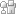« Prev Next »
Most students learn at an early age that organisms can be broadly divided into two types: prokaryotes and eukaryotes. In primary school, children are taught that the main difference between these organisms is that eukaryotic cells contain membrane-bound organelles, such as the nucleus, while prokaryotic cells do not. There is much more to the story, however, particularly with regard to chromosomal structure and organization.
E. coli: A Model Prokaryote
Much of what is known about prokaryotic chromosome structure was derived from studies of Escherichia coli, a bacterium that lives in the human colon and is commonly used in laboratory cloning experiments. In the 1950s and 1960s, this bacterium became the model organism of choice for prokaryotic research when a group of scientists used phase-contrast microscopy and autoradiography to show that the essential genes of E. coli are encoded on a single circular chromosome packaged within the cell nucleoid (Mason & Powelson, 1956; Cairns, 1963).
Prokaryotic cells do not contain nuclei or other membrane-bound organelles. In fact, the word "prokaryote" literally means "before the nucleus." The nucleoid is simply the area of a prokaryotic cell in which the chromosomal DNA is located. This arrangement is not as simple as it sounds, however, especially considering that the E. coli chromosome is several orders of magnitude larger than the cell itself. So, if bacterial chromosomes are so huge, how can they fit comfortably inside a cell—much less in one small corner of the cell?
DNA Supercoiling
The answer to this question lies in DNA packaging. Whereas eukaryotes wrap their DNA around proteins called histones to help package the DNA into smaller spaces, most prokaryotes do not have histones (with the exception of those species in the domain Archaea). Thus, one way prokaryotes compress their DNA into smaller spaces is through supercoiling (Figure 1). Imagine twisting a rubber band so that it forms tiny coils. Now twist it even further, so that the original coils fold over one another and form a condensed ball. When this type of twisting happens to a bacterial genome, it is known as supercoiling. Genomes can be negatively supercoiled, meaning that the DNA is twisted in the opposite direction of the double helix, or positively supercoiled, meaning that the DNA is twisted in the same direction as the double helix. Most bacterial genomes are negatively supercoiled during normal growth.
Proteins Involved in Supercoiling
During the 1980s and 1990s, researchers discovered that multiple proteins act together to fold and condense prokaryotic DNA. In particular, one protein called HU, which is the most abundant protein in the nucleoid, works with an enzyme called topoisomerase I to bind DNA and introduce sharp bends in the chromosome, generating the tension necessary for negative supercoiling. Recent studies have also shown that other proteins, including integration host factor (IHF), can bind to specific sequences within the genome and introduce additional bends (Rice et al., 1996). The folded DNA is then organized into a variety of conformations (Sinden & Pettijohn, 1981) that are supercoiled and wound around tetramers of the HU protein, much like eukaryotic chromosomes are wrapped around histones (Murphy & Zimmerman, 1997).
Once the prokaryotic genome has been condensed, DNA topoisomerase I, DNA gyrase, and other proteins help maintain the supercoils. One of these maintenance proteins, H-NS, plays an active role in transcription by modulating the expression of the genes involved in the response to environmental stimuli. Another maintenance protein, factor for inversion stimulation (FIS), is abundant during exponential growth and regulates the expression of more than 231 genes, including DNA topoisomerase I (Bradley et al., 2007).
Accessing Supercoiled Genes
Supercoiling explains how chromosomes fit into a small corner of the cell, but how do the proteins involved in replication and transcription access the thousands of genes in prokaryotic chromosomes when everything is packaged together so tightly? It has been determined that prokaryotic DNA replication occurs at a rate of 1,000 nucleotides per second, and prokaryotic transcription occurs at a rate of about 40 nucleotides per second (Lewin, 2007), so bacteria must have highly efficient methods of accessing their DNA strands. But how?
Researchers have noted that the nucleoid usually appears as an irregularly shaped mass within the prokaryotic cell, but it becomes spherical when the cell is treated with chemicals to inhibit transcription or translation. Moreover, during transcription, small regions of the chromosome can be seen to project from the nucleoid into the cytoplasm (i.e., the interior of the cell), where they unwind and associate with ribosomes, thus allowing easy access by various transcriptional proteins (Dürrenberger et al., 1988). These projections are thought to explain the mysterious shape of nucleoids during active growth. When transcription is inhibited, however, the projections retreat into the nucleoid, forming the aforementioned spherical shape.
Because there is no nuclear membrane to separate prokaryotic DNA from the ribosomes within the cytoplasm, transcription and translation occur simultaneously in these organisms. This is strikingly different from eukaryotic chromosomes, which are confined to the membrane-bound nucleus during most of the cell cycle. In eukaryotes, transcription must be completed in the nucleus before the newly synthesized mRNA molecules can be transported to the cytoplasm to undergo translation into proteins.
Variations in Prokaryotic Genome Structure
Recently, it has become apparent that one size does not fit all when it comes to prokaryotic chromosome structure. While most prokaryotes, like E. coli, contain a single circular DNA molecule that makes up their entire genome, recent studies have indicated that some prokaryotes contain as many as four linear or circular chromosomes. For example, Vibrio cholerae, the bacteria that causes cholera, contains two circular chromosomes. One of these chromosomes contains the genes involved in metabolism and virulence, while the other contains the remaining essential genes (Trucksis et al., 1998). An even more extreme example is provided by Borrelia burgdorferi, the bacterium that causes Lyme disease. This organism is transmitted through the bite of deer ticks (Figure 2), and it contains up to 11 copies of a single linear chromosome (Ferdows & Barbour, 1989). Unlike E. coli, Borrelia cannot supercoil its linear chromosomes into a tight ball within the nucleoid; rather, these strands are diffused throughout the cell (Hinnebusch & Bendich, 1997).
Other organisms, such as Bacillus subtilis, form nucleoids that closely resemble those of E. coli, but they use different architectural proteins to do so. Furthermore, the DNA molecules of Archaea, a taxonomic domain composed of single-celled, nonbacterial prokaryotes that share many similarities with eukaryotes, can be negatively supercoiled, positively supercoiled, or not supercoiled at all. It is important to note that archaeans are the only group of prokaryotes that use eukaryote-like histones, rather than the architectural proteins described above, to condense their DNA molecules (Sandman et al., 1990). The acquisition of histones by archaeans is thought to have paved the way for the evolution of larger and more complex eukaryotic cells (Minsky et al., 1997).
Other DNA Differences Between Prokaryotes and Eukaryotes
Most prokaryotes reproduce asexually and are haploid, meaning that only a single copy of each gene is present. This makes it relatively easy to generate mutations in the lab and study the resulting phenotypes. By contrast, eukaryotes that reproduce sexually generally contain multiple chromosomes and are said to be diploid, because two copies of each gene exist—with one copy coming from each of an organism's parents.
Yet another difference between prokaryotes and eukaryotes is that prokaryotic cells often contain one or more plasmids (i.e., extrachromosomal DNA molecules that are either linear or circular). These pieces of DNA differ from chromosomes in that they are typically smaller and encode nonessential genes, such as those that aid growth in specific conditions or encode antibiotic resistance. Borrelia, for instance, contains more than 20 circular and linear plasmids that encode genes responsible for infecting ticks and humans (Fraser et al., 1997). Plasmids are often much smaller than chromosomes (i.e., less than 1,500 kilobases), and they replicate independently of the rest of the genome. However, some plasmids are capable of integrating into chromosomes or moving from cell to cell.
Perhaps due to the space constraints of packing so many essential genes onto a single chromosome, prokaryotes can be highly efficient in terms of genomic organization. Very little space is left between prokaryotic genes. As a result, noncoding sequences account for an average of 12% of the prokaryotic genome, as opposed to upwards of 98% of the genetic material in eukaryotes (Ahnert et al., 2008). Furthermore, unlike eukaryotic chromosomes, most prokaryotic genomes are organized into polycistronic operons, or clusters of more than one coding region attached to a single promoter, separated by only a few base pairs. The proteins encoded by each operon often collaborate on a single task, such as the metabolism of a sugar into by-products that can be used for energy (Figure 3).
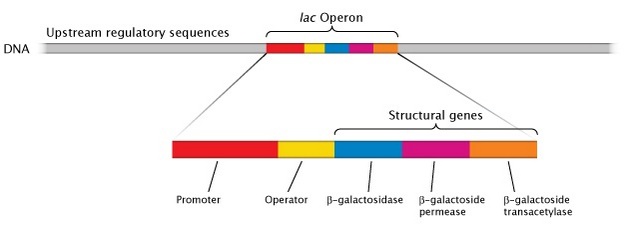
The organization of prokaryotic DNA therefore differs from that of eukaryotes in several important ways. The most notable difference is the condensation process that prokaryotic DNA molecules undergo in order to fit inside relatively small cells. Other differences, while not as dramatic, are summarized in Table 1.
Table 1: Prokaryotic versus Eukaryotic Chromosomes
| Prokaryotic Chromosomes | Eukaryotic Chromosomes |
|
|
References and Recommended Reading
Abbott, A. Lyme disease: Uphill struggle. Nature 439, 524–525 (2006) doi:10.1038/439524a (link to article)
Ahnert, S. E., et al. How much non-coding DNA do eukaryotes require? Journal of Theoretical Biology 252, 587–592 (2008)
Bendich, A. J., & Drlica, K. Prokaryotic and eukaryotic chromosomes: What's the difference? Bioessays 22, 481–486 (2000)
Bradley, M. D., et al. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 153, 2922–2940 (2007)
Cairns, J. The chromosome of Escherichia coli. Cold Spring Harbor Symposia on Quantitative Biology 28, 43–46 (1963)
Dürrenberger, M., et al. Intracellular location of the histonelike protein HU in Escherichia coli. Journal of Bacteriology 170, 4757–1768 (1988)
Endy, D., & Brent, R. Modelling cellular behaviour. Nature 409, 391–395 (2001) doi:10.1038/35053181 (link to article)
Ferdows, M. S., & Barbour, A. G. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proceedings of the National Academy of Sciences 86, 5969–5973 (1989)
Fraser, C. M., et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390, 580–586 (1997) doi:10.1038/37551 (link to article)
Hinnebusch, B. J., & Bendich, A. J. The bacterial nucleoid visualized by fluorescence microscopy of cells lysed within agarose: Comparison of Escherichia coli and spirochetes of the genus Borrelia. Journal of Bacteriology 179, 2228–2237 (1997)
Jacob, F., & Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. Journal of Molecular Biology 3, 318–356 (1961)
Lewin, B. Genes IX (Sudbury, MA, Jones and Bartlett, 2007)
Mason, D. J., & Powelson, D. M. Nuclear division as observed in live bacteria by a new technique. Journal of Bacteriology 71, 474–479 (1956) (link to article)
Minsky, A., et al. Nucleosomes: A solution to a crowded intracellular environment. Journal of Theoretical Biology 188, 379–385 (1997)
Murphy, L. D., & Zimmerman, S. B. Isolation and characterization of spermidine nucleoids from Escherichia coli. Journal of Structural Biology 119, 321–335 (1997)
Rice, P. A., et al. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell 87, 1295–1306 (1996)
Robinow, C., & Kellenberger, E. The bacterial nucleoid revisited. Microbiology and Molecular Biology Reviews 58, 211–232 (1994)
Sandman, K., et al. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proceedings of the National Academy of Sciences 87, 5788–5791 (1990)
Sandman, K., et al. Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome. Cellular and Molecular Life Sciences 54, 1350–1364 (1998)
Sauvonnet, N., et al. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO Journal 19, 2221–2228 (2000) doi:10.1093/emboj/19.10.2221
Sinden, R. R., & Pettijohn, D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proceedings of the National Academy of Sciences 78, 224–228 (1981)
Snyder, L., & Champness, W. Molecular Genetics of Bacteria, 2nd ed. (Washington, DC, ASM Press, 2003)
Trucksis, M., et al. The Vibrio cholerae genome contains two unique circular chromosomes. Proceedings of the National Academy of Sciences 95, 14464–14469 (1998)
Willenbrock, H., & Ussery, D. W. Chromatin architecture and gene expression in Escherichia coli. Genome Biology 5, 252 (2004)
Yasuzawa, K., et al. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene 122, 9–15 (1992).



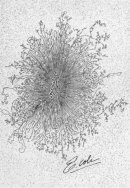
 Figure 1
Figure 1