« Prev Next »
Alu elements are a type of "jumping gene," or transposable element (TE), that exists only in primates. Like all TEs, they are discrete DNA sequences that move, or "jump," from one place on the genome to another, sometimes inserting copies of themselves directly into the middle of protein-coding genes. Although Alu elements have long been considered "junk" DNA, scientists are beginning to question whether these elements might serve important biological functions after all. While much remains to be discovered about the role of these prolific transposons, researchers now recognize that Alu elements are major players in human evolution, as well as useful tools for molecular genetic and forensic applications.
Short Interspersed Elements (SINEs): The Alu Example
Alu elements are highly repetitive DNA sequences that can be classified as SINEs (short interspersed elements), which are themselves a type of "nonautonomous" retrotransposon. (Retrotransposons are TEs that move about the genome via an RNA intermediary.) An Alu element is transcribed into messenger RNA by RNA polymerase and then converted into a double-stranded DNA molecule by reverse transcriptase. The new double-stranded DNA molecule is then inserted into a new location in the genome. Because they are nonautonomous, like all SINEs, Alu elements don't have the genetic capacity to produce DNA copies of themselves or to integrate into new chromosomal locations. For those activities, they rely on another type of transposon, called L1. Most Alu elements are approximately 300 base pairs long, with considerable sequence variation.
Alu elements frequently duplicate when they jump, and scientists estimate that the human genome acquires one new Alu insert in approximately every 200 births. In other words, the number of Alu elements in the human population (and in other primate populations) tends to increase over time. Alu TEs are believed to have emerged in primates around 65 million years ago. Today, they are the most abundant type of human TE, making up an amazing 10% of the (diploid) human genome. Thus, in a mere 65 million years, these transposons have gone from zero to about 1 million copies per cell! Indeed, part of what makes Alu elements so important is that they are found in nearly every human (and other primate) gene. These elements are spread throughout the genome and occur at varying densities in different loci.
Transposable Elements Are More Than Just “Jumping Genes”
The fact that other nonprimate mammals function quite well without Alu elements confirms that these elements are not required for the basic function of cellular processes. Yet, because primate genomes have so many Alu elements, many biologists speculate that these TEs may have played an important role in our species' evolution, and that nature may have taken advantage of these elements in some way, such as by putting them to work in generating new proteins. This conflicts with the long-held belief that all transposable elements are just "junk," existing for no purpose other than self-replication.
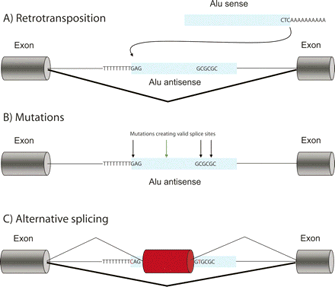
Although they have yet to gather much data to support their arguments, scientists have hypothesized various roles for Alu elements, such as stimulation of protein translation, particularly under stressful cellular circumstances; this hypothesis is rooted in the fact that many Alu RNAs are expressed during times of stress. However, the hypothesis attracting the most attention of late is the one stating that Alu elements may have served an important evolutionary role by contributing to the formation of new genes or gene combinations, or perhaps by adding new functionality to already-existing genes through a process known as exonization (Sela et al., 2007). Exonization is the creation of a new exon from an Alu-containing (or any TE-containing) intron. (i.e., they are "spliced" out). The process of exonization is illustrated in Figure 1. First, the Alu TE is inserted into an intron of a gene. Initially, the Alu is spliced out of the mRNA as part of the intron. Over time (as many as several million years), however, mutations accumulate, leading to the formation of active splice sites on either end of the Alu element, and the Alu TE is thus eventually recognized as an exon, at least in some molecules of mRNA.
Scientists have reported thousands of Alu exonization events among primates. In humans, for example, Alu TEs are responsible for an estimated 62% of all new exons (Zhang & Chasin, 2006). This raises an important question: How could the insertion of so many new exons into vital genes not be detrimental? As Rotem Sorek of the Lawrence Berkeley National Laboratory in California, explains, "The answer lies in alternative splicing" (Sorek, 2007). With alternative splicing, some initial RNA transcripts that contain a new exon arising from Alu are spliced to include the new exon in the final mRNA, while other identical transcripts are spliced differently, thereby removing the Alu exon. As a result, some transcripts lack the new exon and produce the same "old" protein, while others produce entirely new proteins or protein variants. Some of these new proteins might be deleterious; others might be selectively neutral (neither harmful nor beneficial to the organism or cell); and some could very well be beneficial, thereby giving an organism (or a cell or gene) a selective edge while not eliminating the "old" protein altogether.
Scientists are only beginning to explore this possibility, and a few observations do seem to support the idea. For instance, not only do newly born exons undergo alternative splicing more frequently than old exons, but, according to Sorek (2007), almost all exonized Alu elements in the human genome are alternatively spliced. Therefore, even though the original transcript with the newly exonized Alu element is intact and could encode potentially deleterious RNA and protein products, alternative splicing leads to the production of multiple transcripts, multiple mRNAs, and, ultimately, multiple proteins. In fact, some scientists speculate that Alu elements have played a key role in the evolution of the primate brain, and that all of the protein possibilities afforded by Alu movement and exonization have contributed to the complexity of human neuronal circuitry and "higher-order" cognition (e.g., rational thought) (Mattick & Mehler, 2008).
On the other hand, scientists have associated some individual Alu insertions with human disease. The simple insertion of an Alu TE in the wrong place can often wreak molecular havoc. For example, in 1991, researchers discovered a case of neurofibromatosis caused by an Alu sequence inserted in an intron of the NF1 gene (Wallace et al., 1991). The presence of the Alu sequence caused a splicing error, which in turn caused one of the exons to be left out of the transcribed mRNA, thereby leading to a shift in the reading frame and production of an abnormal protein. Since then, researchers have reported a growing number of Alu-disease associations in disorders ranging from hemophilia to breast cancer. According to one estimate, about 0.4% of all human genetic disorders are caused by or associated with Alu TEs. In many cases, it is not clear whether the association is causal (i.e., whether the Alu insertion actually causes the disease), but the lack of any other noticeable mutation often leads researchers to suspect that it is.
Evidence of Alu Jumps in the Genome Serve as Important Genetic Markers
Regardless of what they do to the genome (and the cell and organism) when they jump, Alu TEs have, at the very least, served science well. Today, molecular biologists use Alu TEs as research tools, evolutionary biologists use them to resolve questions about primate systematics and human history, and forensic investigators use then to home in on possible perpetrators of unsolved crimes.
Use of Alu Elements in Molecular Biology
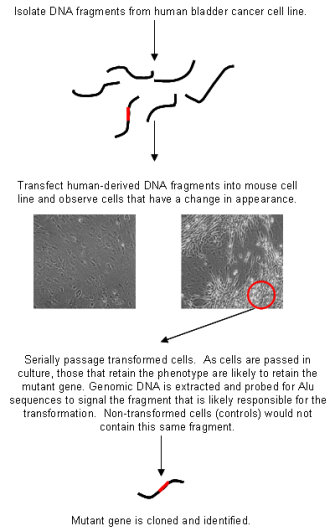
In molecular biology, the fact that Alu elements are widely dispersed throughout the primate genome but absent in other animals makes these elements a useful tool for identifying human DNA sequences that have been inserted into cells of other animals in what are known as marker-rescue experiments. A classic Alu marker-rescue experiment was the 1982 discovery of the first human oncogene by Robert Weinberg and his trainee Chiaho Shih (Shih & Weinberg, 1982). Knowing that human genes can be associated with Alu sequences, the researchers suspected that if they were to transfer a human oncogene into a mouse cell, the oncogene would most likely take some Alu sequences along for the ride. Thus, Weinberg and Shih isolated DNA from a human bladder cancer cell and added it to some mouse cells growing in culture. Many of the mouse cells took up different pieces of this human DNA and added it to their own DNA. In fact, a small number of the mouse cells took up the human oncogene (i.e., the mutated gene responsible for transforming the normal bladder cell into a cancer cell). These particular mouse cells were transformed, becoming visibly different from their counterparts that had not taken up the oncogenic DNA (Figure 2). Moreover, when injected into mice, these cells could form tumors.
Upon observing this sort of tumor formation, Weinberg and Shih were faced with the challenge of finding the lone human oncogene in the middle of billions of base pairs of mouse DNA. Luckily, Alu elements provided the answer. Weinberg and Shih isolated DNA from the newly transformed (i.e., cancerous) mouse cells and added it to other mouse cells growing in culture, again transforming a few of these cells. The scientists did this repeatedly, and after each round, they removed a small sample of DNA for use in a Southern blot analysis, tracking the number of copies of Alu elements to measure how much human DNA remained in the mouse cells. Because the mouse genome does not have Alu sequences naturally, the sequences that the researchers found in the transformed cells could be inferred to be associated with the mutated human gene. The scientists ended these serial "transfections" only when the Southern blot analysis revealed a single band of Alu—that is, only one small region of the original human DNA remained, and, presumably, this region contained the oncogene. (Because this small region had been initially isolated from a human tumor cell and was associated with tumor formation in every generation of mouse cells, it was reasonable to conclude that whatever region of human DNA remained contained the oncogene.) Weinberg and Shih then probed this remaining bit of DNA with various other DNA sequences known to cause cancer (all from animal viruses) to identify the specific oncogene.
Use of Alu Elements in Evolutionary Biology
In evolutionary biology, scientists take advantage of some key features of Alu elements that set them apart as genetic polymorphic loci (i.e., DNA sequences that vary within the population) and make them ideal for answering certain types of questions about genetic ancestry and relatedness. For example, the probability that two different Alu elements will insert independently in the same place is practically zero. Moreover, most Alu elements, once inserted, are rarely removed. Combined, these facts mean that two individuals who share the same Alu sequence in the same spot on the genome most likely had a common ancestor who had that same Alu sequence in that same spot in his or her genome. In contrast, with other types of loci, it is often difficult to know whether two individuals share the same genetic marker because they acquired it from a common ancestor or developed the change independently by chance.
Thus, scientists have used Alu element analyses to confirm, for example, that flying lemurs are not part of the order Primata, because Alu elements have been detected in all primates tested but not in flying lemurs (Xing et al., 2007). Alu elements have also been used in attempts to resolve what is known as the "trichotomy problem": the evolutionary relationship among humans, chimpanzees, and gorillas. In one study, the distribution of more than 100 different Alu elements suggested that humans and chimpanzees are a sister clade and that gorillas are the out-group (Salem et al., 2003). In other words, this study suggests that humans are more closely related to chimpanzees than to gorillas.
Uses of Alu Elements in Forensic Science
In forensics, Alu elements can be used to determine the geographical ancestry of a DNA sample, such as whether it originated from sub-Saharan African, Western European, Indian, or East Asian "stock." While these may seem like broad categories, having this kind of "continental" information can help investigators further refine their analyses (i.e., the identification of unknown samples) by using only those markers known to yield useful information for a particular population group. As Ray et al. (2005) explain, "The inference of an individual's geographic ancestry or origin can be critical in narrowing the field of potential suspects in a criminal investigation."
Summary
References and Recommended Reading
Deininger, P. L., & Batzer, M. A. Alu repeats and human disease. Molecular Genetics and Metabolism 67, 183–193 (1999)
Mattick, J. S., & Mehler, M. F. RNA editing, DNA recoding, and the evolution of human cognition. Trends in Neurosciences 31, 227–233 (2008)
Ray, D. A., et al. Inference of human geographic origins using Alu insertion polymorphisms. Forensic Science International 153, 117–124 (2005)
Salem, A. H., et al. Alu elements and hominid phylogenetics. Proceedings of the National Academy of Sciences 100, 12787–12791 (2003)
Sela, N., et al. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biology 8, R127 (2007) doi:10.1186/gb-2007-8-6-r127
Shih, C., & Weinberg, R. A. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell 29, 161–169 (1982)
Sorek, R. The birth of new exons: Mechanisms and evolutionary consequences. RNA 13, 1603–1608 (2007)
Wallace, M. R., et al. A de novo Alu insertion results in neurofibromatosis type 1. Nature 353, 864–866 (1991) (link to article)
Xing, J., et al. Mobile elements in primate and human evolution. Yearbook of Physical Anthropology 50, 2–19 (2007)
Zhang, X. H., & Chasin, L. H. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proceedings of the National Academy of Sciences 103, 13427–13432 (2006)


 Figure 1: Schematic model for exonization of an Alu element.
Figure 1: Schematic model for exonization of an Alu element.