Abstract
Reduced processing speed is a core deficit in major depressive disorder (MDD) and has been linked to altered structural brain network connectivity. Ample evidence highlights the involvement of genetic-immunological processes in MDD and specific depressive symptoms. Here, we extended these findings by examining associations between polygenic scores for tumor necrosis factor-α blood levels (TNF-α PGS), structural brain connectivity, and processing speed in a large sample of MDD patients. Processing speed performance of n = 284 acutely depressed, n = 177 partially and n = 198 fully remitted patients, and n = 743 healthy controls (HC) was estimated based on five neuropsychological tests. Network-based statistic was used to identify a brain network associated with processing speed. We employed general linear models to examine the association between TNF-α PGS and processing speed. We investigated whether network connectivity mediates the association between TNF-α PGS and processing speed. We identified a structural network positively associated with processing speed in the whole sample. We observed a significant negative association between TNF-α PGS and processing speed in acutely depressed patients, whereas no association was found in remitted patients and HC. The mediation analysis revealed that brain connectivity partially mediated the association between TNF-α PGS and processing speed in acute MDD. The present study provides evidence that TNF-α PGS is associated with decreased processing speed exclusively in patients with acute depression. This association was partially mediated by structural brain connectivity. Using multimodal data, the current findings advance our understanding of cognitive dysfunction in MDD and highlight the involvement of genetic-immunological processes in its pathomechanisms.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) is considered one of the most prevalent and debilitating mental illnesses worldwide, with a lifetime prevalence of approximately 10.8% [1]. Impairments in processing speed (i.e., the ability to quickly acquire, process, and respond to information) are among the core features of acute MDD [2, 3] and have been associated with less favorable clinical trajectories [4] and poor treatment response [5]. Moreover, reduced processing speed is a major predictor of poor psychosocial functioning with adverse consequences for work, family, friends, and health [2, 4]. As most cognitive domains depend on rapid information transfer [6], dysfunctions in processing speed can lead to impairments in several other cognitive domains, such as attention, concentration, and memory [7, 8]. Some of these cognitive deficits persist even after remission [9] and may promote vulnerability to relapses [10]. Given all these findings, it is of particular clinical and scientific interest to understand the pathogenic mechanisms underlying reduced processing speed in MDD.
A promising way to gain more insights into these mechanisms is to study the neurobiological correlates of processing speed. Cognitive processes are thought to arise from a multitude of interacting brain regions rather than from individual brain regions alone [11, 12]. The human connectome, i.e., the network of all brain regions and their white matter (WM) connections, can be examined using diffusion-weighted imaging and network analyses [13]. Applying these methods, neuroimaging studies in healthy individuals have revealed robust associations between brain structural connectivity and cognitive performance, including processing speed [14,15,16], thus linking intact network connectivity to healthy cognitive functioning. Conversely, disruptions in network connectivity have been demonstrated in several mental disorders, including MDD [17,18,19,20], with deficits most pronounced in acutely depressed individuals [18]. These findings lead to the assumption that MDD-related alterations may also affect those brain networks associated with cognitive performance. Indeed, Gruber and colleagues [21] previously identified a structural subnetwork of frontotemporal fiber tracts that was positively related to processing speed. Within this network, the researchers further demonstrated associations between connectivity strength, processing speed, and depression severity, indicating that alterations in subnetwork-specific connectivity may provide a structural basis for impaired processing speed in MDD. However, despite recent advances in network neuroscience, little is known about the biological mechanisms preceding these brain structural alterations.
Brain structural connectivity, processing speed, and depression have been associated with dysregulations of the immune system [22,23,24,25,26], characterized by chronic production and expression of chemokines, acute phase proteins, and proinflammatory cytokines. Along with interleukin-1β and interleukin-6, tumor necrosis factor-α (TNF-α) is considered one of the key proinflammatory cytokines that has been linked to a number of somatic and mental diseases [27]. This is supported, for example, by studies reporting elevated TNF-α levels in peripheral blood and cerebrospinal fluid of MDD patients [28, 29], and anti-depressant effects of drug-induced TNF-α synthesis inhibition [30]. TNF-α interacts with the brain in a complex, bidirectional manner via cellular, neuronal, or endocrine pathways, where it can trigger neurotoxic processes, including inhibition of growth factors [31] or disruption of WM microstructure [32, 33]. While these immunological processes can be caused by exposure to environmental stressors (e.g., childhood maltreatment or negative life events) [34, 35], there is also evidence linking genetic variation to immunological dysregulation in MDD [36]. For example, twin studies have shown that the association between depression and increased immune activation can be attributed, at least in part, to shared genetic factors involved in the regulation of inflammatory processes [37, 38]. Furthermore, 34 of the 269 genes associated with depression in a genome-wide association study (GWAS) by Howard and colleagues [39] were found to be implicated in immunological processes, with some of them exhibiting cytokine-related features [40]. Likewise, there is evidence supporting a link between the polymorphism 308 (G/A) in the TNF-α gene and the risk of developing MDD [41, 42], as well as between polygenic scores for TNF-α blood levels and specific depressive symptoms [43]. So far, however, none of these studies have analyzed the associations with structural network connectivity and processing speed.
The main goal of the present study was to investigate the reciprocal relationships between genetic predisposition to TNF-α blood levels, brain structural connectivity, and processing speed in a well-powered sample of patients with MDD and HC. To this end, we first re-established previous findings of processing speed deficits in MDD patients [2, 3] and their association with structural brain connectivity [21]. We then extended these findings by evaluating the involvement of genetic-immunological processes in these associations using a polygenic score (PGS) for TNF-α blood levels. PGS estimate an individual’s genetic susceptibility to a certain trait or disease [44] and have been applied in various clinical and scientific contexts, including genome-wide gene-by-environment [45] and gene-by-gene interactions [46]. Based on the literature outlined above, we expect TNF-α PGS to be negatively associated with processing speed in MDD. In addition, we hypothesize that the association between TNF-α PGS and processing speed is mediated by connectivity strength in processing speed-related networks. Given that TNF-α PGS [43], structural connectome alterations [18, 21], and cognitive deficits [21, 47] have been particularly linked to current depressive symptomatology, we assume these associations to be most pronounced in acutely depressed patients.
Materials and methods
Participants
Our study consisted of N = 659 patients with MDD and N = 743 HC drawn from the ongoing Marburg-Münster Affective Disorders Cohort Study (MACS, (see [48] for the study protocol and [49] for the quality assurance protocol). Participants aged 18–65 years with West-European ancestry were recruited from January 09, 2015 to May 11, 2018 via newspaper advertisements or local psychiatric hospitals. MDD patients were included in our analyses if they were diagnosed with an acute (MDDa, n = 284) or a partially (MDDpr, n = 177) or fully remitted (MDDfr, n = 198) major depressive disorder. HC were included if they did not report a lifetime diagnosis of any psychiatric disorder. The Structured Clinical Interview for DSM-IV-TR [50] was used by trained interviewers to validate the diagnosis or lack thereof and to determine remission status (for more details see Supplement 1). See Supplement 2 for details on exclusion criteria and sample selection process and Supplement 3 for patients’ medication and comorbidities. Sample characteristics are provided in Table 1. Study procedures were approved by the Ethics Committees of the University of Münster (2014-422-b-S) and Marburg (07/14) following the Declaration of Helsinki. All participants signed informed consent before participation and received financial compensation.
Assessment of processing speed and clinical characteristics
Processing speed was estimated based on the shared variance of five tests from a neuropsychological test battery that assess performance via 1. the time required to solve a certain task or 2. the number of items processed within a given amount of time: the Letter Number Sequencing Test (LNST), the Digit Symbol Substitution Test (DSST), the Trail Making Test (TMT-A & TMT-B), the Corsi Block Tapping Test (forward and backward), and the d2 Attention Test [21] (see Supplement 4).
To adjust for clinical features of MDD, we included the number of prior hospitalizations as a measure of cumulative illness severity and the presence of comorbid mental illness reported by patients in the SCID-I interview. In addition, the type and amount of current medication adherence were assessed using the Medication Load Index (MedIndex) [51]. The severity of current depressive symptomatology was measured in all participants using the Hamilton Depression Rating Scale [52].
MRI data acquisition and preprocessing
MRI data were collected using 3 T whole-body MRI scanners at two scanning sites (Marburg: Tim Trio, Siemens, Erlangen, Germany; Münster: Prisma, Siemens, Erlangen, Germany). Due to a body coil exchange in Marburg, two dummy-coded variables (Marburg pre body-coil, Marburg post body-coil) with Münster as reference category were included as additional covariates in all analyses. See Supplement 5 for detailed information on acquisition parameters and Supplement 6 for preprocessing steps. Anatomical connectome reconstruction was performed using the CATO toolbox [53]. Hundred and fourteen nodes (i.e., cortical brain regions, based on the Cammoun subdivision of Freesurfer’s Desikan-Killiany atlas [54, 55]) were obtained from T1-weighted MRI, while edges (i.e., the mean fractional anisotropy (FA) of a fiber tract connecting a pair of nodes) were reconstructed from diffusion-weighted MRI. FA is an established parameter of WM integrity that has been associated with MDD and cognitive performance in previous neuroimaging studies [18, 56,57,58]. FA can range from 0 to 1, with higher values indicating direct diffusion and intact myelin [59]. Details on the reconstruction and quality assurance procedures are provided in Supplements 7 and 8.
Genetic methods
Genotyping was performed in the MACS cohort with the Illumina Infinium PsychArray BeadChip, followed by quality control and imputation, as described earlier [60,61,62]. Briefly, quality control and population substructure analyses via multidimensional scaling (MDS) were conducted in PLINK v1.90 [63]. Genotype data was imputed to the 1000 Genomes phase 3 reference panel using SHAPEIT and IMPUTE2 [64,65,66]. Participants who were genetically related to other participants (\(\hat{\pi }\ge 12.5\)) in our sample were excluded from downstream analyses. PGS for TNF-α blood levels were computed using summary statistics from a recent GWAS by Ahola-Olli and colleagues [67]. The GWAS sample was independent of the MACS sample included in our study. For PGS calculation, single nucleotide polymorphism weights were estimated using PRS-CS with default parameters. The PRS-CS approach was chosen due to its advantages regarding prediction accuracy between the observed and predicted traits as compared to standard methods such as clumping and thresholding [68, 69]. The global shrinkage parameter set to φ = 1e−6 [68, 70]. The choice of the global shrinkage parameter was based on the assumption of a relatively low polygenicity of blood cytokine levels, including TNF-α, consistent with previous studies [43]. For analyses based on higher values for the shrinkage parameter, see Supplement 9.
Statistical analyses
Demographic characteristics, clinical features, and factor structure of the neuropsychological tests were analyzed using IBM SPSS Statistics (version 28.0; IBM Corporation). Neuroimaging data were analyzed using the Network-based statistic toolbox (NBS [71]) implemented in Matlab 2019b [72]. All statistical models were corrected for sex and age. Models including neuroimaging data were further corrected for scanner settings. For analyses involving TNF-α PGS, body-mass-index (BMI) [67] as well as the first two ancestry components (C1, C2) extracted from the MDS analysis of genotype data based on their relative variance (Supplement 10), were added as covariates. If not otherwise stated, statistical tests were conducted at a two-sided significance level of α = 0.05.
Before testing our hypotheses, we employed a principle component analysis (PCA) to abstract from neuropsychological test scores to underlying cognitive domains. Given our focus on speed tests and based on our previous work [21], we expected a one-factor structure reflecting the underlying capacity of processing speed [6]. The number of factors to be extracted was determined according to the Kaiser-Guttman criterion, the Scree test, and parallel analysis [73]. Component scores were calculated using a linear regression approach. All subsequent analyses were performed with these component scores.
The first part of our analyses focuses on re-establishing processing speed deficits in MDD patients and their relation to structural brain connectivity across MDD and HC, as both results form the basis for our subsequent analyses on the role of TNF-α PGS. Therefore, we first calculated a general linear model (GLM) to examine whether the diagnostic groups (HC, MDDa, MDDpr, MDDfr) differed in their processing speed performance using the extracted component scores as dependent variable. Following our previous findings linking cognition to structural brain connectivity [21], we then used NBS to identify a network associated with processing speed in the whole sample, while accounting for diagnosis on top of the above-mentioned covariates. NBS estimates family-wise error (FWE)-corrected regression models to test the association between edge-wise connectivity strength, as measured by edge-wise mean FA, and the processing speed scores. The significance of an identified network was determined using a permutation test (5000 permutations, for more details, see Supplement 11). In case of significant results, mean FA across all edges from the identified network was extracted to SPSS.
In the second part of our analyses, we sought to investigate the association of TNF-α PGS with processing speed performance and structural brain connectivity. For this purpose, we again calculated a GLM, this time including diagnosis and TNF-α PGS as independent variables and processing speed as dependent variable. The main effect of TNF-α PGS as well as its interaction with diagnosis were analyzed. Finally, using a bootstrapping approach implemented in the SPSS macro PROCESS (http://www.processmacro.org), we tested our mediation hypothesis with TNF-α PGS as predictor variable (X), mean FA as mediator variable (M), and processing speed as outcome variable (Y). Direct as well as indirect effects were estimated. Significance is assumed if the 95% confidence interval (95% CI) does not include zero.
Results
Exploratory principal component analysis for processing speed
As expected, exploratory PCA yielded a single-factor structure that explained 51.05% of the variance of the cognitive tests, with factor loadings ranging from 0.62 to 0.79 (Supplement 12).
Re-establishing processing speed deficits and their association with structural brain connectivity
The diagnostic groups differed significantly in the processing speed factor (F(3,1396) = 39.61, p < 0.001, partial η² = 0.078). Bonferroni-corrected post hoc tests revealed that patients with MDDa scored significantly worse than MDDfr (MD = −0.30, p = 0.001) and HC (MD = −0.61, p < 0.001). HC scored significantly higher compared with MDDpr (MD = 0.43, p < 0.001) and MDDfr (MD = 0.31, p < 0.001). MDDfr and MDDpr as well as MDDa and MDDpr did not differ from each other (all ps \(\ge\) 0.185).
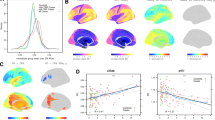
The NBS analysis conducted in the whole sample identified a network of edges whose FA was significantly associated with processing speed (NBS F-threshold = 4.0, 206 edges, 106 nodes, pFWE < 0.001, partial η2 = 0.052). Post-hoc analyses revealed a positive association between processing speed and connectivity strength (i.e., edge-wise FA) (NBS t-threshold=2.0, 121 edges, 89 nodes, pFWE < 0.001, partial η²=0.163, Fig. 1). The network comprised a large proportion of fronto-parietal edges (22%) (Supplement 13). See Supplement 14 for analyses based on different NBS t-thresholds.
A Dorsal, lateral, and medial views of the network of white matter fiber tracts in which connectivity strength, measured with edge-wise fractional anisotropy (FA), was positively associated with processing speed (z-score) across diagnostic groups. The network was identified using the Network-based statistic (NBS) toolbox (NBS t-threshold = 2.0, pFWE < 0.001), while correcting for age, sex, and scanner site. See Supplement 13 for details on participating anatomical brain regions. Network plots were created using BrainNet Viewer. B Scatterplot depicting the positive association between mean FA and processing speed across patients with acute (MDDa), partially (MDDpr), and fully remitted (MDDfr) major depressive disorder and healthy controls (HC). A more positive processing speed score represents higher processing speed performance.
Association between processing speed and TNF-α PGS
The GLM revealed no main effect of TNF-α PGS (p = 0.599). However, we observed a significant TNF-α PGS × diagnosis interaction effect (F(3,1389) = 4.40, p = 0.004, partial η² = 0.009), which was driven by a negative association between TNF-α PGS and processing speed in the MDDa group (B = −2.41, p = .008, partial η² = 0.034; Bonferroni-corrected), whereas no association was found in the MDDpr (p = 0.936), MDDfr (p > 0.999), and HC group (p > 0.999) (Fig. 2).
Note. The figure depicts the significant interaction effect between polygenic score for tumor necrosis factor-α (TNF-α PGS) and diagnosis on processing speed. Patients with acute major depressive disorder (MDDa) showed a negative association between TNF-α PGS and processing speed, whereas no association was found in patients with a partially (MDDpr) or fully remitted (MDDfr) depressive episode and healthy controls (HC). Processing speed (z-score) was calculated from five tests of a neuropsychological test battery using exploratory principal component analysis. A more positive processing speed score represents higher processing speed performance.
Structural brain connectivity as a mediator of the association between TNF-α PGS and processing speed
As we found a significant association between TNF-α PGS and processing speed exclusively in patients suffering from acute depression, the mediation model was tested only in the MDDa subgroup. The analysis showed a significant negative association between TNF-α PGS and processing speed (coeff = −2.45, 95%-CI [−3.97, −0.93], SE = 0.77, t = −3.17, p = 0.002), as well as between TNF-α PGS and mean FA (coeff = −0.04, 95% CI [−0.07, −0.01], SE = 0.02, t = −2.52, p = 0.012). Furthermore, we observed a significant negative indirect (mediated) effect of TNF-α PGS on processing speed through mean FA (coeff = −0.80, 95% CI [−1.45, −0.25], SE = 0.31). Lastly, the model yielded a significant direct effect of TNF-α PGS on processing speed (coeff = −1.65, 95% CI [−3.05, −0.24], SE = 0.71), indicating that the association between TNF-α PGS and processing speed was only partially mediated by mean FA (Fig. 3).
The figure depicts the mediation model with TNF-α PGS as a predictor variable, structural brain connectivity (measured by mean fractional anisotropy) as a mediator variable, and processing speed as an outcome variable in acutely depressed patients. A higher processing speed score represents higher processing speed performance. Unstandardized coefficients and standard errors for each path of the mediation model are presented. Note that c represents the direct effect and c‘ the indirect effect. * indicates significance at p < 0.05.
Robustness checks and exploratory analyses
Correction for influential data points, clinical characteristics within patients, and additional MDS components yielded a comparable pattern of results (Supplement 15). Furthermore, effects did not differ by sex (Supplement 16). Leave-one-site-out cross validation revealed good generalizability and prediction accuracy of the mediation model to unknown data (Supplement 17).
Discussion
To our knowledge, this is the first study to analyze the reciprocal associations between PGS for TNF-α blood levels, brain structural network connectivity, and processing speed in a well-powered sample of MDD patients and HC. Employing state-of-the-art network analyses, we re-established our previous findings on the human connectome, showing a positive association between brain network connectivity strength and processing speed in patients with MDD and HC [21]. The present study extends these findings by providing evidence for a genetic-immunological mechanism that may underlie these structural links. More specifically, we demonstrated a negative association between TNF-α PGS and processing speed in patients suffering from acute MDD, while no association was found in partially or fully remitted patients and HC. Of note, this association was mediated, at least partially, by structural brain connectivity. Although effect sizes were small to moderate, they are consistent with effect sizes found in previous neuroimaging [74] and genetic studies [75]. Our results appear to be robust, as they remained unchanged when clinical characteristics in the MDD sample were taken into account. Overall, the current data suggest genetic-immunologic mechanisms to play a role in impaired processing speed in patients with acute depression.
Corroborating previous findings [2, 3], we demonstrated slowed processing speed in patients with MDD compared to HC. The deficits were most pronounced in patients with acute MDD but still detectable in partially or fully remitted patients, underscoring the persistence of cognitive dysfunction in depression [9]. According to established cognitive theories, processing speed serves as a basic mental capacity that affects performance - and thus deficits - in several other cognitive domains, such as attention, concentration, and memory [7, 8]. Given the detrimental impact of persistent cognitive dysfunction on disease outcomes [4] and psychosocial functioning [2, 4], its effective and targeted treatment in the context of antidepressant therapy is of high clinical relevance.
On a neuronal level, cognitive dysfunction in MDD has been associated with alterations in the structural connectome [21], that is, the network of all brain regions and their WM connections [13]. In line with this evidence, we identified a structural network, whose WM fiber connections were positively associated with processing speed in patients with MDD and HC. The network comprised a large proportion of fronto-parietal fibers and hubs (i.e., brain regions with the highest degree), such as the inferior parietal cortex and parahippocampal gyrus, consistent with the involvement of these brain areas in cognitive processes [76, 77]. Along with previous findings [18, 56], we propose that reduced FA, as an indicator of decreased information exchange between these brain areas and impaired network architecture, might provide a structural basis for processing speed deficits in MDD and HC.
While previous network neuroscience has mainly focused on the mere association between cognitive (dys-)function and structural brain alterations, the present study adds a possible biological mechanism for those alterations. More specifically, we demonstrated a triad of genetic predisposition to TNF-α blood levels, structural brain connectivity, and processing speed in patients with acute MDD. TNF-α is a proinflammatory cytokine that plays a critical role in the development and maintenance of depressive symptoms, including cognitive dysfunction [28, 31]. According to established theories [78], chronically elevated levels of proinflammatory cytokines, such as TNF-α, could lead to prolonged hypothalamic–pituitary–adrenal (HPA) axis activation via stimulating cortisol release and counteracting its negative feedback functions. In fact, HPA axis dysfunction, including hypercortisolemia, is a common feature of MDD that has been linked to its pathophysiology [79, 80], including cognitive dysfunction [79]. Cortisol and other glucocorticoids have been found to alter oligodendrocyte function by inhibiting their differentiation and myelogenesis [81]. These neurotoxic processes might affect WM microstructure [82, 83], leading to alterations in the structural connectome [84]. However, given the complexity of the (bidirectional) relationship between cytokines and HPA-axis [85], the cross-sectional nature of the current study, and the lack of state-dependent immunological and endocrinological markers, this conclusion remains speculative. Future longitudinal studies should extend our findings by adding biological serum markers in repeated-measures designs.
Nevertheless, the genetic perspective allows us to draw tentative conclusions about the causal directions of the effects observed since we can assume that, during periods of acute depression, TNF-α PGS leads to changes in the brain and in processing speed and not vice versa. Indeed, MDD is a multifactorial disorder that arises and is maintained by a complex interaction between environmental and genetic factors [86]. Increasing evidence from genetic research suggests that certain cytokine polymorphisms, including TNF-α, may be involved in the pathogenesis of MDD and related deficits [40, 43, 87]. Consistent with this assumption, we showed that TNF-α PGS is associated with impaired processing speed and that this association is partially mediated by structural brain connectivity. Notably, the effects appeared to be exclusive to patients suffering from acute MDD, as no associations have been found in (partially) remitted patients and HC. This might be explained by the fact that acute patients showed a greater variance in their processing speed performance compared with the other diagnostic groups. Similarly, this finding could indicate that gene × environment interactions are more likely to result in the observed cognitive deficits rather than genetic mechanisms alone. Future studies should consider additional measures (e.g., questionnaires, serum cortisol) in their analyses to evaluate the contribution of environmental factors, especially stress-related factors, to this association.
While our analyses focus on TNF-α as a key proinflammatory cytokine linked to the development and persistence of MDD [27, 88], we also recognize the involvement of other inflammatory markers in depressive symptoms, including cognitive impairment. For instance, previous studies have demonstrated associations between several proinflammatory interleukins such as IL-6 and IL-1β and reduced processing speed performance [89] and structural brain alterations [25], making generalizations of our results to other inflammatory markers likely. On the other hand, evidence also suggests the specificity of inflammatory markers in depressive symptoms by indicating that these associations are not universally applicable across all cytokines [43, 90]. Therefore, future studies are warranted to clarify the specificity of cytokines and their genetic predispositions implicated in the complex interplay between structural brain connectivity and processing speed performance in acute MDD.
The main strengths of the current study are the large and well-characterized sample and the integration of multimodal, i.e., genetic, cognitive, and magnetic imaging-derived brain network data. Nonetheless, some limitations should be noted. As mentioned above, our conclusions must be interpreted in light of the lack of state-dependent immunologic measures and the cross-sectional nature. In particular, mediation models imply causal relationships between variables. As we analyzed both cognitive and imaging data cross-sectionally, we cannot exclude the possibility of reverse causal relationships or the influence of omitted variables [91]. Another limitation arises from the operationalization of processing speed, as the included neuropsychological tests might also tap into other cognitive domains [92]. Regarding genetics, it must be acknowledged that the calculation of PGS was based on a relatively small GWAS consisting of Finns [67]. Since Finns have a Siberian ancestry [93], this may have led to a divergence from the Western-European ancestry of our MACS sample [43]. Further, our interpretations rely on the assumption of low polygenicity of TNF-α blood levels. While the interaction effect between diagnosis and TNF-α PGS on processing speed became significant for two of the three tested shrinkage parameters, mediation analysis yielded less consistent results. This might limit the generalizability of our findings. Finally, we did not directly measure TNF-α blood levels, nor did our PRS consider environmental factors, which also play an important role in the multifactorial nature of inflammation, depression, and cognitive performance [23, 94]. For instance, in our previous work, we showed that childhood maltreatment - one of the most important environmental risk factors for depression - and polygenic risk for MDD were independently associated with cognitive dysfunction [95]. Although we acknowledge these limitations, our approach does shed new light on a possible genetic pathway contributing to the link between structural brain connectivity and processing speed. This, in turn, might represent a starting point for future studies that should replicate our findings and extend their analyses to include serum markers of proinflammatory cytokines.
In conclusion, the current study provides evidence of a shared link between genetic predisposition to TNF-α blood levels, altered brain structural connectivity, and processing speed deficits in patients suffering from acute depression. Based on genetic-immunological, cognitive, and magnetic imaging-derived brain network data, we demonstrated that TNF-α PGS is associated with reduced processing speed in patients with acute MDD and that this association is partially mediated by brain structural connectivity. The current findings advance our understanding of cognitive dysfunction in MDD and could provide novel targets for the development of therapeutic interventions. Longitudinal studies are required to further unravel the genetic, immunological, environmental, and neural mechanisms of cognitive deficits and their interaction in MDD.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Analyses were performed in Matlab 2019b. Codes will be available upon request and communication with the corresponding author.
References
Lim GY, Tam WW, Lu Y, Ho CS, Zhang MW, Ho RC. Prevalence of depression in the community from 30 countries between 1994 and 2014. Sci Rep. 2018;8:2861.
Lam RW, Kennedy SH, McIntyre RS, Khullar A. Cognitive dysfunction in major depressive disorder: effects on psychosocial functioning and implications for treatment. Can J Psychiatry. 2014;59:649–54.
McDermott LM, Ebmeier KP. A meta-analysis of depression severity and cognitive function. J Affect Disord. 2009;119:1–8.
McIntyre RS, Cha DS, Soczynska JK, Woldeyohannes HO, Gallaugher LA, Kudlow P, et al. Cognitive deficits and functional outcomes in major depressive disorder: determinants, substrates, and treatment interventions. Depress Anxiety. 2013;30:515–27.
Dawson EL, Caveney AF, Meyers KK, Weisenbach SL, Giordani B, Avery ET, et al. Executive Functioning at Baseline Prospectively Predicts Depression Treatment Response. Prim Care Companion CNS Disord. 2017;19. https://doi.org/10.4088/PCC.16m01949.
Harvey PD. Domains of cognition and their assessment. Dialogues Clin Neurosci. 2019;21:227–37.
Kail R, Salthouse TA. Processing speed as a mental capacity. Acta Psychol. 1994;86:199–225.
Salthouse TA. The processing-speed theory of adult age differences in cognition. Psychol Rev. 1996;103:403–28.
Semkovska M, Quinlivan L, O’Grady T, Johnson R, Collins A, O’Connor J, et al. Cognitive function following a major depressive episode: a systematic review and meta-analysis. Lancet Psychiatry. 2019;6:851–61.
Gonda X, Pompili M, Serafini G, Carvalho AF, Rihmer Z, Dome P. The role of cognitive dysfunction in the symptoms and remission from depression. Ann Gen Psychiatry. 2015;14:27.
McIntosh AR. Towards a network theory of cognition. Neural Netw. 2000;13:861–70.
Barbey AK. Network neuroscience theory of human intelligence. Trends Cogn Sci. 2018;22:8–20.
van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci. 2013;17:683–96.
Ponsoda V, Martínez K, Pineda-Pardo JA, Abad FJ, Olea J, Román FJ, et al. Structural brain connectivity and cognitive ability differences: a multivariate distance matrix regression analysis. Hum Brain Mapp. 2017;38:803–16.
Wiseman SJ, Booth T, Ritchie SJ, Cox SR, Maniega SM, Hernandez M del CV, et al. Cognitive abilities, brain white matter hyperintensity volume and structural network connectivity in older age. Hum Brain Mapp. 2018;39:622–32.
Zimmermann J, Griffiths JD, McIntosh AR. Unique mapping of structural and functional connectivity on cognition. J Neurosci. 2018;38:9658–67.
Myung W, Han CE, Fava M, Mischoulon D, Papakostas GI, Heo J-Y, et al. Reduced frontal-subcortical white matter connectivity in association with suicidal ideation in major depressive disorder. Transl Psychiatry. 2016;6:e835.
Repple J, Mauritz M, Meinert S, de Lange SC, Grotegerd D, Opel N, et al. Severity of current depression and remission status are associated with structural connectome alterations in major depressive disorder. Mol Psychiatry. 2020;25:1550–8.
Repple J, Gruber M, Mauritz M, de Lange SC, Winter NR, Opel N, et al. Shared and specific patterns of structural brain connectivity across affective and psychotic disorders. Biol Psychiatry. 2022. https://doi.org/10.1016/j.biopsych.2022.05.031.
Sacchet MD, Prasad G, Foland-Ross LC, Thompson PM, Gotlib IH. Support vector machine classification of major depressive disorder using diffusion-weighted neuroimaging and graph theory. Front Psychiatry. 2015;6:21.
Gruber M, Mauritz M, Meinert S, Grotegerd D, de Lange SC, Grumbach P, et al. Cognitive performance and brain structural connectome alterations in major depressive disorder. Psychol Med. 2023;53:6611–22.
Allison DJ, Ditor DS. The common inflammatory etiology of depression and cognitive impairment: a therapeutic target. J Neuroinflammation. 2014;11:151.
Miller AH, Raison CL. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol. 2016;16:22–34.
Thomann AK, Reindl W, Wüstenberg T, Kmuche D, Ebert MP, Szabo K, et al. Aberrant brain structural large-scale connectome in Crohn’s disease. Neurogastroenterol Motil. 2019;31:e13593.
Goldsmith DR, Bekhbat M, Mehta ND, Felger JC. Inflammation-related functional and structural dysconnectivity as a pathway to psychopathology. Biol Psychiatry. 2023;93:405–18.
Siddiqi SH, Kletenik I, Anderson MC, Cavallari M, Chitnis T, Glanz BI, et al. Lesion network localization of depression in multiple sclerosis. Nat Mental Health. 2023;1:36–44.
Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta Mol Cell Res. 2014;1843:2563–82.
Liu Y, Ho RC-M, Mak A. Interleukin (IL)-6, tumour necrosis factor alpha (TNF-α) and soluble interleukin-2 receptors (sIL-2R) are elevated in patients with major depressive disorder: A meta-analysis and meta-regression. J Affect Disord. 2012;139:230–9.
Osimo EF, Pillinger T, Rodriguez IM, Khandaker GM, Pariante CM, Howes OD. Inflammatory markers in depression: A meta-analysis of mean differences and variability in 5,166 patients and 5,083 controls. Brain Behav Immun. 2020;87:901–9.
Uzzan S, Azab AN. Anti-TNF-α compounds as a treatment for depression. Molecules. 2021;26:2368.
Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56.
Lim J, Sohn H, Kwon M-S, Kim B. White matter alterations associated with pro-inflammatory cytokines in patients with major depressive disorder. Clin Psychopharmacol Neurosci. 2021;19:449–58.
Sugimoto K, Kakeda S, Watanabe K, Katsuki A, Ueda I, Igata N, et al. Relationship between white matter integrity and serum inflammatory cytokine levels in drug-naive patients with major depressive disorder: diffusion tensor imaging study using tract-based spatial statistics. Transl Psychiatry. 2018;8:141.
Coelho R, Viola TW, Walss-Bass C, Brietzke E, Grassi-Oliveira R. Childhood maltreatment and inflammatory markers: a systematic review. Acta Psychiatr Scand. 2014;129:180–92.
Kautz MM, Coe CL, McArthur BA, Mac Giollabhui N, Ellman LM, Abramson LY, et al. Longitudinal changes of inflammatory biomarkers moderate the relationship between recent stressful life events and prospective symptoms of depression in a diverse sample of urban adolescents. Brain Behav Immun. 2020;86:43–52.
Beurel E, Toups M, Nemeroff CB. The bidirectional relationship of depression and inflammation: double trouble. Neuron. 2020;107:234–56.
Su S, Miller AH, Snieder H, Bremner JD, Ritchie J, Maisano C, et al. Common Genetic Contributions to depressive symptoms and inflammatory markers in middle-aged men: the Twins Heart Study. Psychosom Med. 2009;71:152–8.
Vaccarino V, Brennan M-L, Miller AH, Bremner JD, Ritchie JC, Lindau F, et al. Association of major depressive disorder with serum myeloperoxidase and other markers of inflammation: a twin study. Biol Psychiatry. 2008;64:476–83.
Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Tubbs JD, Ding J, Baum L, Sham PC. Immune dysregulation in depression: Evidence from genome-wide association. Brain Behav Immun Health. 2020;7:100108.
Cerri AP, Arosio B, Viazzoli C, Confalonieri R, Vergani C, Annoni G. The —308 (G/A) single nucleotide polymorphism in the TNF-α gene and the risk of major depression in the elderly. Int J Geriatr Psychiatry. 2010;25:219–23.
Cerri AP, Arosio B, Viazzoli C, Confalonieri R, Teruzzi F, Annoni G. -308(G/A) TNF-α gene polymorphism and risk of depression late in the life. Arch Gerontol Geriatr. 2009;49:29–34.
Kappelmann N, Czamara D, Rost N, Moser S, Schmoll V, Trastulla L, et al. Polygenic risk for immuno-metabolic markers and specific depressive symptoms: a multi-sample network analysis study. Brai, Behav Immun. 2021;95:256–68.
Choi SW, Mak TS-H, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15:2759–72.
Mullins N, Power RA, Fisher HL, Hanscombe KB, Euesden J, Iniesta R, et al. Polygenic interactions with environmental adversity in the aetiology of major depressive disorder. Psychol Med. 2016;46:759–70.
Agerbo E, Sullivan PF, Vilhjálmsson BJ, Pedersen CB, Mors O, Børglum AD, et al. Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiatry. 2015;72:635–41.
Conradi HJ, Ormel J, de Jonge P. Presence of individual (residual) symptoms during depressive episodes and periods of remission: a 3-year prospective study. Psychol Med. 2011;41:1165–74.
Kircher T, Wöhr M, Nenadic I, Schwarting R, Schratt G, Alferink J, et al. Neurobiology of the major psychoses: a translational perspective on brain structure and function-the FOR2107 consortium. Eur Arch Psychiatry Clin Neurosci. 2019;269:949–62.
Vogelbacher C, Möbius TWD, Sommer J, Schuster V, Dannlowski U, Kircher T, et al. The Marburg-Münster Affective Disorders Cohort Study (MACS): a quality assurance protocol for MR neuroimaging data. NeuroImage. 2018;172:450–60.
Wittchen H-U, Wunderlich U, Gruschwitz S, Zaudig M SKID I. Strukturiertes Klinisches Interview für DSM-IV. Achse I: Psychische Störungen. Interviewheft und Beurteilungsheft. Eine deutschsprachige, erweiterte Bearb. d. amerikanischen Originalversion des SKID I. 1997.
Redlich R, Almeida JR, Grotegerd D, Opel N, Kugel H, Heindel W, et al. Brain morphometric biomarkers distinguishing unipolar and bipolar depression: a voxel-based morphometry–pattern classification approach. JAMA Psychiatry. 2014;71:1222–30.
Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62.
de Lange SC, Helwegen K, van den Heuvel MP. Structural and functional connectivity reconstruction with CATO - A Connectivity Analysis TOolbox. Neuroimage. 2023;273:120108.
Cammoun L, Gigandet X, Meskaldji D, Thiran JP, Sporns O, Do KQ, et al. Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods. 2012;203:386–97.
Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80.
Meinert S, Nowack N, Grotegerd D, Repple J, Winter NR, Abheiden I, et al. Association of brain white matter microstructure with cognitive performance in major depressive disorder and healthy controls: a diffusion-tensor imaging study. Mol Psychiatry. 2022;27:1103–10.
van Velzen, Kelly LS, Isaev S, Aleman D, Aftanas LI A, Bauer J, et al. White matter disturbances in major depressive disorder: a coordinated analysis across 20 international cohorts in the ENIGMA MDD working group. Mol Psychiatry. 2020;25:1511–25.
Flinkenflügel K, Meinert S, Thiel K, Winter A, Goltermann J, Strathausen L, et al. Negative stressful life events and social support are associated with white matter integrity in depressed patients and healthy controls: a diffusion tensor imaging study. Biol Psychiatry. 2023. https://doi.org/10.1016/j.biopsych.2023.03.022.
Alexander AL, Hurley SA, Samsonov AA, Adluru N, Hosseinbor AP, Mossahebi P, et al. Characterization of cerebral white matter properties using quantitative magnetic resonance imaging stains. Brain Connect. 2011;1:423–46.
Andlauer TFM, Buck D, Antony G, Bayas A, Bechmann L, Berthele A, et al. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Sci Adv. 2016;2:e1501678.
Meller T, Schmitt S, Stein F, Brosch K, Mosebach J, Yüksel D, et al. Associations of schizophrenia risk genes ZNF804A and CACNA1C with schizotypy and modulation of attention in healthy subjects. Schizophr Res. 2019;208:67–75.
Pelin H, Ising M, Stein F, Meinert S, Meller T, Brosch K, et al. Identification of transdiagnostic psychiatric disorder subtypes using unsupervised learning. Neuropsychopharmacol. 2021;46:1895–905.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:s13742-015-0047-0048.
Howie B, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLOS Genetics. 2009;5:e1000529.
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9.
Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6.
Ahola-Olli AV, Würtz P, Havulinna AS, Aalto K, Pitkänen N, Lehtimäki T, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100:40–50.
Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776.
Ni G, Zeng J, Revez JA, Wang Y, Zheng Z, Ge T, et al. A comparison of ten polygenic score methods for psychiatric disorders applied across multiple cohorts. Biol Psychiatry. 2021;90:611–20.
Andlauer TFM, Nöthen MM. Polygenic scores for psychiatric disease: from research tool to clinical application. Medizinische Genetik. 2020;32:39–45.
Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. Neuroimage. 2010;53:1197–207.
MATLAB. 9701190202 (R2019b). http://citebay.com/how-to-cite/matlab/. Accessed 21 November 2022.
Horn JL. A rationale and test for the number of factors in factor analysis. Psychometrika. 1965;30:179–85.
Marek S, Tervo-Clemmens B, Calabro FJ, Montez DF, Kay BP, Hatoum AS, et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603:654–60.
Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM. Research review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014;55:1068–87.
Aminoff EM, Kveraga K, Bar M. The role of the parahippocampal cortex in cognition. Trends Cogn Sci. 2013;17:379–90.
Marek S, Dosenbach NUF. The frontoparietal network: function, electrophysiology, and importance of individual precision mapping. Dialogues Clin Neurosci. 2018;20:133–40.
Schiepers OJG, Wichers MC, Maes M. Cytokines and major depression. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:201–17.
Keller J, Gomez R, Williams G, Lembke A, Lazzeroni L, Murphy GM, et al. HPA axis in major depression: cortisol, clinical symptomatology and genetic variation predict cognition. Mol Psychiatry. 2017;22:527–36.
Pariante CM, Lightman SL. The HPA axis in major depression: classical theories and new developments. Trends in Neurosciences. 2008;31:464–8.
Favrais G, van de Looij Y, Fleiss B, Ramanantsoa N, Bonnin P, Stoltenburg-Didinger G, et al. Systemic inflammation disrupts the developmental program of white matter. Ann Neurol. 2011;70:550–65.
Miguel-Hidalgo JJ, Carter K, Deloach PH, Sanders L, Pang Y. Glucocorticoid-induced reductions of myelination and connexin 43 in mixed central nervous system cell cultures are prevented by mifepristone. Neuroscience. 2019;411:255–69.
Zia MTK, Vinukonda G, Vose LR, Bhimavarapu BBR, Iacobas S, Pandey NK, et al. Postnatal glucocorticoid-induced hypomyelination, gliosis, and neurologic deficits are dose-dependent, preparation-specific, and reversible. Neurodegeneration. 2015;263:200–13.
Nguyen L, Kakeda S, Watanabe K, Katsuki A, Sugimoto K, Igata N, et al. Brain structural network alterations related to serum cortisol levels in drug-naïve, first-episode major depressive disorder patients: a source-based morphometric study. Sci Rep. 2020;10:22096.
Pariante CM. Why are depressed patients inflamed? A reflection on 20 years of research on depression, glucocorticoid resistance and inflammation. Eur Neuropsychopharmacol. 2017;27:554–9.
Lopizzo N, Bocchio Chiavetto L, Cattane N, Plazzotta G, Tarazi FI, Pariante CM, et al. Gene-environment interaction in major depression: focus on experience-dependent biological systems. Front Psychiatry. 2015;6:68.
Bufalino C, Hepgul N, Aguglia E, Pariante CM. The role of immune genes in the association between depression and inflammation: A review of recent clinical studies. Brain Behav Immu. 2013;31:31–47.
Rani T, Behl T, Sharma N, Makeen HA, Albratty M, Alhazmi HA, et al. Exploring the role of biologics in depression. Cell Signal. 2022;98:110409.
Goldsmith DR, Haroon E, Woolwine BJ, Jung MY, Wommack EC, Harvey PD, et al. Inflammatory markers are associated with decreased psychomotor speed in patients with major depressive disorder. Brain, Behav Immu. 2016;56:281–8.
Krogh J, Benros ME, Jørgensen MB, Vesterager L, Elfving B, Nordentoft M. The association between depressive symptoms, cognitive function, and inflammation in major depression. Brain Behav Immunity. 2014;35:70–76.
MacKinnon DP, Pirlott AG. Statistical approaches for enhancing causal interpretation of the M to Y relation in mediation analysis. Pers Soc Psychol Rev. 2015;19:30–43.
Jaeger J, Zaragoza S. The digit symbol substitution test (DSST): psychometric properties and clinical utility in major depressive disorder. Eur Neuropsychopharmacol. 2016;26:S341.
Lamnidis TC, Majander K, Jeong C, Salmela E, Wessman A, Moiseyev V, et al. Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe. Nat Commun. 2018;9:5018.
Raison CL, Miller AH. Is depression an inflammatory disorder? Curr Psychiatry Rep. 2011;13:467–75.
Goltermann J, Redlich R, Grotegerd D, Dohm K, Leehr EJ, Böhnlein J, et al. Childhood maltreatment and cognitive functioning: the role of depression, parental education, and polygenic predisposition. Neuropsychopharmacology. 2021;46:891–9.
Acknowledgements
This work was funded by the German Research Foundation (DFG, grant FOR2107 DA1151/5-1 and DA1151/5-2 to UD; SFB-TRR58, Projects C09 and Z02 to UD), the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant Dan3/012/17 to UD), IMF Münster RE111604 to RR und RE111722 to RR, IMF Münster RE 22 17 07 to Jonathan Repple and the Deanery of the Medical Faculty of the University of Münster. TH was supported by the German Research Foundation (DFG grants HA7070/2-2, HA7070/3, HA7070/4). MP was supported by an ERC Consolidator grant (ERC-COG 101001062) and a NWO VIDI grant of the Dutch Research Council (Netherlands Organisation for Scientific Research Grant VIDI-452-16-015). SdL received funding from ZonMw Open Competition, project REMOVE 09120011910032. This work is part of the German multicenter consortium “Neurobiology of Affective Disorders. A translational perspective on brain structure and function“, funded by the German Research Foundation (Deutsche Forschungsgemeinschaft DFG; Forschungsgruppe/Research Unit FOR2107). Principal investigators (PIs) with respective areas of responsibility in the FOR2107 consortium are Work Package WP1, FOR2107/MACS cohort and brainimaging: Tilo Kircher (speaker FOR2107; DFG grant numbers KI 588/14-1, KI 588/14-2), Udo Dannlowski (co-speaker FOR2107; DA 1151/5-1, DA 1151/5-2), Axel Krug (KR 3822/5-1, KR 3822/7-2), Igor Nenadic (NE 2254/1-2), Carsten Konrad (KO 4291/3-1). WP2, animal phenotyping: Markus Wöhr (WO 1732/4-1, WO 1732/4-2), Rainer Schwarting (SCHW 559/14-1, SCHW 559/14-2). WP3, miRNA: Gerhard Schratt (SCHR 1136/3-1, 1136/3-2). WP4, immunology, mitochondriae: Judith Alferink (AL 1145/5-2), Carsten Culmsee (CU 43/9-1, CU 43/9-2), Holger Garn (GA 545/5-1, GA 545/7-2). WP5, genetics: Marcella Rietschel (RI 908/11-1, RI 908/11-2), Markus Nöthen (NO 246/10-1, NO 246/10-2), Stephanie Witt (WI 3439/3-1, WI 3439/3-2). WP6, multi-method data analytics: Andreas Jansen (JA 1890/7-1, JA 1890/7-2), Tim Hahn (HA 7070/2-2), Bertram Müller-Myhsok (MU1315/8-2), Astrid Dempfle (DE 1614/3-1, DE 1614/3-2). CP1, biobank: Petra Pfefferle (PF 784/1-1, PF 784/1-2), Harald Renz (RE 737/20-1, 737/20-2). CP2, administration. Tilo Kircher (KI 588/15-1, KI 588/17-1), Udo Dannlowski (DA 1151/6-1), Carsten Konrad (KO 4291/4-1). Anxiety extension project Benjamin Straube (STR 1146/18-1), Tilo Kircher (KI 588/22-1), Udo Dannlowski (DA 1151/11‑1). Martijn van den Heuvel was supported by an ALW open (ALWOP.179) and VIDI (452-16-015) grant from the Netherlands Organization for Scientific Research (NWO) and a Fellowship of MQ. Data access and responsibility: All PIs take responsibility for the integrity of the respective study data and their components. All authors and coauthors had full access to all study data. Acknowledgements and members by Work Package (WP): WP1: Henrike Bröhl, Katharina Brosch, Bruno Dietsche, Rozbeh Elahi, Jennifer Engelen, Sabine Fischer, Jessica Heinen, Svenja Klingel, Felicitas Meier, Tina Meller, Julia-Katharina Pfarr, Kai Ringwald, Torsten Sauder, Simon Schmitt, Frederike Stein, Annette Tittmar, Dilara Yüksel (Dept. of Psychiatry, Marburg University). Mechthild Wallnig, Rita Werner (Core-Facility Brainimaging, Marburg University). Carmen Schade-Brittinger, Maik Hahmann (Coordinating Centre for Clinical Trials, Marburg). Michael Putzke (Psychiatric Hospital, Friedberg). Rolf Speier, Lutz Lenhard (Psychiatric Hospital, Haina). Birgit Köhnlein (Psychiatric Practice, Marburg). Peter Wulf, Jürgen Kleebach, Achim Becker (Psychiatric Hospital Hephata, Schwalmstadt-Treysa). Ruth Bär (Care facility Bischoff, Neukirchen). Matthias Müller, Michael Franz, Siegfried Scharmann, Anja Haag, Kristina Spenner, Ulrich Ohlenschläger (Psychiatric Hospital Vitos, Marburg). Matthias Müller, Michael Franz, Bernd Kundermann (Psychiatric Hospital Vitos, Gießen). Christian Bürger, Katharina Dohm, Fanni Dzvonyar, Verena Enneking, Stella Fingas, Katharina Förster, Janik Goltermann, Dominik Grotegerd, Hannah Lemke, Susanne Meinert, Nils Opel, Ronny Redlich, Jonathan Repple, Katharina Thiel, Kordula Vorspohl, Bettina Walden, Lena Waltemate, Kira Flinkenflügel, Alexandra Winter, Dario Zaremba (Dept. of Psychiatry, University of Münster). Harald Kugel, Jochen Bauer, Walter Heindel, Birgit Vahrenkamp (Dept. of Clinical Radiology, University of Münster). Gereon Heuft, Gudrun Schneider (Dept. of Psychosomatics and Psychotherapy, University of Münster). Thomas Reker (LWL-Hospital Münster). Gisela Bartling (IPP Münster). Ulrike Buhlmann (Dept. of Clinical Psychology, University of Münster). WP2: Marco Bartz, Miriam Becker, Christine Blöcher, Annuska Berz, Moria Braun, Ingmar Conell, Debora dalla Vecchia, Darius Dietrich, Ezgi Esen, Sophia Estel, Jens Hensen, Ruhkshona Kayumova, Theresa Kisko, Rebekka Obermeier, Anika Pützer, Nivethini Sangarapillai, Özge Sungur, Clara Raithel, Tobias Redecker, Vanessa Sandermann, Finnja Schramm, Linda Tempel, Natalie Vermehren, Jakob Vörckel, Stephan Weingarten, Maria Willadsen, Cüneyt Yildiz (Faculty of Psychology, Marburg University). WP4: Jana Freff (Dept. of Psychiatry, University of Münster). Susanne Michels, Goutham Ganjam, Katharina Elsässer (Faculty of Pharmacy, Marburg University). Felix Ruben Picard, Nicole Löwer, Thomas Ruppersberg (Institute of Laboratory Medicine and Pathobiochemistry, Marburg University). WP5: Helene Dukal, Christine Hohmeyer, Lennard Stütz, Viola Lahr, Fabian Streit, Josef Frank, Lea Sirignano (Dept. of Genetic Epidemiology, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University). Stefanie Heilmann-Heimbach, Stefan Herms, Per Hoffmann (Institute of Human Genetics, University of Bonn, School of Medicine & University Hospital Bonn). Andreas J. Forstner (Institute of Human Genetics, University of Bonn, School of Medicine & University Hospital Bonn; Centre for Human Genetics, Marburg University). WP6: Anastasia Benedyk, Miriam Bopp, Roman Keßler, Maximilian Lückel, Verena Schuster, Christoph Vogelbacher (Dept. of Psychiatry, Marburg University). Jens Sommer, Olaf Steinsträter (Core-Facility Brainimaging, Marburg University). Thomas W.D. Möbius (Institute of Medical Informatics and Statistics, Kiel University). CP1: Julian Glandorf, Fabian Kormann, Arif Alkan, Fatana Wedi, Lea Henning, Alena Renker, Karina Schneider, Elisabeth Folwarczny, Dana Stenzel, Kai Wenk, Felix Picard, Alexandra Fischer, Sandra Blumenau, Beate Kleb, Doris Finholdt, Elisabeth Kinder, Tamara Wüst, Elvira Przypadlo, Corinna Brehm (Comprehensive Biomaterial Bank Marburg, Marburg University). The data represent original work. The manuscript has been published on a non-commercial preprint server (https://osf.io/5b6ek/).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Substantial contribution to conception and design: KF, MG, JR and UD. Substantial contribution to the acquisition of the data: KF, MG, SM, KT, AW, JG, PU, KB, FS, AWro, FTO, JKP, FSD, ECB, DG, TH, EJB, KD, JB, AJ, MMN, AJF, HJ, BS, NA, SHW, MR, IN, MvdH, TK, JR, UD. Analysis and interpretation of data: KF, MG, JR, UD. Drafting the article: KF and MG. Revising it critically for important intellectual content: SM, KT, AW, JG, PU, KB, FS, AWro, FTO, JKP, FSD, ECB, DG, TH, EJB, KD, JB, AJ, MMN, AJF, HJ, BS, NA, SHW, MR, IN, MvdH, TK, JR, UD. Revision of original manuscript: KF and MG. JR and UD supervised this review. All authors contributed to the article and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
TK received unrestricted educational grants from Servier, Janssen, Recordati, Aristo, Otsuka, neuraxpharm. This funding is not associated with the current work. On behalf of all other authors, the corresponding author states that there is no conflict of interest and nothing to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Flinkenflügel, K., Gruber, M., Meinert, S. et al. The interplay between polygenic score for tumor necrosis factor-α, brain structural connectivity, and processing speed in major depression. Mol Psychiatry (2024). https://doi.org/10.1038/s41380-024-02577-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-024-02577-7