Abstract
Hypervirulent Klebsiella pneumoniae (hvKp) is a significant cause of severe invasive infections in Vietnam, yet data on its epidemiology, population structure and dynamics are scarce. We screened hvKp isolates from patients with bloodstream infections (BSIs) at a tertiary infectious diseases hospital in Vietnam and healthy individuals, followed by whole genome sequencing and plasmid analysis. Among 700 BSI-causing Kp strains, 100 (14.3%) were hvKp. Thirteen hvKp isolates were identified from 350 rectal swabs of healthy adults; none from 500 rectal swabs of healthy children. The hvKp isolates were genetically diverse, encompassing 17 sequence types (STs), predominantly ST23, ST86 and ST65. Among the 113 hvKp isolates, 14 (12.6%) carried at least one antimicrobial resistance (AMR) gene, largely mediated by IncFII, IncR, and IncA/C plasmids. Notably, the acquisition of AMR conjugative plasmids facilitated horizontal transfer of the non-conjugative virulence plasmid between K. pneumoniae strains. Phylogenetic analysis demonstrated hvKp isolates from BSIs and human carriage clustered together, suggesting a significant role of intestinal carriage in hvKp transmission. Enhanced surveillance is crucial to understand the factors driving intestinal carriage and hvKp transmission dynamics for informing preventive measures. Furthermore, we advocate the clinical use of our molecular assay for diagnosing hvKp infections to guide effective management.
Similar content being viewed by others
Introduction
Klebsiella pneumoniae (Kp) is a Gram-negative pathogen that constitutes a substantial health burden globally. It is estimated that Kp was the second leading bacterial cause of mortality, responsible for more than 200,000 deaths attributed to antimicrobial resistance (AMR) in 20191. There are two distinct pathotypes of Kp: the classical (cKp) and the hypervirulent (hvKp). cKp are a common cause of opportunistic infections in hospital settings, mainly affecting individuals with pre-existing comorbidities. cKp can cause various infections, including pneumonia, bacteremia, and urinary tract infections, but metastatic dissemination is relatively uncommon2,3. Notably, cKp exhibits a wide diversity of K capsular types and a high propensity to develop resistance to commonly used antibiotics, such as cephalosporins, carbapenems, and colistin. This has led to the emergence and worldwide spread of multiple highly resistant lineages (i.e., ST258, ST15, ST11, and ST307)4,5,6. The first report of hvKp was from a patient with pyogenic liver abscesses in Taiwan in 19867. This discovery highlighted a distinctive form of invasive community-acquired Kp infection that can affect healthy individuals without underlying diseases8. Beyond liver abscesses, hvKp infections can manifest as endophthalmitis, meningitis, or necrotizing fasciitis, which are typically uncommon in cKp infections9,10. Most hvKp infections are caused by K1 (i.e., ST23) and K2 (i.e., ST86 and ST65) capsular serotypes11,12. Among these hvKp lineages, ST23/K1 is the predominant lineage responsible for human liver abscesses, comprising 82% of the disease-causing hvKp isolates13.
Although hvKp are more prevalent in Asia, infections caused by these organisms have been increasingly reported in other parts of the world12,14,15. The US, Canada, and several European countries have documented occasional cases of liver abscess caused by hvKp, mainly associated with travelers or migrants from Asia16,17,18. Few studies have also found hvKp isolates in the gastrointestinal tracts of healthy individuals, with the carriage rate ranging from 4 to 5.2% in China, and 11.3 to 16.7% among healthy people in Malaysia19,20. Further molecular investigations have revealed a close genetic relationship between hvKp isolates from human fecal carriage and liver abscess21,22. This suggests that the human gastrointestinal tract may act as a reservoir for transmission and infection. The majority of hvKp strains reported to date have shown susceptibility to antibiotics. However, there have been increasing reports on convergent Kp strains across different lineages that display both multidrug resistance (MDR) and virulence. There are two major mechanisms of convergence: hvKp strains acquiring the AMR genes/plasmids as seen in ST23-blaKPC23ST65-blaCTX-M24 in China, or the MDR cKp strains acquiring the virulence plasmid as observed in ST11-blaKPC in China25, ST231-blaOXA-232 in India26. The global emergence of carbapenem-resistant hypervirulent K. pneumoniae represents a significant and escalating threat to global health27.
While there is no consensus definition of hvKp infections, hvKp isolates typically harbor a large IncFIBK plasmid of about 200 kb28. This plasmid carries a number of virulence factors that are either absent or present at a low prevalence in cKp isolates, including the siderophores: aerobactin (iucABCD, iutA), salmochelin (iroBCDN) and the hypermucoid regulator (rmpA/rmpA2)28. It also possesses several gene clusters encoding toxin-antitoxin systems and resistance to heavy metals, including copper (pcoABCDRS), silver (silABCEPSR), iron (fecABDERI), arsenic (arsABDR), and tellurium (terABCDEFZ)14,29,30. Notably, the virulence plasmid lacks a type IV secretion system and has been experimentally demonstrated to be non-conjugative30,31. hvKp often display a hypermucoid phenotype which has been employed to detect hvKp infection via the string test32. Nonetheless, the string test fails to identify hvKp isolates lacking the capsule regulator rmpA/rmpA2 gene, and its accuracy can be affected by culture conditions33. Currently, molecular detection of plasmid-encoded virulence genes (iucA, iroB, rmpA, and rmpA2) is widely considered the most accurate method for distinguishing hvKp from cKp isolates34,35.
Data from multi-hospital surveillance in Vietnam have shown that Kp are a frequent cause of invasive infections, accounting for 13% of all bacterial isolates from blood and CSF samples36. Nonetheless, there are no published data concerning the molecular epidemiology of hvKp infections in this setting. Here, we conducted whole genome sequencing (WGS) and plasmid analysis of hvKp isolates derived from bloodstream infections between 2010 and 2020, as well as two cohorts of healthy individuals, one including children and the other involving adults. Our study provided the first comprehensive insights into the population structure, transmission dynamics, and antimicrobial resistance profiles of hvKp in Vietnam. We further deciphered the genetic structure and horizontal transfer mechanism of the virulence plasmid between Kp isolates.
Results
Prevalence and sequence type distribution of hypervirulent Klebsiella pneumoniae isolates in Vietnam
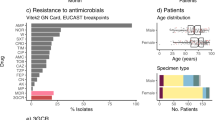
From 1 January 2010 to 31 December 2020, 108,303 blood samples were submitted for culture, yielding a positivity rate of 8.8%. Out of the total 700 Kp isolates responsible for bloodstream infections (BSIs) at HTD between 2010 and 2020, 100 isolates (14.3%) were identified as hvKp based on the results of iucA PCR. The proportion of hvKp infections varied between 11 and 46 cases per 1000 culture-confirmed BSIs, depending on the year (Fig. S1). We identified 13 hvKp isolates from 350 rectal swabs of healthy adult farmers, representing a prevalence of 3.7 percent. Conversely, no hvKp isolates were found in the 500 rectal swabs of healthy children. Most hvKp isolates (89%, 101/113) displayed a hypermucoid phenotype by the string test.
WGS analysis of 113 hvKp isolates revealed that 111 isolates (98.2%) were identified as K. pneumoniae, while the remaining two isolates (1.8%) were K. quasipneumoniae subsp. similipneumoniae. Remarkably, among 113 hvKp isolates, we identified a diverse array of 17 sequence types (STs). The top three most common STs were ST23 (52%), ST86 (11%), and ST65 (8%), followed by ST420 (4%), ST268 (4%), ST25 (4%), and ST660 (3%). Additionally, there were 12 other STs, each represented by one or two isolates (Table 1). Notably, ST23 (n = 57), ST86 (n = 9), and ST65 (n = 9) were the most common genotypes found in patients with BSIs, while ST268 (n = 3), ST23 (n = 2), ST86 (n = 2), and ST420 (n = 2) were prevalent in healthy individuals.
We identified ten distinct K capsular types and seven O antigens. The most common K loci were K1 (51%) and K2 (16%), followed by K20 (10.6%), K5 (8%), K113 (5.3%), and another five K types (9.1%). While K1 was confined to ST23, K2 and K5 were associated with ST86, ST65, and ST25, and K20 was identified in ST420 and ST268. O1 antigen was predominantly found in 87 isolates (77%), spanning various STs: ST23 (n = 50), ST86 (n = 10), ST65 (n = 8), ST420 (n = 5), ST25 (n = 4). This was followed by the O2 antigen identified in 21 isolates (18.6%) across eight different STs. The remaining O antigens (O3, O4, O5) accounted for 4.4% of isolates (Table 1).
Phylogenetic relatedness and genetic profile of hypervirulent Klebsiella pneumoniae isolates
We reconstructed a phylogenetic tree encompassing 111 hvKp isolates (98 from BSIs and 13 from human carriers) and mapped it against the acquired AMR genes, key virulence factors, and plasmid replicon types (Fig . 1). Overall, each ST formed a discernable phylogenetic cluster, except for ST29 and its single-locus variant ST714, which were inferred as a single cluster. Each phylogenetic cluster encompassed isolates spanning multiple years, suggesting the endemic circulation of various hvKp lineages in Vietnam. Within the phylogenetic tree, hvKp isolates from BSIs and human carriers of the same ST clustered together, demonstrating a shared virulence profile and limited genetic variations (mean pairwise SNP distances ranging from 38 SNPs to 290 SNPs). We further investigated the phylogenetic structure of ST23 isolates in a global context to depict the transmission dynamics of this dominant clone. Our analysis demonstrated that ST23 isolates from Vietnam did not originate from a single common ancestor; instead, they were likely associated with multiple strain introductions. Four phylogenetic clusters (C1–C4) were designated, with the major C4 cluster (virulence gene profile: iuc1-iro1-clb2-ybt1-rmp1) spanning from 2011 to 2020, indicating sustained endemic transmission. The C1 and C4 clusters primarily consisted of ST23 isolates from Vietnam, with sporadic isolates from Singapore, Norway, France, and Australia (Fig. S3).
The phylogenetic tree is mid-point rooted. The sequence types (STs) are represented by the colored circles at the terminal nodes of the tree. The years of isolation are shown on the tip labels. The tree scale bar represents the number of SNPs per site. The black stars highlight bootstrap support values ≥80 % on internal nodes, with larger stars indicating higher bootstrap values. The heat maps display the source of isolation (bloodstream infections, human carriage), followed by the presence (in color) or absence (in gray) of different alleles of virulence genes, resistance genes, and plasmid replicon types. The hvKp isolates exhibiting convergent phenotype (AMR and virulence) or carrying the iuc3 plasmids are indicated at the tip labels. Source data for the heat maps are provided as a Source Data file.
Genomic analysis revealed that all isolates possessed at least two plasmid-encoded siderophores biosynthesis genes: iuc (aerobactin) and iro (salmochelin). Additionally, 99 isolates (89%) harbored the hypermucoid regulator rmpA/rmpA2 gene and 95 isolates (86%) contained the chromosomal yersiniabactin locus (ybt). More than half of the isolates (53%) also carried the clb locus encoding colibactin (Table 1). The distribution of key virulence factors displayed distinct patterns both within and between STs. The majority of ST23 isolates (48/59, 81%) displayed the iro1-iuc1-rmpA1(+/-)-ybt1-clb2 virulence pattern, while the remaining isolates (11/59, 19%) exhibited the iro1-iuc1-rmpA1-ybt8 pattern (Fig. 1). Alternatively, seven out of 11 ST86 isolates contained the yersiniabactin-encoding ybt10/22/9 locus but lacked the colibactin-encoding locus; whereas, five out of nine ST65 isolates carried both the yersiniabactin and colibactin-encoding loci (ybt17-clb3). Most hvKp strains (108/111) harbored the iro1-iuc1 virulence profile, with the exception of three isolates carrying the plasmid-encoded iuc3 and the chromosomal ICEKp1 (2 ST25) and IS100 (1 ST828) iro3.
Among 111 hvKp isolates, 108 contained a closely related IncFIBK+/−IncHI1B virulence plasmid, exhibiting significant genetic similarity (73.8–99.9% coverage) to the canonical virulence plasmid pLVPK in hvKp. Conversely, three isolates harbored an IncFIBK-like plasmid with low coverage (ranging from 38.1 to 43.3%). The plasmid profiling and long-read Nanopore sequencing data confirmed the presence of additional plasmids in hvKp isolates, including IncFII+/-IncFIA (10%), IncR (2.7%), IncA/C (0.9%) plasmid and IncFIB phagemid (13.5%). While the majority of hvKp isolates displayed pan-susceptibility to the tested antibiotics and lacked AMR genes, 14 isolates (12.6%) were identified to carry at least one AMR gene. These isolates were obtained from both BSIs and human carriers, encompassing different STs (Table 2). Two of them harbored an AMR gene on the chromosome, including one ST23 isolate (ISEc9-blaCTX-M-14) and one ST86 isolate (ISKpn74-sul1). Five isolates carried a similar array of AMR genes (qnrS1-sul-tetA-dfrA1(+/−)-blaLAP-2(+/−)) on an IncFII+/−IncFIA plasmid. These IncFII plasmids showed genetic variability and were distributed among different STs: ST268 (2), ST23 (1), ST828 (1), and ST25 (1). One ST65 isolate acquired an IncA/C plasmid of 246 kb, carrying 14 different AMR genes (aadA24-aadB-arr2-blaCTX-M-cmlA5-dfrA23-blaLAP-2-blaOXA-10-qnrS1-floR-strA-sul2-tetA-blaVEB-1). Two isolates carried an IncR plasmid with different AMR genes: qnrS1-blaLAP-2-aac3-IId (ST23 isolate) and dfrA1-qnrB4-sul1 (ST25 isolate). The AMR genes were also located in a small col plasmid of an ST25 isolate, IncFIBK-like plasmids of two other ST25 isolates, and two plasmids with unknown replicon types in two ST23 isolates.
Genetic structure of iuc3-carrying virulence plasmids
We sought to determine the genetic structure of the three IncFIBK-like plasmids (designated as pQY1, pQY2, and pQY3) identified in two ST25 isolates and one ST828 isolate. Plasmid nanopore sequencing data revealed that these are multi-replicon IncFIBK-IncFII plasmids, carrying gene features from both the IncFIBK virulence plasmid and IncFII plasmid. The IncFIBK virulence region encompassed the aerobactin biosynthesis (iucABCD-iutA) and heavy metal resistance operons (copper: pcoABCDRS, silver: silABCEFPRS, iron: fecABCDERI, and arsenic: arsABDR). Alternatively, the IncFII region of the iuc3-carrying plasmids contained tra operon encoding for type IV conjugation machinery, with or without AMR gene cassettes (Fig. 2).
The iuc3 virulence plasmid pQY3 is represented by a yellow circle. The GC content and GC skew of the pQY3 plasmid are displayed in the two innermost rings. The gene annotations are displayed as arrows in the outermost ring. The conjugation module is indicated in orange, while the virulence genes are highlighted in green.
The three iuc3-carrying plasmids shared a conserved region of about 139.8 kb (pairwise nucleotide identity >99%) while exhibiting differences in size and gene content. The pQY1 plasmid was 240.6 kb in length, carrying a set of AMR genes, tetA-sul2-strAB-dfrA14-AadA1-blaOXA-10-floR-cmlA-arr. The pQY2 was 194 kb in length and did not carry any AMR genes. The pQY3 was 198 kb and carried tetA-sul2-strAB-floR-aph6-blaLAP-2 (Table 3). Furthermore, plasmid comparative analysis with five publicly available iuc3-carrying plasmids derived from hvKp isolates in Thailand, Laos PDR, and the USA revealed a high degree of genetic similarity (identity: 99.7–99.9%, coverage: 60–97%) (Fig. S4). The iuc3 plasmid from Thailand (CP041094) originated from a pig, while the two iuc3 plasmids from the US (CHS48 and CHS43) were found in two hvKp ST25 isolates. Notably, one iuc3 plasmid from a previously published hvKp strain in Vietnam (p130411) was almost identical to the pQY1 plasmid (identity: 99.9%, coverage: 100%).
Horizontal transfer mechanism of the non-conjugative virulence plasmid
Although the IncFIBK virulence plasmid is not self-transferable due to the lack of T4SS-encoding tra gene cluster, it was found across various sequence types of K. pneumoniae as well as K. quasipneumoniae isolates in this study. We aimed to gain insights into the transmission of the virulence plasmid by examining the congruence between the chromosomal and plasmid phylogenies and conducting a series of conjugation experiments. Apart from the three iuc3-encoding IncFIBK-IncFII virulence plasmids, all 108 iuc1-encoding IncFIBK+/−IncHIB virulence plasmids displayed a high synteny with the reference pLVPK plasmid, with the mapping coverage ranging from 73.8 to 99.9% (Fig. 3). The resulting plasmid phylogeny showed distinct clusters with strong bootstrap support values. We observed several incongruences between the chromosomal and plasmid phylogenies; for instance, ST660 and ST65 formed separate clusters on the chromosomal phylogeny but grouped together on the plasmid phylogeny. Similar discrepancies were identified between ST25 and ST420, as well as between ST218, ST29, ST714, and ST412 (Figs. 1, 3). These findings provided a strong indication that the virulence plasmid has been horizontally transferred across genotypes and Klebsiella species. In contrast, for ST23 and ST86, congruence in the chromosomal and plasmid phylogenetic structures of isolates suggests the virulence plasmid has co-evolved with these host bacteria for some time.
The plasmid phylogenetic tree is mid-point rooted. The sequence types (STs) of isolates are represented by the colored circles at the terminal nodes of the tree. The virulence plasmids that are successfully transferred into the Klebsiella pneumoniae recipient strain via conjugation are marked in red at the tip labels. The blue shaded boxes highlight STs that are associated with incongruences between the chromosomal and plasmid phylogenies. The tree scale bar represents the number of SNPs per site. The numbers on the branches indicate nodes with bootstrap support values ≥ 80 %. The first column shows the percentage of mapping coverage against the reference virulence plasmid. The second column indicates the year of isolation, and the third columns show the source of isolation (light green: blood, dark green: stool). Source data are provided as a Source Data file.
Next, we investigated the horizontal transfer ability of the virulence plasmid between hvKp and cKp via conjugation experiments. Thirty-three hvKp donor strains were chosen to represent all different sequence types and the diversity of non-virulence plasmids. Our conjugation experiments demonstrated the 3 iuc3-encoding IncFIBK:IncF plasmids with intact T4SS were successfully transferred to the recipient Kp strain. Out of 30 hvKp isolates carrying the iuc1-encoding IncFIBK virulence plasmid, five isolates were capable of transferring the virulence plasmid to the recipient Kp strain (Fig. 4). Plasmid Nanopore sequencing confirmed that these isolates contained a conjugative helper plasmid, including an IncFII plasmid with sizes ranging 74 kb to 176 kb (3 ST23 isolates), an IncFII:IncFIA:IncN plasmid of 122 kb (one ST86 isolate) and an IncA/C:IncN plasmid of 246 kb (one ST65 isolate). Furthermore, the predicted nic site (10 bp) in the oriT region of these conjugative helper plasmids exhibited significant sequence similarity with the nic site of the IncFIBK virulence plasmid (Table 4 and Fig. 5). The nic site is a cleaving site of relaxase, a DNA transferase that catalyzes a strand- and sequence-specific cleavage and initiates the plasmid conjugal transfer. This finding signifies that when hvKp isolates acquire a conjugative helper plasmid with a shared nic site in the oriT, the non-conjugative virulence plasmid within these hvKp isolates can be effectively transferred to other classical Kp strains. This notion was further substantiated by the observation that the virulence plasmid remained non-transferable when the hvKp isolates (n = 22) lacked a conjugative helper plasmid or when the hvKp isolates (n = 3) harbored a conjugative helper plasmid with dissimilar nic site regions (Table 4 and Fig. 5). Moreover, by comparing the plasmid profiles of donor, recipient and the randomly selected transconjugants, we found that the virulence plasmid was either transferred alone, or co-transferred with another plasmid (conjugative helper plasmid/phage-like plasmid), or formed a hybrid plasmid with the conjugative helper plasmid (Fig. 4).
This figure illustrates the plasmid profiles of the selective donors (D1–D8), recipient (R), and transconjugants (Transconjugants D1–D8), as well as the confirmation results of successful plasmid transfer by qPCR (iuc gene) and selective media. The arrows are colored according to different plasmids, including white: phagemid (recipient and transconjugants); yellow: helper plasmid, red: iuc1 virulence plasmid, orange: iuc3 virulence plasmid, green: phagemid (donors); and blue: virulence plasmids in transconjugants that differ in sizes compared to the original virulence plasmids in donors. Selective markers: Tellurite resistance (Telr), Tetracycline resistance (Tetr), and Ciprofloxacin resistance (Cipr) are represented with (+/−) to denote the growth or no growth of bacterial strains on selective media. The conjugation experiments were repeated twice with similar results. Source data are provided as a Source Data file.
The inverted-repeat sequences close to the nic site of the plasmids are shown by horizontal arrows and block text highlighted in orange. The nic site are indicated by nucleotides highlighted in different colors, with red representing nucleotide for “A,” blue for “T,” yellow for “G,” and green for “C”.
Discussion
Hypervirulent Klebsiella pneumoniae have emerged as a significant cause of severe community-acquired infections in Asia, with evidence of its global spread20,37,38. Despite its significance, the epidemiological and genetic landscape of hvKp infections in LMICs have been poorly characterized due to the lack of reliable detection methods and comprehensive molecular investigations. This study offers a multiplex real-time PCR assay with melting curve analysis for detecting hvKp isolates based on their typical gene markers, given the absence of a consensus definition for clinical infection. Furthermore, the hvKp isolates identified by PCR showed 100% sensitivity when compared to WGS. This assay can be readily employed in LMICs to identify hvKp infections for rigorous clinical examinations and effective treatment, as well as epidemiological investigations. Given the scarcity of data regarding hvKp infections in Vietnam, our work was first to describe the prevalence, population structure, and virulence plasmid transfer of hvKp in this setting. Akin to previous studies, the preponderance of hvKp infections in Vietnam was attributed to the three prevailing lineages of ST23-K1, ST86-K2, and ST65-K2. Nevertheless, the population structure of hvKp isolates displayed significant diversity, with a multitude of distinct lineages co-circulating throughout the study period. Moreover, hypervirulent Klebsiella infections were not exclusive to K. pneumoniae but were also attributed to two K. quasipneumoniae subsp. similipneumoniae isolates (ST367 and ST816), all of which shared a common virulence plasmid. These findings raise significant concerns regarding the emergence and international spread of novel non-K1 and non-K2 hypervirulent Klebsiella strains, which may exhibit distinct propensities for acquiring antimicrobial resistance genes or differing capabilities in pathogenesis and transmission39,40. The heterogeneous makeup of surface structures, including K capsular types and O antigens, within the hvKp strains also presents a substantial challenge in the development of preventive measures, such as vaccines and innovative therapeutics, targeting these surface antigens. Alternatively, protein-based approaches should be considered for vaccine development as they have the potential to offer broader coverage across various Klebsiella strains.
Previous studies have shown little evidence of K. pneumoniae transmission between humans and non-human sources41,42,43. Meanwhile, hvKp have been found in the gastrointestinal tracts of both healthy individuals and patients19,44. Here, we sought to understand the circulation of hvKp strains in healthy individuals and the extent of genetic sharing with disease-causing organisms. In healthy children, while classical K. pneumoniae strains were identified from rectal swabs, no hvKp strains were detected. We speculate this observation may stem from the frequent exposure of children in our setting to antibiotics45,46, coupled with the prevailing pan-susceptibility of most hvKp strains to these antimicrobial agents. In healthy adults, we found a prevalence of 3.7% of hvKp isolates, exhibiting diverse but significant overlap in sequence types with those found in humans with bloodstream infections. The fact that the carriage and infecting isolates share the same STs and exhibit limited genetic distances strongly suggests that the human gastrointestinal tract constitutes an important reservoir for the transmission of hvKp strains. The biological and epidemiological factors governing the establishment of gut colonization in susceptible hosts and the community transmission of hvKp remain poorly understood. We need to address these knowledge gaps to devise appropriate preventative measures to prevent/reduce risks of gut colonization and infection development in vulnerable populations. Furthermore, although hvKp infections have been rarely reported in children, it is essential to note that such infections can lead to severe illnesses with high mortality rates47. Given that our study primarily centered on bloodstream infections in adult patients, we strongly advocate for further investigations into the community transmission and incidence rates of various hvKp infections across diverse populations (adults vs. children) and settings (community vs. hospital environments).
Although most hvKp isolates were susceptible to antibiotics, the convergence of AMR and virulence genes was observed in 12.6% of isolates. The convergence events were also observed across different STs. The occurrence of MDR-hvKp strains is very concerning as these organisms may gain a competitive advantage and spread in settings with strong antibiotic selection pressures. This trend needs to be closely monitored as such emergence appears unavoidable and could result in substantial global health threats38,48,49. A significant finding from our research lies in the identification of specific conjugative helper plasmids, not only contributing to the MDR phenotype but also offering a means for the horizontal transfer of the non-conjugative virulence plasmid between K. pneumoniae isolates. These helper plasmids shared a homologous oriT region with the virulence plasmid and predominantly belonged to the IncFII group, which is common in human commensal Enterobacteriaceae50,51. Similar findings were reported recently, showing that a blaKPC-positive IncFII plasmid sharing the nic site in the oriT with the virulence plasmid can help to mobilize the virulence plasmid from hvKp to cKp strains49. We surmise that hvKp continues to evolve antibiotic resistance while spreading the virulence phenotype to other niche-sharing classical Kp strains through the acquisition of conjugative helper plasmids. Strikingly, we observed that during the conjugation via the helper plasmids, plasmid profiling showed the differing sizes of virulence plasmids in the transconjugants. This is probably the result of recombination or co-integration between the virulence plasmid and the helper plasmid. Such recombinant/co-integrative events were observed previously, resulting in the formation of hybrid plasmids52,53. These hybrid plasmids can carry a combination of virulence factors, AMR genes, and T4SS, which may facilitate its further spread while reducing the fitness cost associated with the virulence plasmid6. The high prevalence of MDR bacteria in settings like Vietnam may act as a significant gene pool, contributing to the emergence of novel hvKp and MDR-hvKp strains.
In this study, we also found three iuc3-carrying IncFII:IncFIBK virulence plasmids belonging to ST25 and ST828. These plasmids were self-transferable and shared little coverage with the iuc1-carrying IncFIBK virulence plasmid identified in most hvKp isolates. Apart from the aerobactin biosynthesis gene cluster, the iuc3-carrying virulence plasmids commonly contained several AMR gene cassettes and an intact T4SS but lacked rmpA and iroABCD genes. The majority of previously characterized hvKp isolates carrying the iuc3 IncFII:IncFIBK virulence plasmids was traced back to animal sources, thus hinting at a zoonotic reservoir for this plasmid54,55. Notably, ST25 over-represented the hvKp isolates that had been shown to harbor the iuc3-carrying virulence plasmids56,57, suggesting that ST25 is a major vehicle for the transmission of the iuc3 IncFII:IncFIBK virulence plasmid. Here, we found an identical iuc3 IncFII:IncFIBK virulence plasmid in two different STs (ST25 and ST828) of hvKp strains in Vietnam. This finding suggests that once acquired, the iuc3 plasmids can spread further among Kp strains. Furthermore, we found that the hvKp strains carrying the iuc3 virulence plasmids were not hypermucoid due to the absence of the rmpA gene or carried the truncated rmpA, and thus the string test might fail to detect these organisms as hvKp. The lack of hypermucoid phenotype may also alleviate constraints on horizontal gene transfer, promoting DNA uptake in these hvKp strains58.
In conclusion, we present the first genomic description of hypervirulent K. pneumoniae from bloodstream infections and healthy carriage in Vietnam. Our findings reveal a genetically diverse population of seventeen hvKp genotypes, dominated by ST23, ST86, and ST65. We uncovered the emergence of antimicrobial resistance across different hvKp genotypes largely via the acquisition of AMR plasmids/genes. Strikingly, we found that the acquisition of AMR conjugative helper plasmids sharing the common oriT region with the virulence plasmid facilitated the horizontal transfer of the virulence plasmid between K. pneumoniae strains. Carriage and infecting organisms clustered phylogenetically, indicating human intestinal carriage is likely to play a major role in the transmission of hvKp in the community. Enhanced community surveillance to understand factors driving the establishment of intestinal carriage and facilitating the transmission of hypervirulent K. pneumoniae is instrumental to identify appropriate preventative and control measures. Furthermore, molecular testing should be applied in clinical settings to rapidly identify infections caused by hypervirulent K. pneumoniae for thorough evaluation and effective management.
Methods
Bacterial and data collection
A total of 700 Kp isolates were yielded from a retrospective study of bloodstream infections at the Hospital for Tropical Diseases (HTD) between 2010 and 2020. These isolates were identified from routine blood cultures and stored at the Microbiology Department. For blood culture, an aliquot of 5–8 mL (for adults) or 2–5 mL (for children) of venous blood were inoculated into BACTEC plus aerobic bottles. Inoculated bottles were incubated at 37 °C in a BACTEC 9050 automated analyser for up to 5 days and subcultured on fresh sheep blood agar, chocolate agar, and MacConkey agar when the machine indicated a positive signal. Organisms were identified by standard methods, including API20E and API20EN identification kits (Bio-Mérieux) or MALDI-TOF mass spectrometry (Bruker). Antimicrobial susceptibility testing of Kp was performed by VITEK automated machine and, when required, by minimum inhibitory concentrations (MICs) by E-test or the disk diffusion method using the guidelines established by the Clinical and Laboratory Standards Institute (CLSI). The study has been granted ethics approval from HTD’s Ethic Committee (approval number: 1711/QĐ-BVBNĐ).
The rectal swabs/stools were sourced from two cohort studies. The first cohort involved 500 healthy children in Ho Chi Minh City, Vietnam, between 2014 and 2016 to longitudinally assess the incidence rate of dysentery59. Rectal swabs/stools from the final visit of each child were used for this study. Ethical approval for this study was granted by the Oxford University Tropical Research Ethics Committee (OxTREC No. 1058-13). The second cohort included healthy farmers in Dong Thap province (125 km WSW of HCMC) to assess the prevalence of AMR carriage before and after an intervention consisting of removing antimicrobial use in chicken farms60. A total of 350 rectal swabs were collected from farm households during 2020. All the samples originated from healthy adults. The study received ethics approval from the Oxford University Tropical Research Ethics Committee (OxTREC No. 503-20).
Identification of hypervirulent K. pneumoniae isolates
To identify hvKp isolates from 700 Kp BSI isolates, a colony multiplex PCR assay with melting curve analysis was developed to target four virulence genes: iucA, iroB, rmpA, and rmpA2. Each target was associated with a specific melting temperature (Fig. S2 and Table S1). PCR amplifications were run on a LightCycler 480II (Roche applied sciences, UK) with the following thermal conditions: initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation, 95 °C, for 10 s; annealing, 60 °C for 30 s; extension, 72 °C for 30 s. After PCR amplification, the melting curve analysis (MCA) was conducted in the same thermocycler at 65 °C to 95 °C and cooling cycle at 37 °C for 30 s. Fluorescence was continuously measured, and the melting temperature (Tm) was calculated by plotting the negative derivative of fluorescence over temperature versus temperature (−dF/dT versus T). All Kp isolates harboring the iucA gene with/without other virulence genes were classified as hvKp20. For rectal swabs/stools, samples were initially plated on MacConkey agar containing 3 mg/L potassium tellurite to distinguish hypervirulent Klebsiella colonies from other bacteria. K. pneumoniae colonies were later confirmed by MALDI-TOF and underwent colony multiplex PCR for the detection of hvKp isolates. All the hvKp isolates were subjected to the string test and WGS.
Whole genome sequencing
Genomic DNA was extracted using the Wizard Genomic DNA Extraction Kit (Promega, US) following the manufacturer’s protocol. The Nextera XT Library Preparation Kit was used to prepare DNA sequencing libraries. WGS was performed on an Illumina MiSeq platform to generate 250 bp paired-end reads.
Plasmid DNA extraction was performed using a QIAGEN Plasmid MIDI kit (Qiagen) following the manufacturer’s instructions. Nanopore sequencing libraries were generated from 200 ng of plasmid DNA using the SQK-RBK004 Rapid Barcoding Kit and then sequenced using the FLOW-FLG001 flow cell R9.4.1 (Oxford Nanopore Technologies). Nanopore sequencing was performed using MinKNOW software v1.13 to produce raw sequences in Fast5 format, which were subsequently subjected to base calling and de-multiplexing with Guppy v. 4.061.
Genomic analysis
Raw Illumina reads were subjected to quality control using FastQC v0.11.562. De novo assembly of Illumina reads, and hybrid assembly of Illumina and Nanopore reads were performed using Unicycler v.0.4.8b with default parameters63. Assembled sequences were annotated using Bakta v.1.0.464. Kleborate v2.2.065 was used to determine the multi-locus sequence types (MLSTs) with BIGSdb database66, acquired AMR genes with CARD database67, virulence genes with VFDB database68, and K capsular types and O antigens with Kaptive database v.2.0.069. SRST2 v0.2.070 was used to identify plasmid replicon types with PlasmidFinder database71 and Abricate v1.0.0 (https://github.com/tseemann/abricate) was used to identify the plasmid origin of transfer site (oriT) using oriTDB database developed by Li and collaborators72. For plasmids that failed to identify oriT within the oriTDB database, we searched for putative uncharacterized oriT regions. Briefly, oriTfinder (http://bioinfo-mml.sjtu.edu.cn/oriTfinder) was used to identify relaxase-encoding regions within these plasmids. Subsequently, RepeatFinder in Geneious v2022.1.1 (https://www.geneious.com/plugins/repeat-finder/) was used to identify the putative inverted repeats (IRs) neighboring the relaxase-encoding region, and MEME-MAST73 was used to identify the conserved nick regions. The comparative genomics of iuc3-carrying plasmids were performed using Blast Ring Image Generator (BRIG)74.
SNP calling and phylogenetic analysis
Raw Illumina reads of all hvKp isolates were mapped against the reference genome of hvKp strain NTUH-K2044 (accession number: AP006725.1) using the RedDog pipeline v1.10b (https://github.com/katholt/RedDog). Briefly, RedDog used Bowtie2 v2.2.3 to map all raw reads to the reference sequence, and high-quality SNPs with Phred quality score ≥30 were extracted using SAMtools v1.1.3.175. SNPs were subsequently filtered to remove those with fewer than five supporting reads or with >2.5 times the mean read depth (representing putative repeated sequences), or with ambiguous base calls. Core SNPs found in greater than 95% of isolates were extracted, resulting in a final alignment of 109,991 SNPs. Phylogenetic reconstruction was performed with IQ-TREE v.2.076, using the maximum likelihood (ML) method and the best-fit model GTR + ASC + G4, as determined by the software. Support for the ML tree was assessed through 1000 bootstrap replicates. The pairwise genetic distances (measured as a difference in the number of SNPs) between acute and carriage isolates within the same sequence type (STs) were calculated from the above SNP alignment using ape v4.1 and adegenet v2.0.1 packages in R v3.3.2.
To examine the phylogeny of ST23 hvKp isolates on a global scale, we integrated 59 ST23 genomes obtained from this study with an additional 176 ST23 genomes sourced from blood isolates available at the time of access from the Enterobase (https://enterobase.warwick.ac.uk/) (Supplementary Data 1). Raw reads were downloaded from the Enterobase using the ENAdatabase-Downloader script (https://github.com/chauvinhtth13/ENAdatabase-Downloader). Mapping analysis and SNP calling were performed as described above, using the same reference genome of hvKp strain NTUH-K2044 (ST23) and the RedDog pipeline v.1b.11. The snpTable2GenomeAlignment.py script from RedDog was employed to infer a pseudo-genome alignment, which was subsequently subjected to Gubbins v.2.34 to identify recombinant regions77. SNP-sites were used to extract SNPs from the recombination-free multi-FASTA alignment, resulting in an alignment of 8201 SNPs78. IQ-TREE was employed to reconstruct an ML tree from the SNP alignment with the best-fit model TVM + F + ASC + G4 and bootstrap analysis with 1000 replicates.
To explore the transmission dynamics of the virulence plasmid IncFIBK, we reconstructed a mapping-based plasmid phylogeny and assessed its congruence with the chromosomal phylogeny. Plasmid phylogenetic analysis employed the same mapping and SNP calling methods as described for the chromosomal phylogeny. The virulence plasmid pLVPK (accession number AY378100.1) served as the reference plasmid genome for this analysis. The final alignment, comprising 1568 SNPs, was utilized to construct a phylogeny using IQ-TREE, employing the best-fit model TVM + G4.
Plasmid profiling
Crude plasmid extraction was performed for all hvKp isolates, using a modified Kado and Liu method79. The resulting plasmid DNA was subjected to electrophoresis in 0.7% agarose gel at 90 Voltage for 3 h, stained with Nancy-520 (2.5 mg/mL), and photographed. E. coli strain 39R861 carrying three plasmids with known sizes (36 kb, 63 kb, and 147 kb) was used as a marker80.
Transferability of the virulence plasmid
To investigate the transferability of the virulence plasmid, a conjugation experiment was performed between each of the 33 ciprofloxacin-susceptible hvKp strains (donor) and a ciprofloxacin-resistant clinical Kp ST15 strain (recipient). The selection of 33 donors aimed to encompass various sequence types (STs), both with and without the presence of additional non-virulence plasmids. Among 33 donors, 30 strains were resistant to tellurite (MIC >32 mg/L) and three were susceptible to tellurite (MIC <1 mg/L) but resistant to tetracycline (MIC >32 mg/L). The recipient Kp strain was identified from a patient with bloodstream infection in Vietnam, carrying a small Col plasmid of 2328 bp and a non-AMR phagemid of 109,938 bp. This strain exhibited chromosomal ciprofloxacin resistance (MIC = 16 mg/L), intermediate tetracycline resistance (MIC = 8 mg/L), and susceptibility to tellurite (MIC <1 mg/L). The donor and recipient were combined in a 1:1 ratio in LB broth and conjugated at 37 °C for 18 h. The transconjugants were selected on Mueller Hinton agar supplemented with ciprofloxacin (6 mg/L) plus tellurite (8 mg/L) or ciprofloxacin (6 mg/L) plus tetracycline (16 mg/L). Successful transfer of the virulence plasmid was verified by comparing the plasmid profiles of the donor, recipient, and transconjugants. Furthermore, the presence of plasmid-mediated virulence genes in the transconjugants was confirmed by qPCR.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data used in this work are publicly available in the European Nucleotide Archive (ENA) under project numbers PRJEB64408 and PRJEB26814 (https://www.ebi.ac.uk/ena/browser/view/ PRJEB26814)37. Source data are provided with the paper. Source data are provided with this paper.
Code availability
All software utilized for data analysis is open source.
References
Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Lin, Y.-C. et al. Successful treatment of hypervirulent Klebsiella pneumoniae bacteremia with combination carbapenem and rifampicin. IDCases 26, e01276 (2021).
Taraghian, A., Nasr Esfahani, B., Moghim, S. & Fazeli, H. Characterization of hypervirulent extended-spectrum β-lactamase-producing Klebsiella pneumoniae among urinary tract infections: the first report from Iran. Infect. Drug Resist. ume 13, 3103–3111 (2020).
Tzouvelekis, L. S. et al. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob. Agents Chemother. 57, 5144–5146 (2013).
Jia, H. et al. Outbreak of multidrug-resistant OXA-232-producing ST15 Klebsiella pneumoniae in a teaching hospital in Wenzhou, China. Infect. Drug Resist. ume 14, 4395–4407 (2021).
Wang, Q. et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin. Infect. Dis. 67, S196–S205 (2018).
Liu, Y.-C. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913 (1986).
Shon, A. S., Bajwa, R. P. S. & Russo, T. A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence 4, 107–118 (2013).
Zhao, B. et al. Pyogenic liver abscess and endogenous endophthalmitis due to K64-ST1764 hypervirulent Klebsiella pneumoniae: a case report. Infect. Drug Resist. ume 14, 71–77 (2021).
Ng, D. & Frazee, B. Necrotizing fasciitis caused by hypermucoviscous Klebsiella pneumoniae in a Filipino female in North America. West. J. Emerg. Med. 16, 165–168 (2015).
Remya, P., Shanthi, M. & Sekar, U. Occurrence and characterization of hyperviscous K1 and K2 serotype in Klebsiella pneumoniae. J. Lab Physicians 10, 283–288 (2018).
Surgers, L., Boyd, A., Girard, P.-M., Arlet, G. & Decré, D. ESBL-producing strain of hypervirulent Klebsiella pneumoniae K2, France. Emerg. Infect. Dis. 22, 1687–1688 (2016).
Lam, M. M. C. et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 9, 2703 (2018).
Andrade, L. N. et al. Expansion and evolution of a virulent, extensively drug-resistant (Polymyxin B-Resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J. Clin. Microbiol. 52, 2530–2535 (2014).
Lam, M. M. C. et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 10, 77 (2018).
Siu, L. K., Yeh, K.-M., Lin, J.-C., Fung, C.-P. & Chang, F.-Y. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12, 881–887 (2012).
Leigh, J. & Naghdi, R. Cryptogenic hypervirulent Klebsiella pneumoniae pyogenic liver abscess: a case report. Am. J. Case Rep. 24, e939322 (2023).
Hallal Ferreira Raro, O., Nordmann, P., Dominguez Pino, M., Findlay, J. & Poirel, L. Emergence of carbapenemase-producing hypervirulent Klebsiella pneumoniae in Switzerland. Antimicrob. Agents Chemother. https://doi.org/10.1128/aac.01424-22 (2023)
Yang, J. et al. The human gut serves as a reservoir of hypervirulent Klebsiella pneumoniae. Gut Microbes 14, 2114739 (2022).
Russo, T. A. & Marr, C. M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001–e00019 (2019).
Chung, D. R. et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 31, 481–486 (2012).
Fung, C.-P. et al. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg. Infect. Dis. 18, 1322–1325 (2012).
Zhang, R. et al. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob. Agents Chemother. 60, 709–711 (2016).
Fu, Y., Xu, M., Liu, Y., Li, A. & Zhou, J. Virulence and genomic features of a bla CTX-M-3 and bla CTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65. Infect. Drug Resist. 12, 145–159 (2019).
Li, P. et al. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: an epidemiological, genomic and functional study. Virulence 12, 377–388 (2021).
Nagaraj, G. et al. High-resolution genomic profiling of carbapenem-resistant Klebsiella pneumoniae isolates: a multicentric retrospective Indian study. Clin. Infect. Dis. 73, S300–S307 (2021).
Lan, P., Jiang, Y., Zhou, J. & Yu, Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. https://doi.org/10.1016/j.jgar.2021.02.020 (2021).
Chen, Y.-T. et al. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337, 189–198 (2004).
Lam, M. M. C. et al. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 74, 1218–1222 (2019).
Gu, D. et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46 (2018).
Tian, D. et al. Genetic diversity and evolution of the virulence plasmids encoding aerobactin and salmochelin in Klebsiella pneumoniae. Virulence 12, 1323–1333 (2021).
Hagiya, H., Watanabe, N., Maki, M., Murase, T. & Otsuka, F. Clinical utility of string test as a screening method for hypermucoviscosity-phenotype Klebsiella pneumoniae. Acute Med. Surg. 1, 245–246 (2014).
Neumann, B. et al. Detection and characterization of putative hypervirulent Klebsiella pneumoniae isolates in microbiological diagnostics. Sci. Rep. 13, 19025 (2023).
Bulger, J., MacDonald, U., Olson, R., Beanan, J. & Russo, T. A. Metabolite transporter PEG344 Is required for full virulence of hypervirulent Klebsiella pneumoniae strain hvKP1 after pulmonary but not subcutaneous challenge. Infect. Immun. 85, e00093–17 (2017).
Russo, T. A. et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 56, e00776–18 (2018).
Vu, T. V. D. et al. Antimicrobial susceptibility testing results from 13 hospitals in Viet Nam: VINARES 2016–2017. Antimicrob. Resist. Infect. Control 10, 78 (2021).
Wyres, K. L. et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 12, 11 (2020).
Wu, Y. et al. Global evolution and geographic diversity of hypervirulent carbapenem-resistant Klebsiella pneumoniae. Lancet Infect. Dis. 22, 761–762 (2022).
Wu, C. et al. Co-occurrence of OXA-232, RmtF-encoding plasmids, and pLVPK-like virulence plasmid contributed to the generation of ST15-KL112 hypervirulent multidrug-resistant Klebsiella pneumoniae. Front. Microbiol. 14, 1133590 (2023).
Cai, Z. et al. Clinical and molecular analysis of ST11-K47 carbapenem-resistant hypervirulent Klebsiella pneumoniae: a strain causing liver abscess. Pathogens 11, 657 (2022).
Calland, J. K. et al. Population structure and antimicrobial resistance among Klebsiella isolates sampled from human, animal, and environmental sources in Ghana: a cross-sectional genomic One Health study. Lancet Microbe 4, e943–e952 (2023).
Thorpe, H. A. et al. A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings. Nat. Microbiol. 7, 2054–2067 (2022).
Dereeper, A. et al. Limited transmission of Klebsiella pneumoniae among humans, animals, and the environment in a Caribbean Island, Guadeloupe (French West Indies). Microbiol. Spectr. 10, e0124222 (2022).
Siu, L. K. et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J. Clin. Microbiol. 49, 3761–3765 (2011).
Minh, N. N. Q. et al. Antibiotic use and prescription and its effects on Enterobacteriaceae in the gut in children with mild respiratory infections in Ho Chi Minh City, Vietnam. A prospective observational outpatient study. PLoS ONE 15, e0241760 (2020).
Nguyen, T. T. P. et al. A national survey of dispensing practice and customer knowledge on antibiotic use in Vietnam and the implications. Antibiotics 11, 1091 (2022).
Du, Q. et al. Nosocomial dissemination of hypervirulent Klebsiella pneumoniae with high-risk clones among children in Shanghai. Front. Cell Infect. Microbiol. 12, 984180 (2022).
Chen, L. & Kreiswirth, B. N. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect. Dis. 18, 2–3 (2018).
Tian, D. et al. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg. Microbes Infect. 11, 1936–1949 (2022).
Xu, Y. et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13, 119 (2021).
Tian, C. et al. Emergence of IS26-mediated pLVPK-like virulence and NDM-1 conjugative fusion plasmid in hypervirulent carbapenem-resistant Klebsiella pneumoniae. Infect. Genet. Evol. 113, 105471 (2023).
Shankar, C. et al. Hybrid plasmids encoding antimicrobial resistance and virulence traits among hypervirulent Klebsiella pneumoniae ST2096 in India. Front. Cell Infect. Microbiol. 12, 875116 (2022).
Xie, M. et al. A hybrid plasmid formed by recombination of a virulence plasmid and a resistance plasmid in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 23, 466–470 (2020).
Kaspersen, H. et al. Highly conserved composite transposon harbouring aerobactin iuc3 in Klebsiella pneumoniae from pigs. Micro. Genom. 9, 1–12 (2023).
Leangapichart, T. et al. Characterization of Klebsiella pneumoniae complex isolates from pigs and humans in farms in Thailand: population genomic structure, antibiotic resistance and virulence genes. J. Antimicrob. Chemother. 76, 2012–2016 (2021).
Li, J. et al. Isolation and characterization of a sequence type 25 carbapenem-resistant hypervirulent Klebsiella pneumoniae from the mid-south region of China. BMC Microbiol. 19, 219 (2019).
Zhao, J. et al. Emergence of a NDM-1-producing ST25 Klebsiella pneumoniae strain causing neonatal sepsis in China. Front. Microbiol. 13, 980191 (2022).
Wyres, K. L. et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 15, e1008114 (2019).
Thi Quynh Nhi, L. et al. Excess body weight and age associated with the carriage of fluoroquinolone and third-generation cephalosporin resistance genes in commensal Escherichia coli from a cohort of urban Vietnamese children. J. Med. Microbiol. 67, 1457–1466 (2018).
Phu, D. H. et al. Reducing antimicrobial usage in small-scale chicken farms in Vietnam: a 3-year intervention study. Front. Vet. Sci. 7, 612993 (2021).
Wick, R. R., Judd, L. M. & Holt, K. E. Performance of neural network basecalling tools for Oxford nanopore sequencing. Genome Biol. 20, 129 (2019).
FastQC. https://qubeshub.org/resources/fastqc (2015).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, 1–22 (2017).
Schwengers, O. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Micro. Genom. 7, 000685 (2021).
Lam, M. M. C. et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188 (2021).
Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124 (2018).
Alcock, B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699 (2023).
Chen, L. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328 (2004).
Lam, M. M. C., Wick, R. R., Judd, L. M., Holt, K. E. & Wyres, K. L. Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Micro. Genom. 8, 000800 (2022).
Inouye, M. et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6, 90 (2014).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Li, X. et al. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234 (2018).
Wang, J. T., Shapiro, B. A., & Shasha, D. E. (Eds). in Pattern Discovery in Biomolecular Data Ch. 3 (New York, 1999; online edn, Oxford Academic, 2020).
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011).
Holt, K. E. et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl Acad. Sci. USA 112, E3574–E3581 (2015).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15–e15 (2015).
Page, A. J. et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Micro. Genom. 2, e000056 (2016).
Kado, C. I. & Liu, S. T. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145, 1365–1373 (1981).
Moran, R. A., Anantham, S. & Hall, R. M. An improved plasmid size standard, 39R861. Plasmid 102, 6–9 (2019).
Acknowledgements
We would like to thank the healthcare staff at the Hospital for Tropical Diseases, Ho Chi Minh City, Vietnam, for their assistance in data collection for this study. The research was funded by the Wellcome International Training Fellowship (grant number: 222983/Z/21/Z) awarded to P.T.D. and John Fell Oxford University Press Research Fund (reference: 0010734) awarded to M.C., M.A.R., and P.T.D. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Q.N. and N.T.P.Y. conducted experiments, performed data analysis and interpreted the results under the scientific guidance from P.T.D. Q.N. and N.T.P.Y. drafted and edited the paper and P.T.D. contributed in structuring and editing of the paper. P.T.D. provided oversight of the project. H.T.T., T.T.N.D., V.V.P., C.V. and N.T.N.T. conducted experiments and edited the paper. T.V.T., N.L.N.P., L.T.Q.N., N.P.H.L. and J.J.C. contributed to data acquisition in the field, and editing of the paper. S.B., G.T., M.A.R., M.C. and C.T.H. contributed to the data interpretation and editing of the paper. All authors read and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nguyen, Q., Nguyen, Y.T.P., Ha, T.T. et al. Genomic insights unveil the plasmid transfer mechanism and epidemiology of hypervirulent Klebsiella pneumoniae in Vietnam. Nat Commun 15, 4187 (2024). https://doi.org/10.1038/s41467-024-48206-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48206-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.