Abstract
The human cytochrome oxidase complex is a multisubunit assembly in the inner mitochondrial membrane responsible for the terminal event in electron transport in which molecular oxygen is reduced. Various phenotypic forms of cytochrome oxidase deficiency have been recognized, the major varieties involving degeneration of the brain stem and basal ganglia (Leigh syndrome) and lactic acidemia. Others include a fatal infantile form, a benign reversible form, and forms with cardiomyopathy. Early recognition of complementation groups within, for instance, the Leigh syndrome group has recently been followed up with a description of the gene defect for three of the nuclear-encoded forms of cytochrome c oxidase (COX) deficiency. The three genes indicted, SURF1 for Leigh syndrome, COX 10 for leukodystrophy and tubulopathy, and SCO2 for the cardiomyopathic form, all have a role in the assembly of the mature cytochrome oxidase complex. The description of these gene defects and the role these genes play are discussed in terms of what can be learned about COX assembly and about the etiology of the different phenotypic forms of the disease.
Similar content being viewed by others
CYTOCHROME OXIDASE DEFICIENCY: CLINICAL PHENOTYPES
Until recently, the genetic basis for the nuclear defects associated with the mitochondrial respiratory chain remained undefined. Nuclear defects resulting in severe lactic acidemia or progressive neurologic deterioration have been well documented on a clinical and biochemical basis for some time.
Human COX deficiency is one of the many defects associated with lactic acidemia and occurs in several phenotypic forms. Although this group of defects has been biochemically defined for some time, the molecular and genetic nature of these defects has been something of a mystery until recently. The most common form is associated with Leigh disease, progressive deterioration of the basal ganglia and brain stem. The defect leads to an 80 to 90% decrease in COX activity and seems to affect all tissues (1–5). There are variations in the rate of progression of the neurodegeneration, but it is always fatal with death occurring from between 6 mo and 12 y of age (1–5). The Saguenay-Lac-St. Jean form of cytochrome oxidase (SLSJ-COX) deficiency is a unique form of the defect with a less severe form of Leigh disease than the classic form. This form occurs only in the Charlevoix and Saguenay-Lac-St. Jean region of Québec where the carrier frequency rate is one in 23 for the defect (6). Children with this defect are mildly psychomotor retarded and function reasonably well, but many die between 3 and 10 y of age, death occurring as the result of a sudden onset of fulminant lactic acidosis (6, 7). In biochemical terms, the liver and brain are severely affected as in classic Leigh syndrome with COX deficiency, but the kidney and heart have almost normal cytochrome oxidase activity whereas skeletal muscle has approximately 50% (7) (Fig. 1). Both in the SLSJ-COX and classic COX with Leigh syndrome, a failure of assembly of the complex is evident (6–8).
The different clinical phenotypes associated with nuclear-encoded COX deficiency. The defects are arranged as to their major clinical presentation, severity of lactic acidosis, tissue specificity, and genes identified as causative. Tissue specificity is indicated by the abbreviations H, heart;K, kidney;B, brain;S, skeletal muscle;L, liver; and F, skin fibroblasts, with the most severely affected to the least affected tissue going from left to right.
SLSJ-COX and classic COX deficiency presenting with Leigh disease, as above, are two of five phenotypic forms that are recognized to make up the spectrum of inherited COX deficiency. These basic types and the tissues known to be affected are shown in Figure 1. Fatal infantile COX deficiency is the third and most severe type and causes death from lactic acidosis in the newborn period and presents with a renal Fanconi syndrome and myopathy (9–13). The brain and liver appear to be spared in this type, having normal or close to normal activity of cytochrome oxidase but with severe deficiency in kidney and muscle. A subtype of this form may lack the overt kidney involvement, whereas there is a separate form with tubulopathy combined with leukodystrophy (9–14). Another phenotype in which the brain and liver often are spared is a category of myopathy with cardiomyopathy (1, 4, 15, 16). That this is not a variation of the fatal infantile form is shown by the biochemical data in fibroblasts. Whereas the former type is not detectable at all in fibroblasts, the cardiomyopathy type displays a raised lactate to pyruvate ratio and COX activity of 35 to 45% (16, 17). A distinct reversible muscle form of the disease has been well documented by DiMauro et al. (18). In this type, there is a presentation of lactic acidosis, hypotonia, hyporeflexia, and generalized weakness in the neonatal period associated with deficient muscle cytochrome oxidase activity. Reversion of the defect takes place gradually over a 2-y period, COX activities return to normal, and the lactic acidosis goes away (18).
RESOLUTION OF THE GENETIC DEFECTS
Sequencing of the nuclear-encoded subunits of COX by a number of groups did not throw any light on the genetic origin of the cytochrome oxidase deficiencies because, with few exceptions, the results showed no change (19, 20). At the same time, three complementation groups were shown to exist for classic COX deficiency with Leigh syndrome (21). This strongly suggested that the genes responsible were coding for factors involved in assembly of the COX multisubunit enzyme, so approaches were used to narrow down the search area by the use of single chromosome transfer (22, 23), sequencing candidate genes (24), and linkage analysis (25, 26).
The first report defining a genetic defect associated with the COX-Leigh syndrome group of patients was that of Zhu et al. (22). Using single chromosome transfer from microcells generated from a mouse-human chromosome hybrid cell panel, this group found chromosome 9 to correct the COX defect in these patient cell lines. Further analysis with markers delineated an area that contained the gene SURF1, a human homologue of Shy1, a known yeast COX assembly factor (22). Tiranti et al. (23) achieved a similar result by fusing whole mouse cell nuclei containing human chromosomes, including chromosome 9, to deficient human skin fibroblasts; only chromosome 9 nuclei corrected the defect. Although this latter result suggests that mouse SURF1 is unable to complement the human gene defect because mouse cells with no human chromosome were noncomplementing, both groups presented evidence of mutated SURF1 genes in patients (22, 23, 27–29). There appears to be at least one common mutation in Caucasian patients that may account for 35 to 50% of all alleles (27–29).
Linkage analysis was used to obtain a locus for COX deficiency with leukodystrophy and tubulopathy in an African kindred by Valnot et al. (25). This group showed that a marker on chromosome 17 was common to all affected patients. The COX10 gene that codes for a protein catalyzing the addition of a farnesyl group to the heme to produce heme a also mapped to this locus, and missense mutations were discovered in the cDNA for this gene (25).
The third gene to be pinpointed in recent months was the SCO2 gene as the mutant gene in the cardiomyopathic form of COX. This gene was evaluated by the group of Papadopolou et al. (24) as a candidate for COX deficiency phenotypes, and mutations were found in the cDNA for SCO2 in some of the cardiomyopathic COX patients. Subunits I and II were both decreased remarkably in titer in muscle, indicating failure of assembly of the complex. Interestingly, for two patients with identical mutations in SCO2 (Q53X/E140 K compounds), the residual activity of COX in liver was 16 and 52% of normal, respectively. This points to the possibility that either modifying genes or modifying environmental factors, e.g. daily Cu intake, may have a bearing on the eventual outcome of any one SCO2 genotype, but extensive data on tissue residual activity of COX and Cu content are currently lacking. To understand how these defective genes bring about COX deficiency depends on knowledge of the structure and assembly of COX.
STRUCTURE OF THE CYTOCHROME OXIDASE COMPLEX
The human cytochrome oxidase complex (complex IV) is one of five mitochondrial multisubunit complexes responsible for the process of oxidative phosphorylation. Its function is to take electrons from cytochrome c generated from substrate oxidation for the immediate reduction of molecular oxygen to water. In the process, a contribution to the electrochemical gradient across the mitochondrial membrane is made that is used to drive ATP synthesis via the F1F0-ATPase (complex V).
Complex IV itself is a multicomponent membrane protein complex of molecular size 200 kD. It is made up of 13 protein components, 10 of which are encoded in the nucleus and three of which are encoded in mitochondrial DNA (30). The core of the cytochrome oxidase complex is made up from the protein products of the COX I, II, and III genes that are encoded in mitochondrial DNA. Many bacterial oxidases have little more than these three subunits to constitute a fully functional complex. However, both mammalian and yeast systems have difficulty assembling a functional complex unless the 10 additional nuclear subunits are present (31, 32). Subunit I contains both the hemes and one of the Cu ions (CuB), whereas subunit II contains a binuclear Cu assembly (CuA) (30) (Fig. 2). Subunit III has no prosthetic groups and does not appear to be involved in pumping protons, so its role may be structural (33). An Mg2+ ion is coordinated between elements of subunits I and II and is close to the CuA site (34). A Zn2+ ion is coordinated by elements of Vb, but the function is unknown (34). Limited gene mapping has been done in humans, the complicating factor being that for nearly all the nuclear-encoded subunits, there are one to seven pseudogenes (35–37). Perhaps the most interesting subunits as far as human disease is concerned are those that exist as isoenzymes in different tissues. The subunits VIa and VIIa have heart- and liver-specific isoforms so that heart and skeletal muscle express the heart form, whereas brain and liver express the liver form with kidney, lymphoblasts, and fibroblasts in between (30, 35, 38–41).
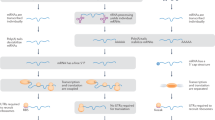
Factors involved in COX assembly. The diagram shows the approximate position of the two hemes (a and a3) in the cytochrome oxidase complex located in the inner mitochondrial membrane. The cytochrome oxidase complex is made up of the mtDNA-encoded COX I, COX II, and COX III subunits that form the core of the complex. This is represented in the diagram by the green oval structure, as it contains the important functional groups. There are two Cu centers, CuA close to the outer surface of the membrane held by subunit COX II and CuB close to the heme a3 where oxygen becomes Fe coordinated. The Cu ions require delivery to the outside face of the inner mitochondrial membrane by COX 17p and incorporation into the membrane by SCO1p or SCO2p. Further delivery to its site in the complex for CuB may depend on COX 11p. The SURF1 protein is required for assembly of the proteins COX I, COX II, and COX III, whereas other genes such as COX 10 are required for heme a synthesis, heme modification, and heme incorporation.
Electron transfer into cytochrome oxidase proceeds initially by the binding of cytochrome c to subunit II on the outer face of the inner mitochondrial membrane. Electrons are then transferred to the bimetallic CuA site just inside the plane of the membrane and then on to heme a, then heme a3, which then reacts with the CuB center to reduce molecular oxygen (30, 34). During this process, one proton is pumped across the membrane for each electron transferred (42). The x-ray crystal structure has been determined at the level of 2,8 Å for bovine cytochrome oxidase (34). The structure resolution so far published is confined to the core of the complex and metal-binding functions, though more data are now emerging on the nuclear-encoded subunits. The conformation of subunit I was essentially as predicted in that it is composed of 12 transmembrane helices that house the two heme groups and the CuB site within the confines of the mitochondrial inner membrane (34). The disposition of the CuA site was surprising in that it was configured as a [2Cu-2S] center similar to the [2Fe-2S] centers found in the proximal areas of the respiratory chain. The CuB was shown to be coordinated by the three histidines of subunit I, 240, 290, and 291, whereas Tyr 244, whose OH group is hydrogen bonded to the hydroxyfarnesylethyl side chain of heme a3, is ideally positioned also to coordinate with the CuB in a way that suggests involvement in proton pumping (34). The process of proton pumping has recently been resolved into two phases so that half of the protons are pumped during the reduction of the hemes whereas the other half are pumped in response to the oxidative phase of the reaction (43).
THE ASSEMBLY OF COX
A possible picture of how COX is assembled is beginning to emerge, aided by the discovery of these genes and their role in the assembly process. The first subunit to assemble is COX I. This has 12 membrane-spanning segments and because it is translated on mitochondrial ribosomes, it is inserted into the membrane from the matrix side, possibly aided by the Oxa 1 protein (44). In Oxa 1 knockouts of yeast, COX fails to assemble. In addition, substitution of the human Oxa 1 into the yeast knockout produces a working assembled complex, but it is leaky to protons, thought to be a result of misfolding of COX I, which is the proton conduit for cytochrome oxidase (45). The next steps appear to be addition of the two hemes and Cu into the transmembrane region of COX I. This may require the services of the membrane-spanning SCO2, which has a consensus binding site for Cu, and it has been suggested that this protein acts as the recipient of Cu from the COX 17 protein, a binuclear Cu carrier (46). Mutations in SCO2 prevent the Cu from gaining proper access to the mitochondria, though why the heart should be more adversely affected than in other COX defects is not clear (24). The COX 11 protein seems to be necessary for the insertion of CuB into the COX I protein. In COX 11 bacterial knockouts, CuA is incorporated even though CuB is missing, showing that the two processes are independent (47). The farnesylation of heme by the COX 10 protein, which is a 7TM helix protein present in the inner mitochondrial membrane, could take place before or after insertion of the heme into COX I (48). The formylation of heme requires a protein, but the gene that encodes the agent for this is not identified in any organism at this moment. The nuclear subunit COX IV is present in the assembly process at an early stage, has a membrane-spanning segment, and is closely associated with elements of COX I and COX II. At this stage, the absence of SURF1 protein appears to prevent the sequential addition of COX II and COX III again from the matrix, so it is likely that SURF1 with its 2TM helices assists integration of COX II at this point. Because the patient population with SURF1 mutations has null alleles, the COX assembly process gets stalled at this stage, although 10 to 20% of fully assembled COX is still made (26, 27). The addition of CuA to COX II is accomplished possibly by the SCOI protein or perhaps directly from COX 17, because this Cu center lies close to cytochrome c on the outer face of the IMM (49). The 7TM helices of COX III are associated with COX I but not with COX II, so it is possible that a further factor is necessary for this part of the assembly process. Next are added COX Va, Vb, VIa, VIb, VIc, VIIa, and VIII with final addition of VIc and VIIb (50).
ARE THERE MORE COX DEFICIENCY GENES?
Missing from the list of genetic causes is the gene for “fatal infantile” COX, the gene for infantile reversible COX, the gene for Saguenay-Lac-St. Jean COX, and at least one more cause of classic COX with Leigh syndrome. With these defects actively under investigation and the numbers of assembly factors for COX growing, the discovery of the full spectrum of causative nuclear genes cannot be far off.
Abbreviations
- COX:
-
cytochrome
- c :
-
oxidase
- SURF1 :
-
the human gene encoding SURF 1 protein COX (cytochrome oxidase)assembly factor
- SCO2 :
-
the human gene corresponding to yeast SCO1 or SCO2 genes for synthesis of cytochrome oxidase
- SLSJ-COX:
-
Saguenay-Lac-St.Jean cytochrome oxidase
- Shy1 :
-
the yeast gene for the COX assembly factor equivalent to the human SURF 1 gene
- IMM:
-
inner mitochondrial membrane
References
Robinson BH 1993 Lactic acidemia. Biochim Biophys Acta 1182: 231–234.
Willems JL, Monnens LA, Trijbels JMF, Veerkamp JH, Meyer AE, van Dam K, van Haelst U 1977 Leigh's encephalomyelopathy in a patient with cytochrome c oxidase deficiency in muscle tissue. Pediatrics 60: 850–857.
DiMauro S, Servidei S, Zeviani M, DiRocco M, DeVivo DC, DiDonato S, Uziel G, Berry K, Hoganson G, Johnsen SD, Johnsen DC 1987 Cytochrome c oxidase deficiency in Leigh syndrome. Ann Neurol 22: 498–506.
Van Coster R, Lombes A, DeVivo DC, Chi TL, Dodson WE, Rothman S, Orrechio EJ, Grover W, Berry FT, Schwartz JF, Habib A, DiMauro S 1991 Cytochrome c oxidase-associated Leigh syndrome: phenotypic features and pathogenetic speculations. J Neurol Sci 104: 97–111.
Lombes A, Nakase H, Tritschler HJ, Kadenbach B, Bonilla E, DeVivo DC, Schon E, DiMauro S 1991 Biochemical and molecular analysis of cytochrome c oxidase deficiency in Leigh's syndrome. Neurol 41: 491–498.
Morin C, Mitchell G, Larochelle J, Lambert M, Ogier H, Robinson BH, DeBraekeleer M 1993 Clinical, metabolic, and genetic aspects of cytochrome c oxidase deficiency in Saguenay-Lac-Saint-Jean. Am J Hum Genet 53: 488–496.
Merante F, Petrova-Benedict R, MacKay N, Mitchell G, Morin C, DeBraekeleer M, Laframboise R, Gagne R, Robinson BH 1993 A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Québec. Am J Hum Genet 53: 481–487.
Glerum DM, Robinson BH, Spratt C, Wilson J, Patrick D 1987 Abnormal kinetic behavior of cytochrome oxidase in a case of Leigh's disease. Am J Hum Genet 41: 584–593.
Van Biervliet JPGM, Bruinvis L, Ketting D, De Bree PK, Van der Heiden C, Wadman SK 1977 Hereditary mitochondrial myopathy with lactic acidemia, a De Toni-Fanconi-Debré syndrome, and a defective respiratory chain in voluntary striated muscles. Pediatr Res 11: 1088–1093.
DiMauro S, Mendell JR, Sahenk Z, Bachman D, Scarpa A, Scofield RM, Reiner C 1980 Fatal infantile mitochondrial myopathy and renal dysfunction due to cytochrome c oxidase deficiency. Neurol 30: 795–804.
Heiman-Patterson TD, Bonilla E, DiMauro S, Foreman J, Schotland DL 1982 Cytochrome c oxidase deficiency in a floppy infant. Neurol 32: 898–901.
Minchom PE, Dormer RL, Hughes IA, Stansbie D, Cross AR, Hendry GA, Jones OT, Johnson MA, Sherratt HS, Turnbull DM 1983 Fatal infantile mitochondrial myopathy due to cytochrome c oxidase deficiency. J Neurol Sci 60: 453–463.
Miyabayashi S, Ito T, Abukawa D, Narisawa K, Tada K, Tanaka M, Ozawa T, Droste M, Kadenbach B 1987 Immunochemical study in three patients with cytochrome c oxidase deficiency presenting Leigh's encephalomyelopathy. J Inherit Metab Dis 10: 289–292.
DiMauro S, Zeviani M, Bonilla E, Bresolin N, Nakagawa M, Miranda AF, Moggio M 1985 Cytochrome c oxidase deficiency. Biochem Soc Trans 13: 651
Robinson BH 1989 Lacticacidemia: biochemical, clinical, and genetic considerations. In: Harris H, Hirschhorn K (eds) Advances in Human Genetics. Plenum Press, New York, 151–179.
Glerum DM, Robinson BH, Capaldi RA 1989 Fibroblasts and cytochrome c oxidase deficiency. In: Azzi A, Drahota Z, Papa S (eds) Molecular Basis of Membrane Associated Diseases. Springer-Verlag, Berlin, 228–238.
Robinson BH, Glerum DM, Chow W, Petrova-Benedict R, Lightowlers R, Capaldi R 1990 The use of skin fibroblast cultures in the detection of respiratory chain defects in patients with lacticacidemia. Pediatr Res 28: 549–555.
DiMauro S, Nicholson JF, Hays AP, Eastwood AB, Papadimitriou A, Koenigsberger R, DeVivo DC 1983 Benign infantile mitochondrial myopathy due to reversible cytochrome c oxidase deficiency. Ann Neurol 14: 226–234.
Adams PL, Lightowlers RN, Turnbull DM 1997 Molecular analysis of cytochrome c oxidase deficiency in Leigh's syndrome. Ann Neurol 41: 268–270.
Jaksch M, Hofmann S, Kleinle S, Liechti-Gallati S, Pongratz DE, Muller-Hocker J, Jedele KB, Meitinger T, Gerbitz KD 1998 A systematic mutation screen of 10 nuclear and 25 mitochondrial candidate genes in 21 patients with cytochrome c oxidase (COX) deficiency shows tRNA(Ser)(UCN) mutations in a subgroup with syndromal encephalopathy. J Med Gen 35: 895–900.
Brown RM, Brown GK 1996 Complementation analysis of systemic cytochrome oxidase deficiency presenting as Leigh syndrome. J Inherit Metab Dis 19: 752–760.
Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA 1998 SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20: 337–343.
Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviani M 1998 Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am J Hum Genet 63: 1609–1621.
Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, VanCoster RV, Lyon G, Scalais E, Lebel R, Kaplan P, Shanske S, DeVivo D, Bonilla E, Hirano M, DiMauro S, Schon EA 1999 Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet 23: 333–337.
Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rötig A 2000 Mutation in the farnesyl: heme A transferase (COX 10) gene in a family. Hum Mol Gen 9: 1245–1249.
Lee HRN, Rioux JD, Daly MJ, Lander ES, Hudson TJ, Morin CC, Robinson BH 1998 Genetic mapping of Saguenay-Lac-Saint-Jean cytochrome oxidase deficiency. Am J Hum Genet 63: A296 abstr.
Yao J, Shoubridge EA 1999 Expression and functional analysis of SURF-1 in Leigh syndrome patients with cytochrome c oxidase deficiency. Hum Mol Genet 8: 2541–2549.
Tiranti V, Galimberti C, Nijtmans L, Bovolenta S, Perini MP, Zeviani M 1999 Characterization of SURF-1 expression and SURF-1p function in normal and disease conditions. Hum Mol Genet 13: 2533–2540.
Brown RM, Humphreys CPA, Dorricott HD, Sage EK, Slater JH, Brown GK 1999 SURF1 mutations in cytochrome oxidase-deficient Leigh disease: two prevalent mutations, one with a common genetic origin. Proceedings, Euromit 4 Meeting, Cambridge, p 90( abstr)
Capaldi RA 1992 Structure and function of cytochrome oxidase. Ann Rev Biochem 59: 569–596.
Taanman JW, Capaldi RA 1992 Purification of yeast cytochrome c oxidase with a subunit composition resembling the mammalian enzyme. J Biol Chem 267: 22481–22485.
Kadenbach B, Merle P 1981 On the function of multiple subunits of cytochrome c oxidase from higher eukaryotes. FEBS Lett 135: 1–11.
Haltia T, Saraste M, Wikstrom M 1991 Subunit III of cytochrome oxidase is not involved in proton translocation: a site-directed mutagenesis study. EMBO J 10: 2015–2021.
Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S 1995 Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science 269: 1069–1074.
Merante F, Ling M, Duncan AMV, Duff C, Robinson BH 1997 Cloning, characterization, and chromosomal localization of COX Vla-L related genes. Genome 40: 325–331.
Merante F, Duncan AMV, Mitchell G, Duff C, Rommens J, Robinson BH 1997 Chromosomal localization of the human liver-form cytochrome c oxidase subunit Vlla gene. Genome 40: 318–324.
Lomax M, Hsieh CL, Darras BT, Franke U 1991 Structure of the human cytochrome c oxidase subunit Vb gene and chromosomal mapping of the coding gene and seven pseudogenes. Genomics 10: 1–9.
Van Kuilenburg ABP, Van Beeumen JJ, Van der Meer NM, Muijsers AO 1992 Subunits VllA,B,c of human cytochrome c oxidase. Identification of both “heart-type” and “liver-type” isoforms of subunit Vlla in human heart. Eur J Biochem 203: 193–199.
Ewart GW, Zhang YZ, Capaldi RA 1991 Switching of bovine cytochrome c oxidase subunit VIa isoforms in skeletal muscle during development. FEBS Lett 292: 79–84.
Capaldi R 1990 Structure and assembly of cytochrome c oxidase. Arch Biochem Biophys 280: 252–261.
Gennis R, Ferguson-Miller S 1995 Structure of cytochrome c oxidase, energy generator of life. Science 269: 1063–1064.
Poyton RO, Trueblood CE, Wright RM, Farrel LE 1988 Expression and function of cytochrome c oxidase subunit isologues. Ann N Y Acad Sci 550: 289–307.
Verkhovsky MI, Jasaitis A, Verkovskya ML, Morgan JE, Wikstrom M 1999 Proton translocation by cytochrome oxidase. Nature 400: 480–483.
Bonnefoy N, Kermorgant M, Groudinsky O, Minet M, Slonomski PP, Dujardin G 1994 Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1 mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 91: 11978–11982.
Bonnefoy N, Chalvet F, Hamel P, Slonomski PP, Dujardin G 1994 OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol 239: 201–212.
Srinivasan C, Posewitz M, George GN, Winge DR 1998 Characterization of copper chaperone COX 17 of Saccharomyces cerevisiae. Biochem 37: 7572–7577.
Hiser L, Di Valentin M, Hamer AG, Hosler JP 2000 Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J Biol Chem 275: 619–623.
Tzagoloff A, Nobrega M, Gorman N, Sinclair P 1993 On the functions of the yeast COX10 and COX11 gene products. Biochem Mol Biol Int 31: 593–598.
Glerum DM, Shtanko A, Tzagoloff A 1996 SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J Biol Chem 271: 20531–20535.
Nijtmans LGJ, Taanman JW, Musjers AO, Speijer D, Van den Bogert C 1998 Assembly of cytochrome oxidase in cultured human cells. Eur J Biochem 254: 389–394.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Robinson, B. Human Cytochrome Oxidase Deficiency. Pediatr Res 48, 581–585 (2000). https://doi.org/10.1203/00006450-200011000-00004
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/00006450-200011000-00004
This article is cited by
-
Construction and verification of a gene knockout system in the nematophagous fungus, Purpreocillium lilacinum (Hypocreales: Ophiocordycipitaceae)
Applied Entomology and Zoology (2016)
-
A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle
Nature Communications (2013)