Abstract
Background:
CTX-M-15 is the dominant type of extended-spectrum β-lactamase in clinical isolates. This enzyme constitutes the most widespread enzymes in Tunisia. In this study, we were interested to understand the causes of the evolutionary success of CTX-M-15 in a Tunisian university hospital.
Methods:
A total of of 72 cefotaxime-resistant Enterobacteriaceae were isolated from newborn patients at the hospital Taher sfar Mahdia in Tunisia and characterized their genetic support by means of molecular techniques.
Results:
Isolates were clustered into various clonal groups, although most isolates belonged to sequence types ST39 (Klebsiella pneumoniae) and ST131 (Escherichia coli). F replicons (FIA, FIB, and FII) were the most frequently detected replicon types in our collection (91.66%).
Conclusion:
This is the first report of QnrB- and CTX-M-15-encoding large IncF-type conjugative plasmids in Tunisia.
Similar content being viewed by others
Main
Most countries have recently experienced the rapid dissemination of Enterobacteriaceae isolates producing extended spectrumBackground: β-lactamases (ESBLs) (1,2). During the past decade, CTX-M-type enzymes have represented the most rapidly growing group of ESBLs, and CTX-M-15 (a variant of CTX-M-3, resulting in a single Asp-240-Glu substitution) has recently emerged as the dominant type of plasmid-borne acquired cefotaximase in Gram-negative pathogens, which caused outbreaks in both nosocomial and community settings (3,4). Clonal outbreaks of CTX-M-15-producing Enterobacteriaceae have been reported in many countries, including France, Italy, Spain, Portugal, Austria, Norway, the United Kingdom, Tunisia, South Korea, and Canada, being Escherichia coli is the most frequently involved organism (5). Plasmid carrying the blaCTX-M-15 gene are mostly incompatibility group FII plasmids (6,7). Furthermore, different genetic elements have been shown to be involved in the mobilization of blaCTX-M-15 including ISEcp1-like insertion sequences, which are the most commonly reported (8).
In the present study, we report the emergence of CTX-M-15-producing isolates from a recent collection of Enterobacteriaceae strains from newborn and characterized their genetic support by means of molecular techniques to better understand the causes of the evolutionary success of this β-lactamase in Tunisia.
Results
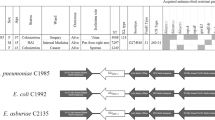
From January to July 2011, 72 multiresistant ESBL-producing clinical isolates of Enterobacteriaceae were collected from the Microbiology Laboratories at the University Hospital of Mahdia in Tunisia. The isolates were recovered from various pathological specimens ( Table 1 ).
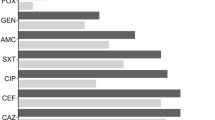
ESBL detection performed by the double-disk diffusion test revealed synergies between amoxicilin–clavulanic acid and oxyiminocephalosporins (cefotaxime, ceftazidime) containing disks with higher level of resistance to cefotaxime for all isolates, suggesting the production of a cefotaxime-type extended-spectrum β-lactamase. The most isolates showed similar susceptibility profiles, characterized by high minimum inhibitory concentration value (>1,024 μg/ml) for cefotaxime and ticarcillin, while being resistant to ceftazidime, cefepime, and aztreonam. Furthermore, some of the strains were also found to be resistant to ciprofloxacin ( Table 2 ).
Polymerase chain reaction (PCR) analysis for β-lactamase genes of the family TEM, SHV, and CTX-M showed amplification products only for blaTEM and blaCTX-M genes. DNA sequence analysis of these PCR products revealed that all isolates carried both the blaCTX-M-15 and the blaTEM-1 genes class. Detection of qnrA, qnrB, and qnrS quinolone resistance determinants revealed that 63 of 72 isolates carried the qnrB1 gene, while qnrB4 was detected in a single isolate of Klebsiella oxytoca isolate ( Table 1 ). The copresence of qnrB4 and CTX-M-15 genes was not, at our best knowledge, previously reported in Tunisia. PCR mapping of the genetic environment surrounding the blaCTX-M-15 identified the same ISEcp1 elements in the upstream region of all resistant strains. The presence of the ISEcp1 element in the vicinity of blaCTX-M genes was analyzed as reported previously (9). It was located 48 bp upstream from blaCTX-M-15 demonstrating that this gene was present in widespread ISEcp1 transposition modules (10).
PFGE analysis of the 72 CTX-M-15-producing isolates showed two different types of two E. coli, three clone of Klebsiella pneumoniae, and two clone of Serratia marcescens isolates. Isolates belonging to the same clonal group were recovered from the same ward, indicating the local simultaneous dissemination of several enterobacterial strains/clones rather than the spread of a single clone. Isolates of all PFGE types were subjected to multilocus sequence typing ( Table 1 ). The K. pneumoniae isolates represented sequence types ST39 and ST4, which are identified the first time in Tunisia. The E. coli isolates represented STs belonging to international clones from humans, ST131 and the ST69, which have been identified as an important cause of infection outbreaks in different communities (11,12,13) and the most prevalent clonal groups among CTX-M-15-producing E. coli isolates in Europe, Asia, North America, and Africa (11,14).
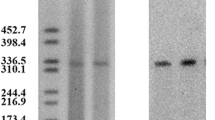
Cefotaxime and ceftazidime resistance was successfully transferred by conjugation from all CTX-M-15-producing strains representative of the different clones. Production of ESBLs was detected in all transconjugants by the double-disk synergy test and the presence of the CTX-M-15 was confirmed by sequence analysis of PCR product from these transconjugants. These findings indicate that the CTX-M-15 was located in conjugative plasmids and confer resistance to cefotaxime and ceftazidime. Southern hybridization analysis revealed that each clinical isolate carried a single plasmid encoding blaCTX-M-15 with size of 100 kb and over (data not shown). Classification based on plasmid incompatibility showed that the F replicons (FIA, FIB, FIC, and FII) were the most frequently detected replicon types in our collection (91.66%). These results are in accordance with those of other authors (15,16).
Discussion
This study demonstrates the predominant presence of CTX-M-15-producing Enterobacteriaceae among ESBL-positive isolates recovered from hospitalized newborns in Hospital Tahar Sfar in Tunisia. The most of the plasmids transferred from the clonal strains studied, belonged to the incompatibility (Inc) group. This, in addition to the presence on the same DNA fragment of the particular set of resistance genes described above, including blaCTX-M-15, suggested the presence of a multidrug resistance region similar to that described for plasmid pC15-1a by Lavollay et al. (17). Plasmid pC15-1a is a circular molecule of 92,353 bp consisting of two distinct regions. The first is a 64-kb region that is essentially homologous to the non-R-determinant region of plasmid R100 except for several point mutations, a few small insertions and deletions, and the absence of Tn10. The second is a 28.4-kb multidrug resistance region that has replaced the R-determinant region of the R100 progenitor and consists mostly of transposons or partial transposons and five copies of the insertion element IS26 (18). In fact, the majority of the blaCTX-M-15 genes were mobilized on various multiresistance IncF-type plasmids, harboring multiple addiction systems that presumptively contribute to their maintenance (19). These addiction models such as toxin–antitoxin systems modules (pemK–pemI, hok–sok, or ccdA–ccdB) have been described for the first time in Tunisia by mnif et al. (20). Curiously, the L/M replicon was detected in six of the strains (8.33%), although this replicon has been considered to be rare in Enterobacteriaceae isolates from humans (21). The spacer describes above between the ISEcp1 and the start of the gene blaCTX-M-15 carried by L/M plasmid is 48 bp and not 127 bp as decribed for CTX-M-3 and CTX-M-15 in Algeria (22). This group of plasmids was initially responsible for the spread of CTX-M-3 in Poland since common plasmid scaffolds were identified in eight species in 15 hospitals (23). The representative plasmid of that family was pCTX-M-3, first observed in 1996 in C. freundii isolates in which CTX-M-3 had been originally identified (24). IncL/M plasmids carrying the blaCTX-M-3 gene were also reported in Eastern countries, France, Belgium, and South Korea and very often encoded, in addition to CTX-M-3 ESBL, the aminoglycoside resistance gene armA. IncL/M family is also responsible for the spread of the class D carbapenemase OXA-48 that has been identified in many clonally unrelated strains and different enterobacterial species from distantly located geographic areas (25).
This is the first report of a CTX-M-15 occurrence in Tunisia associated with large conjugative plasmids with exclusive IncF replicon-type. IncF plasmids have been reported to be of low copy number with more than one replicon promoting the initiation of replication. The multireplicon status has been described to be one means by which plasmids with a narrow host range can accomplish broad host range replication (26), which explains why they are widely diffused in clinically relevant Enterobacteriaceae, representing one of the most frequently encountered plasmid types. Addiction systems encoded by these plasmids also contributed to the promotion of plasmid spread and adaptation to the host (19). Thus, The IncF plasmid family is clearly playing a major role in the dissemination of antimicrobial resistance in Enterobacteriaceae. Based on these findings, the high prevalence of CTX-M-15 is not only due to the spread of a single clone, mainly the pandemic ST131 clone, but is also due to horizontal transfer of multiresistance IncF, which remains widely scattered across Tunisia.
Methods
Bacterial Isolates
A total of 72 consecutive, nonredundant multidrug-resistant clinical isolates of Enterobacteriaceae were recovered from rectal swabs of newborn hospitalized patients at the University Hospital of Tunisia in the period January–June 2011. All isolates were identified using the VITEK 2 system (bioMérieux, La Balme-les-Grottes, France) and the API 20E system (bioMérieux). The collection consisted of 48 K. pneumoniae, 18 E. coli, 2 Serratia marcescens, 1 Enterobacter aerogenes, 1 Citrobacter freundii, 1 Enterobacter cloacae, and 1 K. oxytoca.
Antimicrobial Susceptibility Testing
All the study isolates were phenotypically screened for the production of ESBLs using the double-disk synergy test (27). The minimum inhibitory concentration was determined by a dilution technique on Mueller-Hinton liquid medium with inoculums of 106 colony-forming units (CFU). The results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (28).
Characterization of β-Lactamase and qnr-Encoding Genes
The detection of ESBL-encoding genes was performed by PCR using primers specific for the amplification of blaTEM, blaSHV, and blaCTX-M genes (29). Detection of qnrA, qnrB, and qnrS quinolone resistance determinants was performed similarly by multiplex PCR (30).
Genetic Environment of blaCTX-M Genes
The genetic organization of blaCTX-M was investigated by PCR and by sequencing the regions surrounding these genes. Primers used to investigate the surrounding regions of the blaCTX-M are performed according to ref. (31).
Molecular Epidemiology
The clonal relation between the different clinical isolates was studied by pulsed-field gel electrophoresis using the XbaI restriction enzyme for 4 h at 37 °C to generate the macrorestriction fragments prior to electrophoretic separation (1% agarose gel). All the different clones were characterized by multilocus sequence typing according to the protocol described previously (32,33).
Transfer of Resistance
Transfer of the gene was studied by conjugation experiments with rifampicin-resistant E. coli MKD-135 recipient strain, as described previously (34). The transconjugants were selected with 2 µg/ml cefotaxime and 400 µg/ml rifampicin and they are subjected to an ESBL screening test and PCR to confirm the possible acquisition of blaCTX-M.
Hybridization
Plasmid DNA extraction was performed by using the Kieser method (35) followed by a Southern transfer of an agarose gel containing plasmid DNA of CTX-M-positive isolates onto a nylon membrane (Hybond N+; GE Healthcare, Orsay, France). Plasmid DNA hybridization was performed as described by Sambrook et al. (36). The probes specific for the CTX-M genes consisted of PCR products generated from respective CTX-M-positive isolates. Labeling of those probes and signal detection were carried out using the electrochemiluminescence nonradioactive labeling and detection kit according to the manufacturer’s instructions (GE Healthcare).
Identification of Plasmids by PCR-Based Replicon Typing
PCR-based replicon typing was applied to type the resistance plasmids using the major plasmid incompatibility groups among Enterobacteriaceae (37). Eighteen pairs of primers were designed to perform five multiplex- and three simplex-PCRs, recognizing FIA, FIB, FIC, HI1, HI2, I1-Iγ, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIA.
Ethical Information
The study was approved by the Military Hospital of Tunis, Tunisia. Parental informed consent was obtained.
Statement of Financial Support
This work was partially funded by a grant from the Ministry of Scientific Research Technology and Competence Development of Tunisia and by a grant of the Italian “Ministero dell’Istruzione, Università e Ricerca” (MIUR, contract no. 200929YFMK_004).
Disclosure
None.
References
Oteo J, Lázaro E, de Abajo FJ, Baquero F, Campos J ; Spanish members of EARSS. Antimicrobial-resistant invasive Escherichia coli, Spain. Emerg Infect Dis 2005;11:546–53.
Cantón R, Novais A, Valverde A, et al. Prevalence and spread of extended-spectrum β -lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect 2000;14:144–53.
Mesko Meglic K, Koren S, Palepou MF, et al.; Slovenian ESBL Study Group. Nationwide survey of CTX-M-type extended-spectrum beta-lactamases among Klebsiella pneumoniae isolates in Slovenian hospitals. Antimicrob Agents Chemother 2009;53:287–91.
Oteo J, Cuevas O, López-Rodríguez I, et al. Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J Antimicrob Chemother 2009;64:524–8.
Coque TM, Novais A, Carattoli A, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis 2008;14:195–200.
Hopkins KL, Liebana E, Villa L, Batchelor M, Threlfall EJ, Carattoli A . Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob Agents Chemother 2006;50:3203–6.
Novais A, Cantón R, Moreira R, Peixe L, Baquero F, Coque TM . Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob Agents Chemother 2007;51:796–9.
Bonnet R . Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 2004;48:1–14.
Eckert C, Gautier V, Arlet G . DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother 2006;57:14–23.
Poirel L, Decousser JW, Nordmann P . Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase gene. Antimicrob Agents Chemother 2003;47:2938–45.
Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 2008;61:273–81.
Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M . Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 2010;51:286–94.
Tartof SY, Solberg OD, Manges AR, Riley LW . Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol 2005;43:5860–4.
Rogers BA, Sidjabat HE, Paterson DL . Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 2011;66:1–14.
Sherley M, Gordon DM, Collignon PJ . Species differences in plasmid carriage in the Enterobacteriaceae. Plasmid 2003;49:79–85.
Johnson TJ, Wannemuehler YM, Johnson SJ, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 2007;73:1976–83.
Lavollay M, Mamlouk K, Frank T, et al. Clonal dissemination of a CTX-M-15 beta-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob Agents Chemother 2006;50:2433–8.
Boyd DA, Tyler S, Christianson S, et al. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 2004;48:3758–64.
Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N . Characterization of plasmids encoding extended-spectrum β-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J Antimicrob Chemother 2012;67:878–85.
Mnif B, Vimont S, Boyd A, et al. Molecular characterization of addiction systems of plasmids encoding extended-spectrum beta-lactamases in Escherichia coli. J Antimicrob Chemother 2010;65:1599–603.
Elhani D, Bakir L, Aouni M, et al. Molecular epidemiology of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999-2005. Clin Microbiol Infect 2010;16:157–64.
Baba Ahmed-Kazi Tani Z, Decré D, Genel N, Boucherit-Otmani Z, Arlet G, Drissi M . Molecular and epidemiological characterization of enterobacterial multidrug-resistant strains in Tlemcen Hospital (Algeria) (2008-2010). Microb Drug Resist 2013;19:185–90.
Baraniak A, Fiett J, Sulikowska A, Hryniewicz W, Gniadkowski M . Countrywide spread of CTX-M-3 extended-spectrum beta-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob Agents Chemother 2002;46:151–9.
Gołebiewski M, Kern-Zdanowicz I, Zienkiewicz M, et al. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob Agents Chemother 2007;51:3789–95.
Poirel L, Bonnin RA, Nordmann P . Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 2012;56:559–62.
Villa L, García-Fernández A, Fortini D, Carattoli A . Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 2010;65:2518–29.
Jarlier V, Nicolas MH, Fournier G, Philippon A . Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 1988;10:867–78.
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; 20th Informational Supplement M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute, 2010.
Lartigue MF, Zinsius C, Wenger A, Bille J, Poirel L, Nordmann P . Extended-spectrum beta-lactamases of the CTX-M type now in Switzerland. Antimicrob Agents Chemother 2007;51:2855–60.
Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC . qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother 2006;50:2872–4.
Brasme L, Nordmann P, Fidel F, et al. Incidence of class A extended-spectrum beta-lactamases in Champagne-Ardenne (France): a 1 year prospective study. J Antimicrob Chemother 2007;60:956–64.
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S . Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005;43:4178–82.
Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006;60:1136–51.
Pallecchi L, Malossi M, Mantella A, et al. Detection of CTX-M-type beta-lactamase genes in fecal Escherichia coli isolates from healthy children in Bolivia and Peru. Antimicrob Agents Chemother 2004;48:4556–61.
Kieser T . Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 1984;12:19–36.
Sambrook J, Fritsch EF, Maniatis T . Molecular Cloning: A Laboratory Manwal. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 1989.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ . Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 2005;63:219–28.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lahlaoui, H., De Luca, F., Maradel, S. et al. Occurrence of conjugative IncF-type plasmids harboring the blaCTX-M-15 gene in Enterobacteriaceae isolates from newborns in Tunisia. Pediatr Res 77, 107–110 (2015). https://doi.org/10.1038/pr.2014.153
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2014.153
This article is cited by
-
Diversity of clonal types of Klebsiella pneumoniae causing infections in intensive care neonatal patients in a large urban setting
Brazilian Journal of Microbiology (2019)