Abstract
Biofilms are structured and organized communities of microorganisms that represent one of the most successful forms of life on Earth. Bacterial biofilms have been studied in great detail, and many molecular details are known about the processes that govern bacterial biofilm formation, however, archaea are ubiquitous in almost all habitats on Earth and can also form biofilms. In recent years, insights have been gained into the development of archaeal biofilms, how archaea communicate to form biofilms and how the switch from a free-living lifestyle to a sessile lifestyle is regulated. In this Review, we explore the different stages of archaeal biofilm development and highlight similarities and differences between archaea and bacteria on a molecular level. We also consider the role of archaeal biofilms in industry and their use in different industrial processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


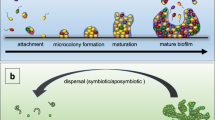
Part a adapted from ref.16, Springer Nature Limited. Image in part b courtesy of C. Moissl-Eichinger, Medical University Graz, Austria, and G. Wanner, University of Munich, Germany. Part c adapted from ref.43, CC-BY-4.0. Part d adapted from ref.15, CC-BY-4.0. Image in part e courtesy of J. Berger, Max Planck Institute for Developmental Biology, Tübingen, Germany. Part f adapted from ref.79, CC-BY-3.0.

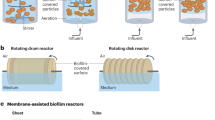
Part a adapted from ref.50, Springer Nature Limited.
Similar content being viewed by others
References
Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002).
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. & Lappin-Scott, H. M. Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995).
Davey, M. E. & O’toole, G. A. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867 (2000).
Flemming, H.-C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Flemming, H.-C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
An, D. & Parsek, M. R. The promise and peril of transcriptional profiling in biofilm communities. Curr. Opin. Microbiol. 10, 292–296 (2007).
O’Toole, G., Kaplan, H. B. & Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000).
Garrett, R. T., Bhakoo, M. & Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 18, 1049–1056 (2008).
Maier, B. & Wong, G. C. L. How bacteria use type IV pili machinery on surfaces. Trends Microbiol. 23, 775–788 (2015).
Solano, C., Echeverz, M. & Lasa, I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 18, 96–104 (2014).
Koerdt, A., Gödeke, J., Berger, J., Thormann, K. M. & Albers, S.-V. Crenarchaeal biofilm formation under extreme conditions. PLoS ONE 5, e14104 (2010).
Megaw, J. & Gilmore, B. F. Archaeal persisters: persister cell formation as a stress response in Haloferax volcanii. Front. Microbiol. 8, 1589 (2017).
Lapaglia, C. & Hartzell, P. L. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl. Environ. Microbiol. 63, 3158–3163 (1997).
DeMaere, M. Z. et al. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc. Natl Acad. Sci. USA 110, 16939–16944 (2013).
Chimileski, S., Franklin, M. J. & Papke, R. T. Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC Biol. 12, 65 (2014).
Wegener, G., Krukenberg, V., Riedel, D., Tegetmeyer, H. E. & Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590 (2015). This study describes direct interspecies electron transfer between bacteria and archaea for the first time.
Probst, A. & Moissl-Eichinger, C. “Altiarchaeales”: uncultivated archaea from the subsurface. Life 5, 1381–1395 (2015).
Moissl-Eichinger, C. et al. Archaea are interactive components of complex microbiomes. Trends Microbiol. 26, 70–85 (2018).
Couradeau, E. et al. Prokaryotic and eukaryotic community structure in field and cultured microbialites from the alkaline Lake Alchichica (Mexico). PLoS ONE 6, e28767 (2011).
Battin, T. J., Wille, A., Sattler, B. & Psenner, R. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67, 799–807 (2001).
Edwards, K. J. et al. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287, 1796–1799 (2000).
Webster, N. S. & Negri, A. P. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 8, 1177–1190 (2006).
Bang, C. et al. Biofilm formation of mucosa-associated methanoarchaeal strains. Front. Microbiol. 5, 353 (2014). This paper presents the first studied biofilm of a human gut archaeon.
Miller, T. L., Weaver, G. A. & Wolin, M. J. Methanogens and anaerobes in a colon segment isolated from the normal fecal stream. Appl. Environ. Microbiol. 48, 449–450 (1984).
Belay, N., Johnson, R., Rajagopal, B. S., Conway de Macario, E. & Daniels, L. Methanogenic bacteria from human dental plaque. Appl. Environ. Microbiol. 54, 600–603 (1988).
Belay, N., Mukhopadhyay, B., Conway de Macario, E., Galask, R. & Daniels, L. Methanogenic bacteria in human vaginal samples. J. Clin. Microbiol. 28, 1666–1668 (1990).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Justice, N. B. et al. Heterotrophic archaea contribute to carbon cycling in low-pH, suboxic biofilm communities. Appl. Environ. Microbiol. 78, 8321–8330 (2012).
Offre, P., Spang, A. & Schleper, C. Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 67, 437–457 (2013).
Boetius, A. et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626 (2000). This study describes a cross-kingdom biofilm that has an important environmental role in the global methane cycle.
Hallam, S. J. et al. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462 (2004).
Wrede, C., Dreier, A., Kokoschka, S. & Hoppert, M. Archaea in symbioses. Archaea 2012, 596846 (2012).
Knittel, K. & Boetius, A. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334 (2009).
Haroon, M. F. et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570 (2013).
Dekas, A. E., Poretsky, R. S. & Orphan, V. J. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426 (2009).
Baker, B. J. et al. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44, 139–152 (2003).
Wilmes, P. et al. Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 3, 266–270 (2009).
Fröls, S., Dyall-Smith, M. & Pfeifer, F. Biofilm formation by haloarchaea. Environ. Microbiol. 14, 3159–3174 (2012). This paper provides an extensive description of the morphology and structure of a large number of haloarchaeal biofilms.
Losensky, G., Vidakovic, L., Klingl, A., Pfeifer, F. & Fröls, S. Novel pili-like surface structures of Halobacterium salinarum strain R1 are crucial for surface adhesion. Front. Microbiol. 5, 755 (2015).
Esquivel, R., Xu, R. & Pohlschroder, M. Novel, archaeal adhesion pilins with a conserved N-terminus. J. Bacteriol. 195, 3808–3818 (2013).
Esquivel, R. N., Schulze, S., Xu, R., Hippler, M. & Pohlschroder, M. Identification of Haloferax volcanii pilin N -glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J. Biol. Chem. 291, 10602–10614 (2016).
Esquivel, R. N. & Pohlschroder, M. A conserved type IV pilin signal peptide H-domain is critical for the post-translational regulation of flagella-dependent motility. Mol. Microbiol. 93, 494–504 (2014).
Liao, Y. et al. Morphological and proteomic analysis of biofilms from the Antarctic archaeon, Halorubrum lacusprofundi. Sci. Rep. 6, 37454 (2016).
Henche, A.-L. et al. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ. Microbiol. 14, 3188–3202 (2012).
Zolghadr, B. et al. Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 192, 104–110 (2010).
Zhang, R. et al. Visualization and analysis of EPS glycoconjugates of the thermoacidophilic archaeon Sulfolobus metallicus. Appl. Microbiol. Biotechnol. 99, 7343–7356 (2015).
Castro, C. et al. Biofilm formation and interspecies interactions in mixed cultures of thermo-acidophilic archaea Acidianus spp. and Sulfolobus metallicus. Res. Microbiol. 167, 604–612 (2016).
Thoma, C. et al. The Mth60 fimbriae of Methanothermobacter thermoautotrophicus are functional adhesins. Environ. Microbiol. 10, 2785–2795 (2008).
Moissl, C., Rachel, R., Briegel, A., Engelhardt, H. & Huber, R. The unique structure of archaeal ‘hami’, highly complex cell appendages with nano-grappling hooks. Mol. Microbiol. 56, 361–370 (2005). This report is the first description of hami, which are archaeal surface structures that enable the formation of a highly structured biofilm.
Orell, A. et al. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. ISME J. 7, 1886–1898 (2013). This study describes the identification of an archaeal biofilm regulator.
Koerdt, A. et al. Macromolecular fingerprinting of sulfolobus species in biofilm: a transcriptomic and proteomic approach combined with spectroscopic analysis. J. Proteome Res. 10, 4105–4119 (2011).
Petrova, O. E. & Sauer, K. Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 194, 2413–2425 (2012).
Antón, J., Meseguer, I. & Rodríguez-Valera, F. Production of an extracellular polysaccharide by Haloferax mediterranei. Appl. Environ. Microbiol. 54, 2381–2386 (1988).
Busch, A. & Waksman, G. Chaperone-usher pathways: diversity and pilus assembly mechanism. Phil. Trans. R. Soc. 367, 1112–1122 (2012).
Schneewind, O. & Missiakas, D. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1843, 1687–1697 (2014).
Blanco, L. P., Evans, M. L., Smith, D. R., Badtke, M. P. & Chapman, M. R. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20, 66–73 (2012).
Giltner, C. L., Nguyen, Y. & Burrows, L. L. Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 (2012).
Pohlschroder, M., Ghosh, A., Tripepi, M. & Albers, S.-V. Archaeal type IV pilus-like structures—evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 14, 357–363 (2011).
Szabo, Z. & Pohlschroder, M. Diversity and subcellular distribution of archaeal secreted proteins. Front. Microbiol. 3, 207 (2012).
Shahapure, R., Driessen, R. P. C., Haurat, M. F., Albers, S.-V. & Dame, R. T. The archaellum: a rotating type IV pilus. Mol. Microbiol. 91, 716–723 (2014).
Albers, S.-V. & Jarrell, K. F. The archaellum: how archaea swim. Front. Microbiol. 6, 23 (2015).
Makarova, K. S., Koonin, E. V. & Albers, S.-V. Diversity and evolution of type IV pili systems in archaea. Front. Microbiol. 7, 667 (2016).
Pohlschroder, M. & Esquivel, R. N. Archaeal type IV pili and their involvement in biofilm formation. Front. Microbiol. 6, 190 (2015).
Jarrell, K. F., Stark, M., Nair, D. B. & Chong, J. P. J. J. Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces. FEMS Microbiol. Lett. 319, 44–50 (2011).
Henche, A. L., Koerdt, A., Ghosh, A. & Albers, S. V. Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ. Microbiol. 14, 779–793 (2012).
Tripepi, M., Imam, S. & Pohlschröder, M. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J. Bacteriol. 192, 3093–3102 (2010).
Bellack, A., Huber, H., Rachel, R., Wanner, G. & Wirth, R. Methanocaldococcus villosus sp. nov., a heavily flagellated archaeon that adheres to surfaces and forms cell-cell contacts. Int. J. Syst. Evol. Microbiol. 61, 1239–1245 (2011).
Nather, D. J., Rachel, R., Wanner, G. & Wirth, R. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J. Bacteriol. 188, 6915–6923 (2006).
Herzog, B. & Wirth, R. Swimming behavior of selected species of archaea. Appl. Environ. Microbiol. 78, 1670–1674 (2012).
Schopf, S., Wanner, G., Rachel, R. & Wirth, R. An archaeal bi-species biofilm formed by Pyrococcus furiosus and Methanopyrus kandleri. Arch. Microbiol. 190, 371–377 (2008).
Esquivel, R. N., Xu, R. & Pohlschroder, M. Novel archaeal adhesion pilins with a conserved N terminus. J. Bacteriol. 195, 3808–3818 (2013).
Perras, A. K. et al. Grappling archaea: ultrastructural analyses of an uncultivated, cold-loving archaeon, and its biofilm. Front. Microbiol. 5, 397 (2014).
Nickell, S., Hegerl, R., Baumeister, W. & Rachel, R. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography. J. Struct. Biol. 141, 34–42 (2003).
Rudolph, C., Wanner, G. & Huber, R. Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl. Environ. Microbiol. 67, 2336–2344 (2001).
Probst, A. J. et al. Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat. Commun. 5, 5497 (2014).
Moissl, C., Rudolph, C. & Huber, R. Natural communities of novel archaea and bacteria with a string-of-pearls-like morphology: molecular analysis of the bacterial partners. Appl. Environ. Microbiol. 68, 933–937 (2002).
Doddema, H. J., Derksen, J. W. M. & Vogels, G. D. Fimbriae and flagella of methanogenic bacteria. FEMS Microbiol. Lett. 5, 135–138 (1979).
Rieger, G., Rachel, R., Hermann, R. & Stetter, K. O. Ultrastructure of the hyperthermophilic archaeon Pyrodictium abyssi. J. Struct. Biol. 115, 78–87 (1995).
Comolli, L. R. & Banfield, J. F. Inter-species interconnections in acid mine drainage microbial communities. Front. Microbiol. 5, 367 (2014).
Koerdt, A. et al. Complementation of Sulfolobus solfataricus PBL2025 with an α-mannosidase: effects on surface attachment and biofilm formation. Extremophiles 16, 115–125 (2012).
Jachlewski, S. et al. Isolation of extracellular polymeric substances from biofilms of the thermoacidophilic archaeon Sulfolobus acidocaldarius. Front. Bioeng. Biotechnol. 3, 123 (2015).
Ellen, A. F., Albers, S.-V. & Driessen, A. J. M. Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles 14, 87–98 (2010).
Zhao, D. et al. Improving polyhydroxyalkanoate production by knocking out the genes involved in exopolysaccharide biosynthesis in Haloferax mediterranei. Appl. Microbiol. Biotechnol. 97, 3027–3036 (2013).
Gloag, E. S. et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl Acad. Sci. USA 110, 11541–11546 (2013).
Dominiak, D. M., Nielsen, J. L. & Nielsen, P. H. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 13, 710–721 (2011).
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C. & Mattick, J. S. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002).
Izano, E. A., Amarante, M. A., Kher, W. B. & Kaplan, J. B. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74, 470–476 (2008).
Ibáñez de Aldecoa, A. L., Zafra, O. & González-Pastor, J. E. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 8, 1390 (2017).
Chimileski, S., Dolas, K., Naor, A., Gophna, U. & Papke, R. T. Extracellular DNA metabolism in Haloferax volcanii. Front. Microbiol. 5, 57 (2014).
Ajon, M. et al. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817 (2011).
van Wolferen, M., Wagner, A., van der Does, C. & Albers, S.-V. The archaeal Ced system imports DNA. Proc. Natl Acad. Sci. USA 113, 2496–2501 (2016).
Fröls, S. et al. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70, 938–952 (2008).
Yoon, M. Y., Lee, K.-M., Park, Y., Yoon, S. S. & McFeters, G. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS ONE 6, e16105 (2011).
López-Ribot, L. J. Candida albicans biofilms: more than filamentation. Curr. Biol. 15, R453–R455 (2005).
Fourie, R. et al. Candida albicans and Pseudomonas aeruginosa interaction, with focus on the role of eicosanoids. Front. Physiol. 7, 64 (2016).
Konstantinidou, N. & Morrissey, J. P. Co-occurence of filamentation defects and impaired biofilms in Candida albicans protein kinase mutants. FEMS Yeast Res. 15, fov092 (2015).
Duggin, I. G. et al. CetZ tubulin-like proteins control archaeal cell shape. Nature 519, 362–365 (2015).
Kjelleberg, S., Barraud, N. & Rice, S. A. in Microbial Biofilms 2nd edn (eds Ghannoum, M., Parsek, M., Whiteley, M. & Mukherjee, P.) 343–362 (American Society of Microbiology, 2015).
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D. & Kjelleberg, S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50 (2012).
Baker-Austin, C., Potrykus, J., Wexler, M., Bond, P. L. & Dopson, M. Biofilm development in the extremely acidophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Extremophiles 14, 485–491 (2010).This study is the first to use omics technologies to investigate archaeal biofilms.
Losensky, G. et al. Shedding light on biofilm formation of Halobacterium salinarum R1 by SWATH-LC/MS/MS analysis of planktonic and sessile cells. Proteomics 17, 1–13 (2017).
Reimann, J. et al. Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius. Mol. Cell. Proteomics 12, 3908–3923 (2013).
Li, L. et al. Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius. Mol. Microbiol. 105, 777–793 (2017).
Pesavento, C. et al. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22, 2434–2446 (2008).
Orell, A. et al. A regulatory RNA is involved in RNA duplex formation and biofilm regulation in Sulfolobus acidocaldarius. Nucleic Acids Res. 46, 4794–4806 (2018).
Ghaz-Jahanian, M. A., Khodaparastan, F., Berenjian, A. & Jafarizadeh-Malmiri, H. Influence of small RNAs on biofilm formation process in bacteria. Mol. Biotechnol. 55, 288–297 (2013).
Mika, F. & Hengge, R. Small regulatory RNAs in the control of motility and biofilm formation in E. Coli and Salmonella. Int. J. Mol. Sci. 14, 4560–4579 (2013).
Prasse, D., Ehlers, C., Backofen, R. & Schmitz, R. A. Regulatory RNAs in archaea: first target identification in Methanoarchaea. Biochem. Soc. Trans. 41, 344–349 (2013).
Hawver, L. A., Jung, S. A. & Ng, W.-L. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752 (2016).
Waters, C. M. & Bassler, B. L. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 (2005).
Parsek, M. R. & Greenberg, E. P. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13, 27–33 (2005).
Davies, D. G. et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 (1998).
Zhang, G. G. et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 6, 1336–1344 (2012). This study describes archaeal AHL-based quorum sensing.
Tommonaro, G., Abbamondi, G. R., Iodice, C., Tait, K. & De Rosa, S. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb. Ecol. 63, 490–495 (2012).
Paggi, R. A., Martone, C. B., Fuqua, C. & De Castro, R. E. Detection of quorum sensing signals in the haloalkaliphilic archaeon Natronococcus occultus. FEMS Microbiol. Lett. 221, 49–52 (2003).
Nichols, J. D., Johnson, M. R., Chou, C.-J. & Kelly, R. M. Temperature, not LuxS, mediates AI-2 formation in hydrothermal habitats. FEMS Micriobiol. Ecol. 68, 173–181 (2009).
Ng, F. S. W., Wright, D. M. & Seah, S. Y. K. Characterization of a phosphotriesterase-like lactonase from Sulfolobus solfataricus and its immobilization for disruption of quorum sensing. Appl. Environ. Microbiol. 77, 1181–1186 (2011).
Hiblot, J., Gotthard, G., Chabriere, E. & Elias, M. Characterisation of the organophosphate hydrolase catalytic activity of SsoPox. Sci. Rep. 2, 779 (2012).
Fuqua, C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188, 3169–3171 (2006).
Rajput, A. & Kumar, M. Computational exploration of putative LuxR solos in archaea and their functional implications in quorum sensing. Front. Microbiol. 8, 798 (2017).
Romling, U. Great times for small molecules: c-di-AMP, a second messenger candidate in bacteria and archaea. Sci. Signal. 1, 39 (2008).
Cowan, D. A. & Fernandez-Lafuente, R. Enhancing the functional properties of thermophilic enzymes by chemical modification and immobilization. Enzyme Microb. Technol. 49, 326–346 (2011).
Orell, A., Navarro, C. A., Arancibia, R., Mobarec, J. C. & Jerez, C. A. Life in blue: copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol. Adv. 28, 839–848 (2010).
Klose, H., Röder, J., Girfoglio, M., Fischer, R. & Commandeur, U. Hyperthermophilic endoglucanase for in planta lignocellulose conversion. Biotechnol. Biofuels 5, 63 (2012).
Anderson, I. et al. Novel insights into the diversity of catabolic metabolism from ten haloarchaeal genomes. PLoS ONE 6, e20237 (2011).
Quehenberger, J., Shen, L., Albers, S.-V., Siebers, B. & Spadiut, O. Sulfolobus — a potential key organism in future biotechnology. Front. Microbiol. 8, 2474 (2017).
Brown, M. N., Briones, A., Diana, J. & Raskin, L. Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol. Ecol. 83, 17–25 (2013).
Barcón, T., Alonso-Gutiérrez, J. & Omil, F. Molecular and physiological approaches to understand the ecology of methanol degradation during the biofiltration of air streams. Chemosphere 87, 1179–1185 (2012).
Gómez-Silván, C. et al. Structure of archaeal communities in membrane-bioreactor and submerged-biofilter wastewater treatment plants. Bioresour. Technol. 101, 2096–2105 (2010).
Morales, M. et al. Bio-oxidation of H2S by Sulfolobus metallicus. Biotechnol. Lett. 33, 2141–2145 (2011).
Valenzuela, L. et al. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnol. Adv. 24, 197–211 (2006).
Vera, M., Schippers, A. & Sand, W. Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation-part A. Appl. Microbiol. Biotechnol. 97, 7529–7541 (2013).
Olson, G. J., Brierley, J. A. & Brierley, C. L. Bioleaching review part B. Appl. Microbiol. Biotechnol. 63, 249–257 (2003).
Rawlings, D. E. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 4, 13 (2005).
Watling, H. R. The bioleaching of sulphide minerals with emphasis on copper sulphides — a review. Hydrometallurgy 84, 81–108 (2006).
Lindström, E. B., Gunneriusson, E. & Tuovinen, O. H. Bacterial oxidation of refractory sulfide ores for gold recovery. Crit. Rev. Biotechnol. 12, 133–155 (1992).
Kurosawa, N. et al. Sulfurisphaera ohwakuensis gen. nov., sp. nov., a novel extremely thermophilic acidophile of the order Sulfolobales. Int. J. Syst. Bacteriol. 48, 451–456 (1998).
Norris, P. R., Clark, D. A., Owen, J. P. & Waterhouse, S. Characteristics of Sulfobacillus acidophilus sp. nov. and other moderately thermophilic mineral-sulphide-oxidizing bacteria. Microbiology 142, 775–783 (1996).
Calderón, K. et al. Archaeal diversity in biofilm technologies applied to treat urban and industrial wastewater: recent advances and future prospects. Int. J. Mol. Sci. 14, 18572–18598 (2013).
Sekiguchi, Y., Kamagata, Y., Nakamura, K., Ohashi, A. & Harada, H. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65, 1280–1288 (1999).
Rademacher, A. et al. Characterization of microbial biofilms in a thermophilic biogas system by high-throughput metagenome sequencing. FEMS Microbiol. Ecol. 79, 785–799 (2012).
Wang, H. et al. Development of microbial populations in the anaerobic hydrolysis of grass silage for methane production. FEMS Microbiol. Ecol. 72, 496–506 (2010).
Le-Clech, P. Membrane bioreactors and their uses in wastewater treatments. Appl. Microbiol. Biotechnol. 88, 1253–1260 (2010).
Calderón, K., Rodelas, B., Cabirol, N., González-López, J. & Noyola, A. Analysis of microbial communities developed on the fouling layers of a membrane-coupled anaerobic bioreactor applied to wastewater treatment. Bioresour. Technol. 102, 4618–4627 (2011).
Yeon, K.-M. et al. Quorum sensing: a new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ. Sci. Technol. 43, 380–385 (2009).
Bang, C. & Schmitz, R. A. Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol. Rev. 39, 631–648 (2015).
Dridi, B., Raoult, D. & Drancourt, M. Archaea as emerging organisms in complex human microbiomes. Anaerobe 17, 56–63 (2011).
Probst, A. J., Auerbach, A. K. & Moissl-Eichinger, C. Archaea on human skin. PLoS ONE 8, e65388 (2013).
Dridi, B., Henry, M., El Khéchine, A., Raoult, D. & Drancourt, M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE 4, e7063 (2009).
Lagier, J.-C., Million, M., Hugon, P., Armougom, F. & Raoult, D. Human gut microbiota: repertoire and variations. Front. Cell. Infect. Microbiol. 2, 136 (2012).
McNeil, N. I. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 39, 338–342 (1984).
Samuel, B. S. et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl Acad. Sci. USA 104, 10643–10648 (2007).
Pimentel, M., Gunsalus, R. P., Rao, S. S. & Zhang, H. Methanogens in human health and disease. Am. J. Gastroenterol. Suppl. 6, 28–33 (2012).
White, J. F. Syntrophic imbalance and the etiology of bacterial endoparasitism diseases. Med. Hypotheses 107, 14–15 (2017).
Bang, C., Weidenbach, K., Gutsmann, T., Heine, H. & Schmitz, R. A. The intestinal archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS ONE 9, e99411 (2014).
Faveri, M. et al. Prevalence and microbiological diversity of Archaea in peri-implantitis subjects by 16S ribosomal RNA clonal analysis. J. Periodontal Res. 46, 338–344 (2011).
Matarazzo, F., Ribeiro, A. C., Feres, M., Faveri, M. & Mayer, M. P. A. Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. J. Clin. Periodontol. 38, 621–627 (2011).
Vianna, M. E., Conrads, G., Gomes, B. P. F. A. & Horz, H. P. T-RFLP-based mcrA gene analysis of methanogenic archaea in association with oral infections and evidence of a novel Methanobrevibacter phylotype. Oral Microbiol. Immunol. 24, 417–422 (2009).
Lepp, P. W. et al. Methanogenic Archaea and human periodontal disease. Proc. Natl Acad. Sci. USA 101, 6176–6181 (2004).
Orell, A., Fröls, S. & Albers, S.-V. Archaeal biofilms: the great unexplored. Annu. Rev. Microbiol. 67, 337–354 (2013).
Comolli, L. R., Baker, B. J., Downing, K. H., Siegerist, C. E. & Banfield, J. F. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 3, 159–167 (2009).
Baker, B. J. et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc. Natl Acad. Sci. USA 107, 8806–8811 (2010).
Macalady, J. L., Jones, D. S. & Lyon, E. H. Extremely acidic, pendulous cave wall biofilms from the Frasassi cave system, Italy. Environ. Microbiol. 9, 1402–1414 (2007).
Rascovan, N., Maldonado, J., Martín, P. V. & María, E. F. Metagenomic study of red biofilms from Diamante Lake reveals ancient arsenic bioenergetics in haloarchaea. Int. Soc. Microb. Ecol. 109, 1–11 (2015).
Brileya, K. A., Camilleri, L. B., Zane, G. M., Wall, J. D. & Fields, M. W. Biofilm growth mode promotes maximum carrying capacity and community stability during product inhibition syntrophy. Front. Microbiol. 5, 693 (2014).
Schrenk, M. O., Kelley, D. S., Delaney, J. R. & Baross, J. A. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69, 3580–3592 (2003).
Auernik, K. S., Maezato, Y., Blum, P. H. & Kelly, R. M. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74, 682–692 (2008).
Kozubal, M. A. et al. Microbial iron cycling in acidic geothermal springs of yellowstone national park: integrating molecular surveys, geochemical processes, and isolation of novel fe-active microorganisms. Front. Microbiol. 3, 109 (2012).
Rinker, K. D. & Kelly, R. M. Growth physiology of the hyperthermophilic archaeon thermococcus litoralis: development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence of biofilm formation. Appl. Environ. Microbiol. 62, 4478–4485 (1996).
Acknowledgements
A.O. received intramural funding from the Max Planck Society, and M.v.W. and S.V.A. were supported by a European Research Council starting grant (52311; ARCHAELLUM).
Author information
Authors and Affiliations
Contributions
M.v.W., A.O. and S.V.A. researched data for the article, substantially contributed to discussion of content, wrote the article and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Sessile communities
-
Immobilized communities of cells attached to a surface.
- Extracellular polymeric substances
-
(EPSs). Extracellular biopolymers (including polysaccharides, nucleic acids, proteins and lipids) that provide structural support to the biofilm and nutrition to the surrounding cells.
- Microcolonies
-
Colonies of cells that can be observed only through a microscope.
- Syntrophy
-
The symbiotic relationship between two different species in which one or both benefit from the other through metabolic interactions.
- Acid mine drainages
-
Discharges of acidic water, often containing toxic compounds, from coal or metal mines.
- Reverse methanogenesis
-
The anaerobic oxidation of methane.
- Biodegradation
-
The breakdown of materials by microorganisms or through other biological means.
- Bioleaching
-
The extraction of metals from their ores by means of microorganisms, which convert insoluble metals into a soluble form.
- Archaella
-
Archaeal motility structures analagous to flagella but homologous in their subunit composition to T4P.
- Cytoplasmic bridges
-
Structures between two cells that connect their cytoplasms.
- Nanowires
-
Electrically conductive appendages of a few nanometres in diameter formed by certain microorganisms.
- Pyrite
-
An iron sulfide with the chemical formula FeS2.
- Ced system
-
A transporter essential for the exchange of chromosomal DNA between connecting Sulfolobus cells. Homologues can be found in several Crenarchaeota.
- Quorum-sensing inducers
-
Signalling molecules produced by microorganisms that allow the communication between cells in response to changes in population density.
- Carboxylated acyl homoserine lactones
-
(AHLs). A class of quorum-sensing signalling molecules.
- Persister cells
-
Dormant cells that are phenotypic variants of the wild-type population. They are highly tolerant to different stresses.
- Autoinducer
-
(AI-2). A class of quorum-sensing signalling molecules.
- Second messenger
-
An intracellular signalling molecule that acts in response to extracellular molecules and triggers signal transduction cascades.
- Grass silage
-
Fermented grass.
Rights and permissions
About this article
Cite this article
van Wolferen, M., Orell, A. & Albers, SV. Archaeal biofilm formation. Nat Rev Microbiol 16, 699–713 (2018). https://doi.org/10.1038/s41579-018-0058-4
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-018-0058-4
This article is cited by
-
Methanogenic partner influences cell aggregation and signalling of Syntrophobacterium fumaroxidans
Applied Microbiology and Biotechnology (2024)
-
Stormwater ponds serve as variable quality habitat for diverse taxa
Wetlands Ecology and Management (2024)
-
Symbiotic Interactions of Archaea in Animal and Human Microbiomes
Current Clinical Microbiology Reports (2023)
-
Antibiofilm properties of cathelicidin LL-37: an in-depth review
World Journal of Microbiology and Biotechnology (2023)
-
Insights into the defensive mechanism of bioleaching microorganisms under extreme environmental copper stress
Reviews in Environmental Science and Bio/Technology (2023)