Abstract
Autism spectrum disorder (ASD) is a neurobehavioral disorder with a heterogeneous genetic etiology. Based on the literature, several single-gene disorders, including Rett syndrome, Smith-Lemli-Opitz syndrome, PTEN hamartoma tumor syndrome and tuberous sclerosis, are associated with a high prevalence of ASD. We estimated the prevalence of these four conditions in a large cohort of patients using whole-exome sequencing data from 2392 families (1800 quads and 592 trios) with ASD from the National Database for Autism Research. Seven patients carried a pathogenic or likely pathogenic variant in either TSC1, TSC2, PTEN, DHCR7 or MECP2, with 6 out of 7 reportable variants occurring in PTEN (1 in 399).
Similar content being viewed by others
Introduction
According to the literature, several well-described disorders are associated with a high prevalence of autism spectrum disorder (ASD), including Rett syndrome, Smith-Lemli-Opitz syndrome, PTEN hamartoma tumor syndrome and tuberous sclerosis (TSC). Patients with TSC, Rett syndrome and Smith-Lemli-Opitz syndrome have a prevalence of ASD in the range of 16–61%, 17.5–58%,1, 2 and 50–60%3, 4 respectively. Interestingly, up to 80–90% of patients with Smith-Lemli-Opitz syndrome appear to fulfill some ASD criteria.5 In the Diagnostic and Statistical Manual of Mental Disorders (DSM 4), Rett syndrome was a subtype of autism. In DSM 5,6 however, an individual with Rett syndrome is not automatically assumed to have a diagnosis of ASD. Due to the essential difficulty to assess the precise prevalence for invisible mental disorder(s), the prevalence of each of these disorders within ASD cohorts is unclear, making the indication for routine molecular testing in all patients with ASD uncertain. We have analyzed whole exome sequencing (WES) data from 2392 families with ASD for variants within the genes causing the above-mentioned conditions. Our goal was to assess the prevalence of patients with genetic alterations in the genes corresponding to the four conditions targeted based on a large cohort of individuals diagnosed with ASD who do not exhibit severe neurological defects.
Materials and methods
Whole-exome data from 2392 families (1800 quads and 592 trios) with ASD, obtained from the National Database for Autism Research7 (NDAR), were analyzed for pathogenic or likely pathogenic variants in TSC1, TSC2, PTEN, DHCR7 and MECP2. After McGill REB approval, variant calls were downloaded from NDAR (study 348) and were annotated using the reference genome hg19/GRCh37. Rare variants (minor allele frequency, MAF,⩽0.005) with a functional impact (defined as ‘missense’, ‘frameshift’, ‘stopgain’, ‘stoploss’, ‘startloss’ and ‘splicing’) were selected and manually inspected using the Integrative Genome Viewer (IGV; study 334). Variant calls that were not supported by visualization in IGV were removed from further analysis. Variant classification was performed in accordance with ACMG guidelines.8 Clarifications and adaptations of the criteria are located in Supplementary Table S1.
Results
Please refer to Table 1 and Supplementary Section for the variants identified.
Discussion
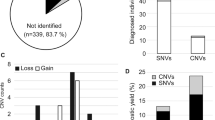
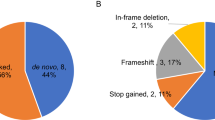
Based on the selection criteria, 148 variants in the genes targeted were classified. Following variant classification, seven patients were found to carry a variant that was likely responsible for the patient’s phenotype in one of these five genes (Table 1), with the majority (6 out of 7) occurring in PTEN. It is possible that this prevalence is an underestimate, as ~10% of Cowden syndrome patients with negative WES and copy number variant (CNV) analysis have been found to harbor pathogenic promoter variants.9 We did not have access to the clinical information of the patients in the cohort for cephalic measurements.
No patients were determined to harbor biallelic DHCR7 variants (Supplementary Table S3). Twenty-eight patients carried a pathogenic or likely pathogenic DHCR7 variant, giving a carrier frequency of 1 in 85. If variants of uncertain significance (VUS) are included, the carrier frequency is ~1 in 40. Carrier frequency estimates for DHCR7 are ~1–2% for individuals of Caucasian ancestry.10 Therefore, it is possible that some of the identified VUS may in fact be disease-causing alleles.
No causative mutations for Rett syndrome were identified in our study. However, one VUS in MECP2 was particularly notable. A male proband was hemizygous for the variant c.691G>A, p.G231R, which has been previously reported in a female patient with seizures and intellectual disability. This variant is absent from population databases.11 While this variant was present in the proband’s unaffected mother and sister, males carrying MECP2 variants have been diagnosed with X-linked mental retardation (OMIM 300260). Therefore, this variant has been classified as a VUS. An additional 38 patients carried one of 36 heterozygous or hemizygous VUS in either PTEN, TSC1, TSC2 or MECP2 (Supplementary Table S2). One patient (12 621) harbored both a likely pathogenic de novo variant and a paternally inherited VUS in TSC2.
De novo status was observed in 3 out of the 6 PTEN variants and the one TSC2 variant. In total, six variants have been previously reported in patients, including four reported in patients with ASD or related phenotypes. The estimated frequency of unselected patients with autism caused by a variant in the PTEN and TSC2 gene is ~1 in 399 and 1 in 2392 respectively. In addition, two of the probands were previously determined to carry large pathogenic de novo duplications of TSC2 (Supplementary Table S4).12 Only 53% of the probands analyzed in the present study had been previously tested for CNVs; therefore, additional probands may also carry pathogenic CNVs. Therefore, at least 3 of 2392 probands (1 in 797) carried a TSC2 pathogenic variant; this appears to be more frequent than in the general population, where TSC has been reported to occur in ~1 in 5800 live births.13
There are several limitations to this study. This analysis was performed on previously obtained research WES data and variants with low coverage could not be confirmed by Sanger sequencing; in addition, some variants may have been missed due to low coverage in certain regions, or due to their presence in regions located outside of the exome. Taking this into consideration, our analysis was not consistent with a high prevalence of the four targeted disorders in ASD patient cohorts. Although this study was not designed to be an evaluation of WES in the clinical setting, this manuscript highlights the issue of uncovering a large burden of VUS in the context of making a small number of clear diagnoses. Despite our analysis being limited to five genes, 54 unique VUS that require further exploration (and resources) were identified. Therefore, preparation for efficient VUS interpretation must be considered to ensure that widespread implementation of WES in ASD will be cost-effective. This study emphasizes the importance of data sharing to further scientific advancement in the field of genomics, as previously published studies using the NDAR data set also illustrate.14, 15, 16, 17 It also emphasizes the need for a large prospective study evaluating the prevalence of different genetic syndromes in a large group of patients with ASD. Ideally, detailed phenotypic information should be available for these patients and individuals with severe neurological deficits should not be excluded, which were inevitable limitations of the current study.
References
Young, D. J., Bebbington, A., Anderson, A., Ravine, D., Ellaway, C., Kulkarni, A. et al. The diagnosis of autism in a female: could it be Rett syndrome? Eur. J. Pediatr. 167, 661–669 (2008).
Wulffaert, J., Van Berckelaer-Onnes, I. A. & Scholte, E. M. Autistic disorder symptoms in Rett syndrome. Autism 13, 567–581 (2009).
Tierney, E., Nwokoro, N. A., Porter, F. D., Freund, L. S., Ghuman, J. K. & Kelley, R. I. Behavior phenotype in the RSH/Smith-Lemli-Opitz syndrome. Am. J. Med. Genet. 98, 191–200 (2001).
Bukelis, I., Porter, F. D., Zimmerman, A. W. & Tierney, E. Smith-Lemli-Opitz syndrome and autism spectrum disorder. Am. J. Psychiatry 164, 11 (2007).
Sikora, D. M., Pettit-Kekel, K., Penfield, J., Merkens, L. S. & Steiner, R. D. The near universal presence of autism spectrum disorders in children with Smith-Lemli-Opitz syndrome. Am. J. Med. Genet. A 140, 1511–1518 (2006).
American Psychiatric A, Force DSMT. in Diagnostic and statistical manual of mental disorders: DSM-5. p57 (American Psychiatric Association, Washington, D.C., USA, 2013).
Hall, D., Huerta, M. F., McAuliffe, M. J. & Farber, G. K. Sharing heterogeneous data: the national database for autism research. Neuroinformatics 10, 331–339 (2012).
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–423 (2015).
Zhou, X. P., Waite, K. A., Pilarski, R., Hampel, H., Fernandez, M. J., Bos, C. et al. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am. J. Human Genet. 73, 404–411 (2003).
Porter, F. D. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Human Genet. 16, 535–541 (2008).
Sundaram, S., Huq, A. H. M., Hsia, T. & Chugani, H. Exome sequencing and diffusion tensor imaging in developmental disabilities. Pediatr. Res. 75, 443–447 (2013).
Krumm, N., Turner, T. N., Baker, C., Vives, L., Mohajeri, K., Witherspoon, K. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
Osborne, J. P., Fryer, A. & Webb, D. Epidemiology of tuberous sclerosis. Ann. N. Y. Acad. Sci. 615, 125–127 (1991).
Sanders, S. J., Murtha, M. T., Gupta, A. R., Murdoch, J. D., Raubeson, M. J., Willsey, A. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012).
O’Roak, B. J., Vives, L., Girirajan, S., Karakoc, E., Krumm, N., Coe, B. P. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012).
Iossifov, I., Ronemus, M., Levy, D., Wang, Z., Hakker, I., Rosenbaum, J. et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012).
Iossifov, I., O’Roak, B. J., Sanders, S. J., Ronemus, M., Krumm, N., Levy, D. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Acknowledgements
Data used in the preparation of this manuscript was obtained from the NIH supported NDAR. Raw data (including VCF files and additional data files) accessed and used in the preparation of this study is available from NDAR (doi: 10.15154/1169318; doi: 10.15154/1169195). These data were submitted to NDAR by Krumm et al. (2015; excess of rare, inherited truncating mutations in autism).12 We thank NDAR and the authors of this study for the WES data. This manuscript reflects the views of the authors and may not reflect the opinions and views of the NIH. We would like to thank Bogdan Istrate, Alexandre Dionne Laporte and Dr Guy Rouleau for bioinformatics support and the Montreal Children’s Hospital Foundation for financial support for VF during the writing of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Saskin, A., Fulginiti, V., Birch, A. et al. Prevalence of four Mendelian disorders associated with autism in 2392 affected families. J Hum Genet 62, 657–659 (2017). https://doi.org/10.1038/jhg.2017.16
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.16
This article is cited by
-
Exploring key genes and pathways associated with sex differences in autism spectrum disorder: integrated bioinformatic analysis
Mammalian Genome (2024)
-
Behavioural and psychological features of PTEN mutations: a systematic review of the literature and meta-analysis of the prevalence of autism spectrum disorder characteristics
Journal of Neurodevelopmental Disorders (2022)
-
Cholesterol modulates presynaptic and postsynaptic properties of excitatory synaptic transmission
Scientific Reports (2020)