello-cortical convergence in ventro-lateral motor thalamus. J. Physiol. 599, 2055–2073 (2021).
Oude Munnink, B. B. et al. Genomic monitoring to understand the emergence and spread of Usutu virus in the Netherlands, 2016–2018. Sci. Rep. 10, 2798 (2020).
Vlaskamp, D. R. et al. First autochthonous human west nile virus infections in the Netherlands, July to August 2020. Eurosurveillance 25, 1–4 (2020).
Langerak, T. et al. Transplacental Zika virus transmission in ex vivo perfused human placentas. PLoS Negl. Trop. Dis. 16, e0010359 (2022).
Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 162, 480–483 (1931).
Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 1–7 (2017).
Author information
Authors and Affiliations
Contributions
Conceptualization: BR and GMGMV; Methodology: EMM & ASR; Formal analysis: EMM & ASR; Investigation: EMM & ASR; Writing Original Draft: EMM & ASR; Writing, Review & Editing: EMM, ASR, MvG, BR & GMGMV; Visualization: EMM & ASR; Supervision: MvG, BR & GMGMV; Project Administration and Funding Acquisition: BR & GMGMV.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Human fetal brain tissues from legally terminated second trimester pregnancies (17–20 weeks) was obtained by the Human Immune System (HIS)-Mouse Facility of Academic Medical Center (AMC; Amsterdam, The Netherlands), after written informed consent of the mothers for the use of tissues in research and with approval of the Medical Ethical Review Board of the AMC (MEC: 03/038) and Erasmus MC (MEC-2017-009). Study procedures were performed according to the Declaration of Helsinki, and in compliance with relevant Dutch laws and institutional guidelines. The tissues obtained were anonymized and non-traceable to the donors. On request by the researchers, only gender and gestational age was provided.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Marshall, E.M., Rashidi, A.S., van Gent, M. et al. Neurovirulence of Usutu virus in human fetal organotypic brain slice cultures partially resembles Zika and West Nile virus. Sci Rep 14, 20095 (2024). https://doi.org/10.1038/s41598-024-71050-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71050-w
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.