Abstract
We aimed to develop a new artificial intelligence software that can automatically extract and measure the volume of white matter hyperintensities (WMHs) in head magnetic resonance imaging (MRI) using only thick-slice fluid-attenuated inversion recovery (FLAIR) sequences from multiple centers. We enrolled 1092 participants in Japan, comprising the thick-slice Private Dataset. Based on 207 randomly selected participants, neuroradiologists annotated WMHs using predefined guidelines. The annotated images of participants were divided into training (n = 138) and test (n = 69) datasets. The WMH segmentation model comprised a U-Net ensemble and was trained using the Private Dataset. Two other models were trained for validation using either both thin- and thick-slice MRI datasets or the thin-slice dataset alone. The voxel-wise Dice similarity coefficient (DSC) was used as the evaluation metric. The model trained using only thick-slice MRI showed a DSC of 0.820 for the test dataset, which is comparable to the accuracy of human readers. The model trained with the additional thin-slice dataset showed only a slightly improved DSC of 0.822. This automatic WMH segmentation model comprising a U-Net ensemble trained on a thick-slice FLAIR MRI dataset is a promising new method. Despite some limitations, this model may be applicable in clinical practice.
Similar content being viewed by others
Introduction
White matter hyperintensities (WMHs) are common neuroimaging findings characterized by high signals either as periventricular hyperintensities (PVHs) or deep subcortical WMHs (DSWMHs) in fluid-attenuated inversion recovery (FLAIR) sequences of head magnetic resonance imaging (MRI)1. Many reports have shown associations of the WMH degree with ischemic stroke, depression, dementia, and emotional disorders2,3,4,5,6,7,8. Although the WMH prevalence is higher in older individuals, studies of healthy younger people have reported that the WMH degree is also correlated with the execution of speed-demanding functions, that is, subfrontal cortical functions, such as word recall6,7,8,9,10.
Brain Dock is a widely implemented brain checkup system that uses MRI and magnetic resonance angiography to detect asymptomatic brain diseases, such as unruptured cerebral aneurysms and WMHs at an early stage, thereby preventing stroke and dementia in healthy individuals in Japan11,12. During a Brain Dock, the evaluation of WMHs is required, using a standard database with the aim of accumulating data on the diagnosis and early detection of disease risk11,12,13. To date, manual interpretation by neuroradiologists has been considered an effective method for quantitatively assessing WMHs14. However, this approach is not only time-consuming and labor-intensive but also has the disadvantage of high interrater variability (10–68%), which can be expected to result in significant bias, making large-scale studies using quantitative evaluation difficult14,15,16,17.
Recently, artificial intelligence (AI) has been used for automated reading and volumetric measurements. Although many publications have reported the use of AI to assess brain atrophy on T1-weighted images (T1WIs), reports on AI algorithms for automated WMH segmentation using only FLAIR images are limited15,18. The size, number, shape, and location of WMHs are heterogeneous and age-dependent, making them difficult to evaluate using AI. Moreover, WMHs are present in a variety of diseases, requiring background information, such as patient history and disease course, and multiple MRI sequences for differentiation15,19,20. Only a few of the reported AI algorithms are applicable in clinical practice because many are limited to specific diseases, have small sample sizes, evaluate multiple sequences, or require a thin slice thickness as the imaging condition19,21,22. To ensure applicability in large-sized clinical practice for brain screening, accurate results must be provided consistently under imaging protocols for various WMHs, often requiring multicenter collaborative studies23,24. Only a few studies to date have validated the AI algorithm for automatic WMH segmentation by FLAIR alone in a multicenter setting25. If AI algorithms for automated WMH segmentation using only FLAIR images were developed and automatically classified in Brain Dock, the scientific contribution to preventive medicine in healthy people would be immeasurable. The initial purpose of this study was to develop a new AI software that can automatically extract and measure WMH volumes on head MR images using only thick-slice FLAIR images from multiple centers, in accordance with the Japan Brain Dock Society standards. In the future, we plan to develop an AI software that can automatically classify WMHs.
Results
According to the five neuroradiologists participating in this study, the average WMH volume was 18.11 ± 22.07 mL, with a maximum of 109.45 mL and a minimum of 0.00 mL indicating the absence of a WMH. The distribution of WMH volumes in the Private Dataset (PR) (Fig. 1a) closely resembled that in the WMH Segmentation Challenge (WMHC) Dataset (Fig. 1b). Given the potential for biased evaluation data to negatively impact generalization performance on unseen data, the PR was expected to evaluate generalization performance with similar accuracy to that of the WMHC Dataset. Moreover, the unannotated images of 885 participants were utilized for pseudo-labeling to enhance model performance.
Histograms showing the WMH volume distributions throughout the annotated Private Dataset (n = 207) and WMHC Dataset (n = 170). The WMH volume histograms of the Private Dataset (a) and WMHC Dataset (b) are shown. The x-axis represents the total WMH volume in each participant, and the y-axis represents the counts. WMH: white matter hyperintensity, WMHC: white matter hyperintensity segmentation challenge.
Quantitative analysis
Table 1 shows the evaluation results of WMH segmentation based on the Dice similarity coefficient (DSC), Recall, precision, and modified Hausdorff distance (95th percentile of the Hausdorff distance in mm) were compared. The test results for the PR and WMHC Dataset are shown separately. The PR model demonstrated high performance with a DSC of 0.8 or higher on the PR, whereas its performance on the WMHC Dataset was not as high. The PR + WMHC model, which included the WMHC Dataset for training, showed improved performance and the ability to handle both thick- and thin-slice MRI datasets simultaneously. However, its performance on the PR improved only marginally, with a DSC increase of only 0.002. This modest degree of improvement might be attributed to domain differences between these datasets, suggesting that the thin-slice dataset does not significantly enhance performance on the thick-slice dataset. Additionally, the model trained only on the WMHC Dataset exhibited overfitting to the test dataset.
Regarding the performance of the existing commercial software that served as the basis for the algorithm in this study, it had a DSC of 0.635 for the PR, whereas the new algorithm showed a DSC of 0.820.
Qualitative analysis
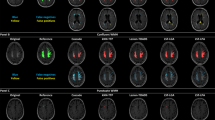
Figure 2 shows the segmentation results of the PR and WMHC Dataset models used with the test dataset. The top row presents a participant with a relatively small WMH volume. All models successfully detected regions that were similar to the ground truth. In a participant with extensive WMHs, as in the second row, both the PR and PR + WMHC models accurately detected the WMH regions, whereas the WMHC model exhibited false negatives (FNs), leading to a lower DSC. However, in a participant with small punctate WMHs, as observed in the right hemisphere in the third row, the WMHC model demonstrated better results compared with the other two models. The differences between the PR and the PR + WMHC models are shown in the last row. False positives (FPs), which occurred in the PR model, were reduced in the PR + WMHC model. For all other participants, the difference between these two models was minimal, which resulted in a slight DSC difference of 0.002.
Segmentation results for Private Dataset and WMHC Dataset models used with the test dataset. The columns from left to right show the original FLAIR image, ground-truth WMH annotations, and segmentation results for the PR, PR + WMHC, and WMHC models. The first row shows a TP case in which all models correctly detected WMHs, the second shows FN regions in the WMHC results, and the third row shows small FN regions in the PR and PR + WMHC results. The last row shows the difference in FP regions between PR and PR + WMHC results. FLAIR: fluid-attenuated inversion recovery, FN: false negative, FP: false positive, PR: Private Dataset, TP: true positive, WMH: white matter hyperintensity, WMHC: WMH Segmentation Challenge.
A tendency toward FPs and FNs was observed in the PR model in areas with small punctate WMHs, near the lateral ventricles, near the septum pellucidum, lacunar infarcts, or where the boundary between WMHs and other structures was ambiguous. As shown in Fig. 2, row 3, the model missed the small punctate WMHs in the left hemisphere. Conversely, in the example from Fig. 2, row 4, punctate FPs can be observed in the left hemisphere. The figure also shows FPs near the surface of the lateral ventricle body, a region where the boundary between a WMH and other structures might be indistinct. In the same example, the FPs were only observed in the PR and PR + WMHC models. For such cases with ambiguous boundaries, the PR model tends to detect slightly excessively, suggesting the possibility of slight over-annotation in the PR for training. While the examples in Fig. 2, rows 1 and 2, are generally accurate, an unintended “hole” appeared in the right hemisphere of Fig. 2, row 2. Furthermore, the recall of the PR model was lower than its precision, indicating that the model tended to have an under-detection bias on the PR.
To better understand variations on a case-by-case basis, the DSC for each case was calculated using the PR model results of the PR. The DSC values ranged from a minimum of 0.158 to a maximum of 0.901. Figure 3a,b show the cases with the lowest and highest DSCs, respectively. The case with the lowest DSC, at 0.158, missed small punctate WMHs, and the missed area accounted for a large portion of the total WMH area. Conversely, the case with the highest DSC (0.901) showed a larger total WMH area. Although some FNs or FPs were observed, their proportions to the total count were limited, resulting in a high DSC.
Segmentation results of the PR model for cases with the lowest and highest DSC values from the Private Dataset. (a) The case with the lowest DSC value of 0.158; (b) the case with the highest DSC value of 0.901. The columns titled “Original FLAIR,” “WMH annotation,” and “PR” display the original FLAIR images; the ground-truth WMH annotations and the segmentation results of the PR model are shown in green and red, respectively. DSC: Dice similarity coefficient, FLAIR: fluid-attenuated inversion recovery, PR: Private Dataset, WMH: white matter hyperintensity.
Processing time analysis
Processing time measurements were conducted for thick-slice MR images, revealing an average processing time of 18.5 s per case.
Discussion
In this study, we evaluated the performance of an AI using automatically extracted volumetric measurement of WMH lesions on head MR images on thick-slice FLAIR images. The results showed that the DSC in our study reached 0.820, which is comparable to human reading accuracy.
Although various algorithms for AI-based automated volumetric measurements of WMHs have been reported in recent years, very few are applicable in clinical practice19,21,22. This is because most previous studies on AI and volumetric measurements of WMHs, many were limited to specific diseases, such as Alzheimer’s disease or multiple sclerosis, and had relatively small sample sizes of approximately 100 patients26,27,28 and an algorithm that used three-dimensional (3D) networks with thin-slice image data with 1–3 mm slice thicknesses27,29,30. Indeed, thin-slice MRI is reportedly superior to thick-slice MRI in detecting lesions in small areas, such as the brainstem, and in segmentation accuracy31. Thick-slice MRI, as used in this study, may reduce the accuracy of volumetric measurements in small lesions. However, thin-slice MRI is only widely applied in some developed countries and is not currently commonly used worldwide and in developing countries; MRI with thick slices of approximately 5 mm, as used in this study, remains the most commonly applied method for routine work14. Because thick-slice data is the mainstream in Brain Dock and we aimed to develop a calculation method based on existing Brain Dock data and the default imaging method, we used thick-slice data in this study. The size, number, shape, and location of WMHs are heterogeneous and age-dependent, making reliable and objective assessments difficult14,24. In the future, if thin-slice FLAIR imaging becomes the mainstream for brain screening, additional considerations may be necessary.
WMHs result primarily from aging processes, such as demyelination and axonal loss, both of which occur as a result of small vessel disease of the brain7. Thus, WMHs are frequently observed in older asymptomatic patients, although they can also occur in a variety of other diseases, including demyelinating diseases such as multiple sclerosis; genetic diseases such as cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy; infectious diseases such as human immunodeficiency virus encephalopathy; psychiatric disorders such as depressive disorders and schizophrenia; and various other diseases such as autoimmune diseases, neoplastic diseases, intoxication, and hypoxic encephalopathies32. Therefore, volumetric detection of WMHs in younger people is an important aspect of brain checkups, and lifestyle improvements are needed in examinees with increased WMH volume over time.
This study proposes a new AI software that solves these problems, and its major feature is the analysis of 5-mm thick-slice FLAIR images. Moreover, this study evaluated both the PR and the WMHC Dataset as thick (5 mm) and thin (1–3 mm) slices, respectively, with a sample size of over 200 participants. The results showed that the WMH segmentation accuracy based on two-dimensional (2D) FLAIR images with 5 mm slices, which is widely used in general hospitals, was comparable to that of previous studies. This suggests that the proposed AI algorithm can be applied in clinical practice.
We only identified 39 publications that analyzed the automatic segmentation of WMHs on head MRI using AI similar to the procedure in the present study. Of these, we reviewed 17 publications since 2018, including the present study (Table 2). The sample size varied from 20 to 1092 overall, with the sample size of this study being the largest. Multiple MRI sequences analyzing both T1WI and FLAIR images were identified in 12 studies20,22,26,33,34,35,36,37,38, and only one study in addition to the present study evaluated only FLAIR images. Regarding the MRI field strength, nearly all studies, including ours, used 3 T MRI. Regarding the segmentation accuracy, the DSC values ranged from 0.41 to 0.83, showing a wide range of accuracy. The DSC of the AI model in this study was 0.820, indicating higher accuracy compared with that of previous studies overall. Therefore, the model developed in the present study demonstrably has a better performance than that of existing AI models.
The unique feature of our study is that we have developed an AI that performs better than previous AI models using “thick slices” and “FLAIR images only” in accordance with the standards of the Japan Brain Dock Society. Notably, the performance of this AI model was tested on the largest sample size in a study of this type of more than 1000 cases, using data from multiple institutions. In addition, the processing time when performing segmentation was as fast as 18.5 s per case for our AI model. When corrected for comparison with a previous study, the equivalent value was approximately 1.8 min. This is faster than the approximate processing time of 5 min using a supervised deep learning-based method reported in the previous study32. Therefore, this AI algorithm seems more appropriate for real-world clinical adaptation. MRI with thick slices and FLAIR imaging are only simple methods for brain checkups but have an advantage in that MRI examinations are easily accessible for patients and versatile. Our findings can also be used as a basis for PVH and DSWMH grading as proposed by the Japan Brain Dock Society. The ultimate goal of our AI model is to classify PVH and DSWMH grading to screen normal participants at risk of early-onset dementia to prevent or delay the onset of dementia through lifestyle guidance.
This study had several limitations. First, approximately 10 physicians diagnosed the images for annotation, possibly leading to individual differences in physician judgment, resulting in diagnostic bias. However, all physicians in this study were board-certified radiologists from the Japan Radiological Society or neuroradiologists with more than 7 years of experience and board-certified neurosurgeons from the Japan Neurosurgical Society; therefore, the diagnostic results should be of a uniformly high standard. Second, because only high-intensity areas on FLAIR images were evaluated in this study, lacunar infarction might not have been clearly distinguished because only two lacunar infarctions were included in the training dataset. We would like to develop another AI to distinguish between WMHs and lacunar infarctions after acquiring a lacunar infarctions dataset in the near future. Third, a detailed analysis differentiating between PVH and DSWMH within WMHs was not conducted in this study; however, our research is currently ongoing. Fourth, although the data in this study was normalized by scaling the data to the range of 0.0–1.0, some bright artifacts might have affected the scaled value of the lesion. Although various methods are used for normalization, we used a simple normalization method and managed the intensity variations through contrast augmentation. Other approaches, such as ensuring uniform contrast during the normalization stage, were not validated in this study. Finally, this study was based only on FLAIR images for our purpose, which were not combined with other sequence images such as T1WIs, T2WIs, diffusion-weighted images, and other images that are useful for differentiation. Because the most clinically important imaging method is FLAIR for the diagnosis of WMHs, we believe that the development of a new AI software automatically extracting and measuring WMH volumes using only thick-slice FLAIR images is most meaningful for initial brain screening. As the next step, we plan to create an AI algorithm that combines FLAIR images with other sequences such as T1WIs to differentiate lesions and classify WMHs in the near future.
In conclusion, the automatic WMH segmentation model based on a U-Net ensemble trained on a thick-slice FLAIR MRI dataset showed a DSC of 0.820 and a promising AI algorithm. This model may be applicable in clinical practice for brain checkups.
Methods
Ethics approval and informed consent
This study was approved by the Institutional Review Board of Hiroshima University (approval number: E2022-0262). All study protocols were developed according to the guidelines of Hiroshima University Hospital. Individual data were anonymized and collected during routine Brain Dock examinations; thus, all participants provided informed consent based on an opt-out method. Moreover, in this study, we were provided with anonymized data; therefore, the authors did not have access to personally identifiable participant information during or after data collection.
Study design
In this study, a new automatic WMH segmentation model was developed based on existing commercial software (EIRL Brain Metry version 1.13.0, LPIXEL Inc., Tokyo, Japan). This software employs U-Net for segmentation. Although the core usage of U-Net remains consistent, we replaced the backbone network with a new model and also explored the model ensembles.
The WMH segmentation model of the software was updated using a newly collected dataset, the PR. This new model was named “PR” as it was trained using the PR. The model performance was also evaluated using the publicly available WMHC Dataset29. The WMHC Dataset has thinner slices than those in the PR.
For validation, two additional models were trained using the same training procedure. One was a model trained using both the PR and the WMHC Dataset (PR + WMHC model) to evaluate the effect of adding an MRI dataset with thin slices to the PR with thick slices. The other was a model trained using only the WMHC Dataset (the WMHC model) to evaluate the performance of a model trained using only a thin-slice MRI dataset.
Private dataset
For this dataset, 1092 FLAIR MR images were collected from three clinics in Japan. To minimize potential biases related to the WMH severity, data were gathered in a manner that ensured a balanced distribution of PVH/DSWMH grades (0–4) as determined by each clinic. The WMH annotation process was restricted to 207 randomly selected participants because of the annotation costs. Nearly all the annotated WMHs were age-related, and 2 of these 207 participants had lacunar infarcts. The slice thickness of the annotated data ranged from 5 to 6 mm, with an average of 5.37 mm for the entire dataset. Detailed spatial characteristics, including slice thickness, are presented in Table 3. The data were acquired using nine different types of scanners, and the detailed MRI parameters are listed in Supplementary Table S1.
Five neuroradiologists shared the annotation efforts of WMHs for the 207 selected participants. To minimize interrater variability and ensure annotation quality, an annotation guideline was developed under the supervision of neuroradiologists. The following guidelines were provided for areas prone to individual variations: (a) the area around the ventricles, sulci, and longitudinal fissure may show a non-WMH signal; therefore, an annotation is only made when a WMH is clearly visible on neighboring upper and lower slices, (b) the septum pellucidum of the lateral ventricles should not be annotated, (c) only WMHs in the cerebrum should be annotated, and (d) lacunar infarcts and enlarged perivascular spaces (EPVSs) should be excluded. The imaging differentiation between lacunar infarcts, EPVSs, and WMHs is that a WMH is depicted as a clear hyperintense lesion on FLAIR, whereas an EPVS is depicted as an iso-to-hypointense lesion and a lacunar infarct is depicted as an iso-to-hyperintense lesion with a hypointense center11,13. In addition, WMHs are found mainly in the cerebral white matter, whereas EPVSs and lacunar infarcts are found in the basal ganglia, in addition to the cerebral white matter.
For the model development, the annotated datasets were randomly divided into training (n = 138) and test (n = 69) datasets. To ensure an accurate evaluation, a second annotation review was conducted on 69 test participants. The review was performed by three neuroradiologists who differed from the five physicians employed in the initial annotation process. The reviewers were asked to modify the previously annotated WMH regions if needed. As a result, several modifications were made to the test dataset, thereby ensuring the reliability of the evaluation criteria.
WMH segmentation challenge dataset
For the evaluation using an external dataset, the WMHC Dataset was employed29. A competition using this dataset was originally held at the 20th International Conference on Medical Image Computing and Computer-Assisted Intervention in 2017 in Quebec City, Quebec, Canada. After the competition, models could be submitted via their website until December 2022. Afterward, the previously undisclosed test dataset was made available. Several previous studies have used this dataset15,26,31,32, and our study assessed performance using the same dataset. Although the dataset included both FLAIR images and T1WIs, only FLAIR images were used in this study.
The WMHC Dataset consisted of 60 training and 110 test participants. These participants were sourced from three institutes located in the Netherlands and Singapore. The data were acquired using five different types of scanners. The average slice thickness of the FLAIR sequences in this dataset was 2.23 mm. Notably, the average slice thickness was approximately 41% that of the PR. The WMH volume averaged 16.92 ± 21.52 mL, with a minimum of 0.78 mL and a maximum of 195.15 mL29.
WMH segmentation model
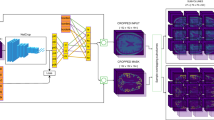
The algorithm for WMH detection in the software comprised a 2D convolutional neural network (CNN) model that processes individual MRI slices sequentially. Initially, pixel values of the input MR images were normalized to fall within the range of 0.0–1.0. Notably, image processing-based bias field corrections were not applied. We made this decision because the effect of the contrast augmentation during training was anticipated to accommodate such variations. Then, the normalized image was resized to 512 × 512 pixels and input into the main CNN model. This CNN model was an ensemble of two U-Net models, each with distinct characteristics. The first U-Net model utilized ResNext50 as the encoder, and the ImageNet pretrained weights were used as the initial weights. The learning rate was set to 0.001, the batch size was set to 15, and the model was trained for 15 epochs using the Adam optimizer. These hyperparameters were obtained using a grid search. During training, the learning rate was decayed using a cosine-annealing learning-rate reducer to facilitate convergence. Matthews correlation coefficient loss was used as the loss function, as originally proposed for skin lesion segmentation39. This loss function incorporates a penalty for misclassification of the true negative pixels and is expected to show robust performance. During the training process, data augmentations such as horizontal flips, vertical flips, shifting, rotation, contrast transformation (changing the window width), and grid distortion were applied to all the slices, regardless of WMH presence, to foster the development of a noise-robust model. The second U-Net model used EfficientNet-B5 as the encoder, which was trained in the same manner as the first U-Net. The output of the main CNN model was a threshold of 0.5, and the predicted binary WMH mask was obtained. The number of ensembles was set to two to obtain a balance between processing time and improved accuracy. Ensembles comprising three or more models increased the processing time by approximately 1.5 times or more, although the enhancement in performance was marginal.
Pseudo-labeling was employed to effectively utilize the unlabeled data in the training of the actual model40. This technique uses the model’s predictions for unlabeled data to generate labels, which are then used to retrain the model, potentially increasing the diversity of the training dataset and improving generalization performance. First, a model for pseudo-labeling was trained using the PR for training only. The model was then used to predict the WMH regions in unannotated images of 885 participants. These predicted WMH masks can be regarded as pseudo-WMH annotations (pseudo-labeling dataset). The pseudo-labeling model showed a DSC of 0.817 for the PR. Although the pseudo-labeling dataset contains both correct and incorrect predictions, increasing the diversity of the training dataset can be expected to improve the generalization performance. Finally, a new model was trained using both the PR and pseudo-labeling dataset.
Evaluation metrics
To evaluate WMH segmentation performance, the voxel-wise DSC was used in this study. The DSC is a measure of the agreement between predicted and ground-truth regions and is defined as DSC = 2 × true positive (TP)/(2 × TP + FP + FN), where TP, FP, and FN are the numbers of TP, FP, and FN voxels, respectively. Moreover, recall and precision were calculated using the following formulas: recall = TP/(TP + FN) and precision = TP/(TP + FP). Since there are significantly more voxels in images without versus with WMHs, resulting in a notably higher number of true negatives versus FPs, specificity tends to approach a value near 1. This characteristic makes it unsuitable for detailed comparisons, and thus, was not used as an evaluation metric in this study.
Processing time measurement
To evaluate the applicability of the model in clinical practice with limited resources, processing time was measured without using a graphics processing unit (GPU). The average of 10 measurements was computed for thick-slice MR images. To facilitate comparison with a previous study32, the measured value was corrected for differences in the number of slices and central processing unit (CPU) clock frequencies as follows. In the prior study, time measurements were obtained using MRI with 192 slices on a computer with a 3.5-GHz CPU, whereas our study used 22 slices on a computer without a GPU, equipped with an Intel Core i5-10500 T processor (Intel Corp., Santa Clara, CA, USA) running at 2.30 GHz with 16 GB of memory. Assuming that processing time decreases proportionally with the CPU clock frequency, the corrected processing time, intended for comparison with the results of the previous study, was calculated as follows: corrected processing time = measured time (min) × (192/22) × (2.3/3.5).
Data availability
Anonymized data from the present study may be shared by the corresponding author upon request from a qualified researcher and upon permission from the institutional review board.
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- AI:
-
Artificial intelligence
- CNN:
-
Convolutional neural network
- CPU:
-
Central processing unit
- DSC:
-
Dice similarity coefficient
- DSWMH:
-
Deep and subcortical white matter hyperintensity
- EPVS:
-
Enlarged perivascular space
- FLAIR:
-
Fluid-attenuated inversion recovery
- FN:
-
False negative
- FP:
-
False positive
- GPU:
-
Graphics processing unit
- MRI:
-
Magnetic resonance imaging
- PR:
-
Private Dataset
- PVH:
-
Periventricular hyperintensity
- T1WI:
-
T1-weighted image
- T2WI:
-
T2-weighted image
- TP:
-
True positive
- WMH:
-
White matter hyperintensity
- WMHC:
-
WMH Segmentation Challenge
References
Wardlaw, J. M. et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 12, 822–838 (2013).
van Dijk, E. J. et al. Progression of cerebral small vessel disease in relation to risk factors and cognitive consequences: Rotterdam Scan study. Stroke 39, 2712–2719 (2008).
Mosley, T. H. et al. Cerebral MRI findings and cognitive functioning: the Atherosclerosis Risk in Communities study. Neurology 64, 2056–2062 (2005).
Doddy, R. S., Massman, P. J., Mawad, M. & Nance, M. Cognitive consequences of subcortical magnetic resonance imaging changes in Alzheimer’s disease: Comparison to small vessel ischemic vascular dementia. Neuropsychiatry Neuropsychol. Behav. Neurol. 11, 191–199 (1998).
O’Brien, J. et al. Severe deep white matter lesions and outcome in elderly patients with major depressive disorder: Follow up study. BMJ 317, 982–984 (1998).
Ter Telgte, A. et al. Cerebral small vessel disease: From a focal to a global perspective. Nat. Rev. Neurol. 14, 387–398 (2018).
Prins, N. D. & Scheltens, P. White matter hyperintensities, cognitive impairment and dementia: an update. Nat. Rev. Neurol. 11, 157–165 (2015).
Simoni, M. et al. Age- and sex-specific rates of leukoaraiosis in TIA and stroke patients: population-based study. Neurology 79, 1215–1222 (2012).
Yamasaki, T. et al. Prevalence and risk factors for brain white matter changes in young and middle-aged participants with Brain Dock (brain screening): A registry database study and literature review. Aging (Albany NY) 13, 9496–9509 (2021).
Breteler, M. M. et al. Cognitive correlates of ventricular enlargement and cerebral white matter lesions on magnetic resonance imaging. The Rotterdam Study. Stroke 25, 1109–1115 (1994).
New Guidelines Development Committee for Brain Dock. [The Guideline for Brain Dock 2019]: Kyobunsha; 2019.
Morita, A. Value of brain dock (brain screening) system in Japan. World Neurosurg. 127, 502 (2019).
Saito, I. The guideline for brain dock 2003. Nihon Rinsho 64(Suppl 7), 297–302 (2006).
Zhu, W. et al. Automatic segmentation of white matter hyperintensities in routine clinical brain MRI by 2D VB-Net: A large-scale study. Front. Aging Neurosci. 14, 915009 (2022).
Joo, L. et al. Diagnostic performance of deep learning-based automatic white matter hyperintensity segmentation for classification of the Fazekas scale and differentiation of subcortical vascular dementia. PLoS One 17, e0274562 (2022).
Zijdenbos, A. P., Forghani, R. & Evans, A. C. Automatic, “pipeline” analysis of 3-D MRI data for clinical trials: Application to multiple sclerosis. IEEE Trans. Med. Imaging 21, 1280–1291 (2002).
Grimaud, J. et al. Quantification of MRI lesion load in multiple sclerosis: A comparison of three computer-assisted techniques. Magn. Reson. Imaging 14, 495–505 (1996).
Røvang, M. S. et al. Segmenting white matter hyperintensities on isotropic three-dimensional fluid attenuated inversion recovery magnetic resonance images: Assessing deep learning tools on a Norwegian imaging database. PLoS One 18, e0285683 (2023).
Ding, Y. et al. Using deep convolutional neural networks for neonatal brain image segmentation. Front. Neurosci. 14, 207 (2020).
Ding, T. et al. An improved algorithm of white matter hyperintensity detection in elderly adults. Neuroimage Clin. 25, 102151 (2020).
Park, G., Hong, J., Duffy, B. A., Lee, J. M. & Kim, H. White matter hyperintensities segmentation using the ensemble U-Net with multi-scale highlighting foregrounds. Neuroimage 237, 118140 (2021).
Liu, L., Kurgan, L., Wu, F. X. & Wang, J. Attention convolutional neural network for accurate segmentation and quantification of lesions in ischemic stroke disease. Med. Image Anal. 65, 101791 (2020).
Le, M. et al. FLAIR2 improves LesionTOADS automatic segmentation of multiple sclerosis lesions in non-homogenized, multi-center, 2D clinical magnetic resonance images. Neuroimage Clin. 23, 101918 (2019).
Heinen, R. et al. Performance of five automated white matter hyperintensity segmentation methods in a multicenter dataset. Sci. Rep. 9, 16742 (2019).
Zhang, Y. et al. A deep learning algorithm for white matter hyperintensity lesion detection and segmentation. Neuroradiology 64, 727–734 (2022).
Park, B. Y. et al. DEWS (DEep White matter hyperintensity Segmentation framework): A fully automated pipeline for detecting small deep white matter hyperintensities in migraineurs. Neuroimage Clin. 18, 638–647 (2018).
Moeskops, P. et al. Evaluation of a deep learning approach for the segmentation of brain tissues and white matter hyperintensities of presumed vascular origin in MRI. Neuroimage Clin. 17, 251–262 (2018).
Gibson, E., Gao, F., Black, S. E. & Lobaugh, N. J. Automatic segmentation of white matter hyperintensities in the elderly using FLAIR images at 3T. J. Magn. Reson. Imaging 31, 1311–1322 (2010).
Kuijf, H. J. et al. Standardized assessment of automatic segmentation of white matter hyperintensities and results of the WMH Segmentation Challenge. IEEE Trans. Med. Imaging 38, 2556–2568 (2019).
Li, H. et al. Fully convolutional network ensembles for white matter hyperintensities segmentation in MR images. Neuroimage 183, 650–665 (2018).
Iwamura, M. et al. Thin-slice two-dimensional T2-weighted imaging with deep learning-based reconstruction: Improved lesion detection in the brain of patients with multiple sclerosis. Magn. Reson. Med. Sci. 23, 184–192 (2024).
Rachmadi, M. F., Valdés-Hernández, M. D. C., Agan, M. L. F., Di Perri, C. & Komura, T. Segmentation of white matter hyperintensities using convolutional neural networks with global spatial information in routine clinical brain MRI with none or mild vascular pathology. Comput. Med. Imaging Graph 66, 28–43 (2018).
Tran, P. et al. Automatic segmentation of white matter hyperintensities: Validation and comparison with state-of-the-art methods on both Multiple Sclerosis and elderly subjects. Neuroimage Clin. 33, 102940 (2022).
Fiford, C. M. et al. Automated white matter hyperintensity segmentation using Bayesian model selection: assessment and correlations with cognitive change. Neuroinformatics 18, 429–449 (2020).
Wu, J., Zhang, Y. & Tang, X. Simultaneous tissue classification and lateral ventricle segmentation via a 2D U-net driven by a 3D fully convolutional neural network. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 5928–5931 (2019).
Wu, D. et al. Multi-atlas based detection and localization (MADL) for location-dependent quantification of white matter hyperintensities. Neuroimage Clin. 22, 101772 (2019).
Ling, Y., Jouvent, E., Cousyn, L., Chabriat, H. & De Guio, F. Validation and optimization of BIANCA for the segmentation of extensive white matter hyperintensities. Neuroinformatics 16, 269–281 (2018).
Manjón, J. V. et al. MRI white matter lesion segmentation using an ensemble of neural networks and overcomplete patch-based voting. Comput. Med. Imaging Graph 69, 43–51 (2018).
Abhishek, K. & Hamarneh, G. Matthews correlation coefficient loss for deep convolutional networks: Application to skin lesion segmentation. in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). https://doi.org/10.1109/ISBI48211.2021.9433782 (2021).
Lee DH. Pseudo-Label: The simple and efficient semi-supervised learning method for deep neural networks. ICML 2013 Workshop: Challenges in Representation Learning (WREPL) https://www.researchgate.net/publication/280581078/ (2013).
Acknowledgements
We thank the radiologists and neurosurgeons for providing the diagnoses used in this study.
Funding
This work was supported by the Japan Society for the Promotion of Science, Grant-in-Aid for Scientific Research, (C)23K08521.
Author information
Authors and Affiliations
Contributions
All authors have made substantial contributions to the intellectual content of the paper, contributed to data interpretation, approved the final manuscript, and agreed to its submission to this journal. M.K. contributed to the study design and concept; funding acquisition; research conduction; data collection, curation, management, and analysis; quality control; statistical analysis; and manuscript drafting. F.I. contributed to the study design and concept; funding acquisition; data collection and analysis; quality control; statistical analysis; and manuscript revision. S.N. and S.K. helped conceive and oversee the study and contributed to the revision of the manuscript. D.I., H.K., T.H., and Y.M. contributed to data collection, curation, and analysis. R.S., S.M., and T.K. helped with data collection and analysis and the revision of the manuscript. Y.S. helped conceive and oversee the study and assisted in data collection. N.H. assisted in the planning and supervision of this study, as well as in the revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Shinji Nakazawa, Ryo Sato, and Shiyuki Maeyama are employees of LPIXEL Inc. Yuki Shimahara owns stock in the company. Taiki Kaneko is currently an employee of Aile Home Clinic, Niigata, Japan. Masashi Kuwabara, Fusao Ikawa, Saori Koshino, Daizo Ishii, Hiroshi Kondo, Takeshi Hara, Yuyo Maeda, and Nobutaka Horie declare no potential conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kuwabara, M., Ikawa, F., Nakazawa, S. et al. Artificial intelligence for volumetric measurement of cerebral white matter hyperintensities on thick-slice fluid-attenuated inversion recovery (FLAIR) magnetic resonance images from multiple centers. Sci Rep 14, 10104 (2024). https://doi.org/10.1038/s41598-024-60789-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-60789-x
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.