Abstract
Endoplasmic reticulum aminopeptidase 1 (ERAP1) is an essential component of the immune system, because it trims peptide precursors and generates the N--restricted epitopes. To examine ERAP1's unique properties of length- and sequence-dependent processing of antigen precursors, we report a 2.3 Å resolution complex structure of the ERAP1 regulatory domain. Our study reveals a binding conformation of ERAP1 to the carboxyl terminus of a peptide and thus provides direct evidence for the molecular ruler mechanism.
Similar content being viewed by others
Introduction
Antigen processing is an integral part of cell-mediated immune surveillance and responses that involve MHC-restriction and T cell recognition1. Processed antigenic peptides, when presented on cell surface by MHC molecules to T cells, serve as identity tags to be monitored and responded to by our immune system. To be presented by MHC class I molecules, endogenous antigens need to be cut into 8 to 11 residues in length in order to fit into the MHC binding groove2,3. The precursors of class I antigenic peptides are generated mainly by proteasomes in the cytosol to have the correct C-terminus but with an N-terminal extension of several amino acids4. These peptide precursors are then transported into the lumen of the endoplasmic reticulum (ER) by transporter associated with antigen processing (TAP), that is an MHC-encoded peptide transporter capable of importing both mature epitopes and N-extended precursors ranging in length from 7 to more than 20 amino acids5,6,7. TAP transports the longer precursors more efficiently than the mature epitopes8. Thus, many peptide precursors need to be further trimmed inside the ER to generate the final N-termini of mature epitopes9,10.
ERAP1 (ER aminopeptidase 1), also named ERAAP (ER aminopeptidase associated with antigen processing), is one of two ER luminal aminopeptidases that make the final N-terminal trimming of class I antigenic peptides11,12,13. A second enzyme is called ERAP2 or L-RAP (leukocyte-derived arginine aminopeptidase)14. These two enzymes are highly homologous and both are induced by interferon-γ, a potent stimulator of MHC class I presentation. ERAP1 is identical to a previously isolated enzyme called adipocyte-derived leucine aminopeptidase15, whose immunological relevance was not previously recognized. As expected for a peptidase involved in generating MHC class I antigens, ERAP1 is expressed in most cells16 and its expression level is strongly upregulated by interferon-γ11,12,13. ERAP1 strongly prefers peptide substrates that are 9–16 residues in length12,13,17,18, which corresponds to the lengths of peptides transported selectively by TAP5,6,7,8. The length preference allows ERAP1 to rapidly trim N-extended peptides to 8–9 residues, but further cleavages occur much more slowly or cease completely12,13,17,18, producing the optimal length of epitopes that are presented by most MHC class I molecules. Moreover, ERAP1 catalysis is activated by an unusual allosteric mechanism, termed the “molecular ruler” mechanism that monitors the substrate length and the identity of the C-terminal amino acids 9–16 residues away from the N-terminal cleavage site18. Thus, the substrate specificity of ERAP1 influences which peptides are trimmed and available for MHC class I presentation and is critical for immunological function.
To gain insights into the molecular ruler mechanism, our group has been pursuing the structure-function studies of the ERAP1 enzyme. During the preparation of this manuscript, two structures of ERAP1 were reported by other groups19,20. However, without a peptide bound in those ERAP1 structures and with relative low resolutions (2.7–3 Å), the ERAP1 mechanism of peptide length- and sequence-dependent activity remained elusive. We report here a 2.3 Å-resolution structure of the ERAP1 regulatory domain in complex with a peptide fragment. This complex structure allows a direct visualization of peptide binding to the ERAP1 regulatory domain and thus provides a structural basis for the molecular ruler mechanism.
Results
ERAP1 C-terminal domain proposed as the putative regulatory domain
There is indication that ERAP1 has a modular organization for its molecular ruler mechanism: binding of peptide's C-terminus to a regulatory site, distinct from the catalytic site, allosterically activates its peptidase activity18. Insights into the structure of the ERAP1 regulatory domain should elucidate its unusual length dependence and substrate specificity, as well as the mechanism of antigen processing. Based on sequence comparisons, we hypothesized that the unique C-terminal half of ERAP1 contains the C-terminus binding groove for antigenic peptide precursors and acts as the regulatory domain for the molecular ruler mechanism. ERAP1 is a zinc-containing metallopeptidase that is a member the “M1/gluzincin” family of peptidases15,21. It is most closely related to two other aminopeptidases of the M1 family: ERAP2 and P-LAP22, with 43–49% sequence identity spread along the entire sequences. There was also evidence to suggest that ERAP2 trims peptides in a way similar to ERAP117. These three enzymes recognize and cleave peptide precursors and belong to the oxytocinase subfamily of the M1 peptidases21. However, sequence alignment with other members of the M1 family that have a broader range of substrates shows that homology is only clustered within the N-terminal half of ERAP1: the M1 homology region. In contrast, there is no significant sequence homology between the C-terminal half of ERAP1 and other broad-substrate-range M1 members. Nonetheless, this C-terminal domain is highly conserved among the three oxytocinase subfamily members that all bind and trim peptide substrates14,21 and thus is highly likely to harbor the regulatory site to sense and recognize peptide's length and sequences near its C-terminus.
Structure of ERAP1 C-terminal domain in complex with a peptide C-terminus
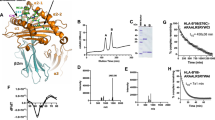
We have determined a 2.3 Å resolution crystal structure of the ERAP1 regulatory domain (a.a. 529–941) in complex with a peptide carboxyl-terminus, with Rcryst = 0.19 and Rfree = 0.26. Other crystallographic data collection and refinement statistics are summarized in Table 1. This ERAP1 regulatory domain is composed of two subdomains: a small beta-sandwich with two β-sheets (residues 530–611) and a larger bowl-shaped alpha-helix domain with 16 α-helices (residues 614–941) forming a concave surface (Fig. 1). The overall structure is similar to the corresponding domain of the full-length ERAP1 structure in the closed form20, with root mean square (r.m.s.) deviations of 1.7 Å for all main-chain atoms; main differences result from a slight hinge movement between these two subdomains. In the crystal, the C-terminus end of a peptide, an engineered His-tag tail from a neighboring molecule (-RMHHHHHH-CO2), is docking into a groove on the concave surface of the alpha-helix domain. Interactions between ERAP1 and the peptide mainly involve hydrophobic and van der Waals contacts, plus a few hydrogen-bond interactions with the carboxylate end of the peptide (Fig. 1b and see below). As oriented in Fig. 1a, this peptide C-terminus binding groove is facing towards the N-terminal catalytic site in an intact ERAP1 protein with a large internal cavity in-between the N-terminal catalytic zinc site and the C-terminal regulatory domains19,20. The location of the C-terminus binding groove, ∼29 Å from the N-terminal catalytic zinc site (Zn in Fig. 1a), is consistent with the expectation for a C-terminus anchoring site of a molecular ruler to monitor the length and sequence of an antigen precursor of nine- to ten-residue in length. Longer antigenic precursors could be accommodated by bulging or zig-zagging in the middle of peptide into the central and widest portion (∼30 Å) of the large substrate cavity.
Structure of the ERAP1 C-terminal regulatory domain in complex with a peptide.
(a) Overall structure of the ERAP1 regulatory domain, with respect to the N-terminal catalytic domain. The β-sandwich and α-helix subdomains of the regulatory domain are shown as a ribbon diagram and colored in blue and green, respectively. Bound peptide is shown as a stick model and colored by atom types. Location of the catalytic zinc site in the N-terminal domain is shown as a grey sphere, with its distance to the peptide C-terminus indicated in angstroms. (b) ERAP1 interactions with bound His-tag peptide (sequence MHHHHHH-CO2). Bound peptide is shown as a thick stick model and colored by atom types: green for main-chain carbons, yellow for side-chain carbons, blue for nitrogens and red for oxygens. Side-chains of selected residues of ERAP1 are also shown and labeled as a thin stick model in gray. Dotted lines denote hydrogen-bond interactions.
Allosterical activation of the ERAP1 aminopeptidase activity by histidine-containing peptides
ERAP1 is likely to bind and process His-containing antigen precursors since histidine is an anchor or preferred residue at various positions of antigen ligands for many MHC class I molecules. Histidine had been reported to be an anchor residue at the P2 position of antigen peptides for HLA-B*3801, at the P2 position for HLA-B*39011 and at the P7 (PC-2, the 2nd position N-terminal to the peptide carboxyl-terminal) position for mouse H Qa-223. It is also a preferred residue at various positions of antigens, including the P9/PC carboxyl-terminal position of antigen peptides for HLA-B*2705, or the P8/PC-1 position for HLA-B8, HLA-B*3701, HLA-Cw*0401, or the P7/PC-2 position for HLA-A*3101, HLA-B8, HLA-B*390223. In addition, HLA-A3 had also been shown to bind peptides with a histidine at the PC position24. Thus binding of the His-tag peptide to the ERAP1 regulatory domain as observed in the crystal structure is likely to reflect a functional conformation of ERAP1's substrate recognition.
More studies are needed to resolve some conflicting results reported for the substrate sequence preference of ERAP1. For example, Arg, Lys and His at the PC position were reported by Chang et al.18 to be suboptimal substrates, yet Evnouchidou et al. concluded that Arg and Lys are the best anchor residues at the PC position25. To verify the functional relevance of the His-tag binding, we performed enzyme activation assays. It has been shown that peptides shorter than 8 residues are not long enough to be efficiently trimmed by ERAP119. However, these short peptides can bind to the regulatory domain of ERAP1 and allosterically activate the hydrolysis of L-AMC (leucine-7-amido-4-methyl-coumarin) by the ERAP1 catalytic site. To demonstrate a functional binding of His-tag peptides, we monitored activation of L-AMC hydrolysis by a series of peptides based on the bound His-tag sequence RMHHHHHH. Consistent with previous studies19,25, ERAP1 hydrolyzes L-AMC with a low basal level in the absence of peptide (open bar in Fig. 2), or in the presence of a predominantly negatively charged peptide (the peptide EEEEGEG); such a negatively charged peptide was considered to be a poor substrate for ERAP125. We then examined activation of L-AMC hydrolysis by peptides containing C-terminal sequences of two naturally processed antigenic peptides that have a histidine at the PC position26: TRYPILAGH (a cytochrome P450 epitope, underlined C-terminal fragment used in Fig. 2) and RRIKEIVKKH (a HSP 86 epitope). In the presence of these two C-terminal fragments of antigenic peptides, a significant increase of ERAP1 aminopeptidase activity on L-AMC over the basal level was detected, suggesting an allosterical binding to the regulatory domain. Similarly, a significant increase of ERAP1 aminopeptidase activity was also detected in the presence of the His-tag peptides MHHHHHH and RMHHHHHH. Activation was further increased by placing an arginine anchor near the C-terminus of the peptide MHHHRHH, which is consistent with ERAP1 specificity studies at this (PC-2) peptide position25 (see below). Altogether, these His-containing peptides can thus bind to the ERAP1 regulatory domain to allosterically activate the aminopeptidase activity at the ERAP1's catalytic zinc site. Since many T-cells reactive to self and prevalent antigens presented by MHC are eliminated during their development in the thymus to avoid autoimmunity27, small but significant effect from minor antigens, similar to those observed in Figure 2, could play critical roles for protection against foreign pathogens.
Allosteric activation of ERAP1 by His-tag peptides.
Activation of L-AMC hydrolysis by a series of peptides with a histidine at the PC position. As negative controls, hydrolysis activities were measured in the absence of a peptide, or with a negatively charged peptide (EEEEGEG) that was considered to be a poor substrate for ERAP125. Peptides YPILAGH and KEIVKKH are derived from two naturally processed antigenic peptides, a cytochrome P450 epitope and a HSP 86 epitope, respectively. The other three peptides, MHHHHHH, RMHHHHHH and MHHHRHH, are derived from the bound C-terminal His-tag determined in the crystal structure. The basal activity in which L-AMC hydrolysis was measured in the absence of added peptide is indicated by the open bar and the horizontal line. Standard deviations of three separate experiments are indicated by error bars.
Molecular modeling of an optimal binding peptide into the ERAP1 regulatory site
To analyze the peptide binding groove and potential pockets or sub-sites for recognizing substrate side-chains and C terminus, we modeled an Arg-Ala-Phe sequence into the last three positions (PC-2, PC-1 and PC) of the bound His-tag peptide (Fig. 3a). This tri-peptide model sequence was based on substrate specificity of ERAP1 at PC-2 and PC positions25; in the crystal the PC-1 side chain points away from the binding groove and thus was reduced to an alanine in the model tri-peptide. To better fit into the binding groove and sub-pockets, small adjustments were made (Fig. 3a): ∼0.9 Å towards Glu831 and a side-chain rotation to allow an aromatic ring interaction between the PC Phe and Phe803. One pocket contains predominantly non-polar atoms located at the floor of the peptide binding cleft, surrounded by Ile 681, Leu733, Leu734, Val 737 and Leu769, with Phe803 ready to make an aromatic ring interaction with the PC phenylalanine side-chain of the model peptide (Figs. 3a&b). Another deep pocket is located in close proximity to the PC-2 arginine side-chain, with the entry wrapped around by residues Leu769, Phe803 and Ser799 and tunnels into the negatively charged residues Glu802 and Glu831. This E802/E831 pocket appears to be ideal for binding to an anchoring side-chain of arginine or lysine of antigenic precursors. As shown in Fig. 2, adding an arginine anchor in the PC-2 position of the peptide substantially increases the peptide's activation capacity. Meanwhile, the carboxylate end of the peptide ligand makes direct or water-mediated contacts with Tyr684, Lys685, Arg807 and Arg841 and is partially exposed to bulk solvents. Model building suggests that small adjustments could be made to allow extensions (e.g. with an amine or glycine) from the current C-terminus location, similar to the C-terminal extension of an MHC class I binding mode28. Most of the residues made up of the two binding pockets for peptide side-chain anchors are polymorphic among ERAP1, ERAP2 and P-LAP22, suggesting different binding specificities for these three enzymes. On the other hand, several residues in and around the carboxylate binding site are conserved among these oxytocinase subfamily members. These conserved residues, which include Tyr684 and Arg841, provide additional contacts to recognize a constant feature of antigenic precursors. In the crystal structure, Tyr684 make a water-mediated contact with the peptide carboxylate end whereas Arg841 makes a direct contact with the main-chain carbonyl of the PC-2 residue (Fig. 1c) and a water-mediated contact with the peptide carboxylate end.
Specificity pockets of ERAP1 for peptide's anchors.
(a) Overlay of a modeled tri-residue peptide (yellow) onto the crystal structure of the last three positions of the bound His-tag peptide (green). The model tri-residue peptide Arg-Ala-Phe was built based on the last three positions (PC-2, PC-1, PC carboxyl-terminal) of the bound His-tag peptide, with minor adjustments for better fit (see text and Methods). Surrounding side-chains of ERAP1 residues are shown and labeled. (b) The C terminus binding groove of ERAP1 is shown as solvent accessible surfaces colored by electrostatic potentials: from red to blue for negatively charged to positively charged areas. The model tri-peptide is shown as a thick stick model colored by atom types: green for main-chain carbons, yellow for side-chain carbons, blue for nitrogens and red for oxygens. Side-chains of selected ERAP1 residues are also shown and labeled in white. (c) Schematic outlines of specificity pockets, with surrounding side-chains of selected ERAP1 residues shown and labeled.
Discussion
The high resolution structure of ERAP1/peptide complex provides detailed insights into the peptide C-terminus recognition and the molecular ruler mechanism. Structural comparisons between the open and closed conformations of ERAP1 suggest that a conformational change with reorientation of a key catalytic residue is involved in the molecular ruler mechanism19,20. Adding the new peptide binding conformation reported here, we propose a detailed model for the ERAP1's molecular ruler mechanism to sense the substrate length and recognize the sequence near the peptide C-terminal end (Fig. 4). In the absence of a peptide anchored at the regulatory pocket, ERAP1 stays in the lower-activity open conformation, resulting in the basal level of L-AMC hydrolysis (Fig. 4a). This lower-activity open conformation could also account for some low level trimming of peptides down to 4 or 5 residues4, by cutting peptide's N-terminus without having its C-terminus in contact with the regulatory pocket. In the presence of a peptide ligand longer than 9 or 10 residues, ERAP1 uses specificity pockets at its regulatory domain to anchor the peptide's side-chains at or near the carboxyl-terminus (Fig. 4b). This anchoring and recognition triggers conformational changes to activate the N-terminal catalytic center located ∼30 Å away. As observed in Figure 2, short peptides can also bind to the regulatory domain of ERAP1 and activate in trans the hydrolysis of L-AMC by the N-terminal catalytic site. Nonetheless, for a peptide precursor to be efficiently processed, it needs to be 9 or 10 residues long to be able to simultaneously place its N- and C-terminal ends into the catalytic zinc site and the C-terminus docking pockets, respectively. Longer peptides could be accommodated by bulging or zig-zagging into the widest portion of the large substrate cavity. Thus, even though the architects of the binding grooves are different, ERAP1 and MHC class I molecules utilize the same strategies to bind a large repertoire of antigenic peptides, by binding at both N- and C-termini of peptides with common feature and side-chain anchors, but allowing flexibility in sequence and length through bulging or zig-zagging in the middle2,3. It is interesting to note that ERAP1 has dual specificities at the PC position of a peptide nonamer series: preferring either a positively charged (R, K) or a hydrophobic side-chain (F, V, M)25. This is likely to result from different specificity pockets involved in anchoring these two types of peptide anchors. As for the model peptide (Fig. 3b), the Phe803 pocket could bind a phenylalanine anchor. However, for a peptide precursor with an arginine at the PC position, it is plausible to insert this positively charged side-chain anchor into the Glu802/Glu831 pocket instead. For the latter group of peptides with a PC arginine, additional affinity could come from the conserved Arg841 to make direct contacts with the peptide's carboxylate end.
Proposed model for the molecular ruler mechanism.
(a) A small substrate (L-AMC shown) cannot concurrently reach the catalytic site and the regulatory pockets. ERAP1 stays in the lower-activity open conformation and inefficiently hydrolyses L-AMC. This could also account for some low level trimming of peptides down to 4 or 5 residues4. (b) A peptide longer than 9 or 10 residues can reach the catalytic zinc site from the regulatory domain binding groove where its carboxyl-terminus is anchored. Triggered by the peptide anchoring into the regulatory pockets (lightening), ERAP1 changes into the higher-activity closed conformation and efficiently trims one residue from the peptide's N-terminal end. Slightly shorter and longer peptides take the less (red) and more (green) bulging paths, respectively. Allosteric activation of ERAP1 on L-AMC hydrolysis by a short peptide can also be achieved in trans (a pair of red slashes) by concurrently occupying the catalytic site and regulatory pockets.
Methods
Protein expression and purification
Baculovirus cDNA encoding for human ERAP1 full-length protein or C-terminal domain was constructed with a C-terminal hexa-histidine tag according to the protocols of the manufacturer (Invitrogen). The presence of ERAP1 protein and the integrity of the purified recombinant bacmid DNA were verified by PCR. To express the protein, the bacmid DNA was transfected into Sf9 insect cells according to the manufacturer's protocols. The protein was expressed and harvested 48 hours after infection by adding the P4 recombinant viral stock into Sf9 insect cells with an MOI of 1 pfu/cell. Protein expression was confirmed by western blot using primary antibody against the hexa-histidine tag.
Cell pellets were re-suspended in 50 mM NaH2PO4, pH 8.0, 300 mM NaCl and 10 mM imidazole and lysed by freeze-thaw cycles and sonication. The supernatant was loaded onto a Ni-NTA column and washed several times with 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl and 10–30 mM imidazole. The protein was then eluted with 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl and 400 mM imidazole. Glycerol was added to the eluted solution to a final concentration of 16% (v/v) and then the concentrated protein was further purified through a Superdex 200 gel filtration column (Amersham Pharmacia) by FPLC system with a buffer containing 10 mM Tris, pH 7.5, 10 mM NaCl. A single peak for ERAP1 enzyme or C-terminal domain was collected and the protein was concentrated to 3–7 mg/ml for crystallization.
Enzyme Activation Assays
Aminopeptidase activity was determined by measuring the fluorescence of 7-amido-4-methylcoumarin (AMC) released by hydrolysis of Leucine-AMC (L-AMC). Assays were performed at 25°C in 200 μl of 50 mM Tris/HCl, pH 8.0, 0.1 M NaCl, containing 0.75 μg ml−1 ERAP1 enzyme in the presence or absence of 100 μM peptides. Hydrolysis of 50 μM Leucine-AMC (L-AMC) was followed for 5 minutes and measured using an excitation wavelength of 380 nm and an emission wavelength of 460 nm. Fluorescence intensities were calibrated using AMC as standard12.
Crystallization, data collection and structure determination
Initial crystallization screening at room temperature used the hanging-drop vapor diffusion technique. Promising conditions were further refined at 4°C. To improve the crystal quality, micro-seeding method was used for final crystallization. The best looking crystal was formed above a well solution containing 100 mM Tris, pH 8.5 and 8% PEG8000 at 4°C in 4 days.
For data collection, the crystal was cryoprotected in solution containing 100 mM Tris-HCl buffer (pH 8.0) and 30% glycerol. X-ray data were collected using the beamline X29 at National Synchrotron Light Source (NSLS). The data was processed with the Mosflm29 and the CCP4 suite30.
The structure of ERAP1 C-terminal domain was determined by molecular replacement method using the ERAP1 intact protein structure (PDB code 2XDT)20 as the starting model, with residues 530–610 deleted and all remaining residues changed to alanines to reduce model bias. Molecular replacement was preformed with Molrep and refinements were performed with the Refmac program. 5% of the total reflection data was excluded from the refinement cycles and used to calculate the free Rfactor (Rfree ) for monitoring refinement progress. Rigid body and subsequent restrained refinements and model building with COOT31 led to the final crystallographic Rwork /Rfree of 19.1%/26.1% at 2.3 Å resolution. The X-ray data and structure refinement statistics are shown in Table 1. A structural alignment of the ERAP1 C-terminal domain with corresponding domain of Tricon Interacting Factor F3 (TIFF3)32 is shown in Supplementary Figure 1. All the figures were drawn using PyMOL (DeLano Scientific) and labels were added using Adobe® Photoshop.
Modeling of the tri-residue peptide Arg-Ala-Phe
The modeled tri-residue peptide Arg-Ala-Phe is based on the last three positions (PC-2, PC-1 and PC) of the experimentally determined His-tag peptide. These three histidine residues were first mutated to Arg, Ala and Phe, respectively. Side chains of the mutated tripeptide were then adjusted to avoid steric clashes with the ERAP1 structure, using the torsion option in Coot31. Additional adjustments were made to better fit into the binding groove and sub-pockets: ∼0.9 Å towards Glu831 and a side-chain rotation of PC Phe to allow an aromatic ring interaction between the PC Phe and Phe803 (Fig. 3a). The modeled tripeptide in complex with ERAP1 domain was further refined with energy minimization protocol with 20 steps of Steepest Descent algorithm in GROMOS96 force field using the SwissPdb Viewer program33.
Accession code
Diffraction data and coordinates are deposited under accession code 3RJO in the Protein Data Bank.
References
Cresswell, P., Ackerman, A. L., Giodini, A., Peaper, D. R. & Wearsch, P. A. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol Rev 207, 145–57 (2005).
Guo, H.-C. et al. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 360, 364–6 (1992).
Fremont, D. H., Matsumura, M., Stura, E. A., Peterson, P. A. & Wilson, I. A. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science 257, 919–27 (1992).
Rock, K. L., York, I. A. & Goldberg, A. L. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol 5, 670–7 (2004).
Momburg, F., Roelse, J., Hammerling, G. J. & Neefjes, J. J. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med 179, 1613–23 (1994).
van Endert, P. M. et al. A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity 1, 491–500 (1994).
Uebel, S. et al. Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. Proc Natl Acad Sci U S A 94, 8976–81 (1997).
Neisig, A. et al. Major differences in transporter associated with antigen presentation (TAP)-dependent translocation of MHC class I-presentable peptides and the effect of flanking sequences. J Immunol 154, 1273–9 (1995).
Fruci, D., Niedermann, G., Butler, R. H. & van Endert, P. M. Efficient MHC class I-independent amino-terminal trimming of epitope precursor peptides in the endoplasmic reticulum. Immunity 15, 467–76 (2001).
Serwold, T., Gaw, S. & Shastri, N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat Immunol 2, 644–51 (2001).
Serwold, T., Gonzalez, F., Kim, J., Jacob, R. & Shastri, N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419, 480–3 (2002).
Saric, T. et al. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol 3, 1169–76 (2002).
York, I. A. et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol 3, 1177–84 (2002).
Tanioka, T. et al. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J Biol Chem 278, 32275–83 (2003).
Hattori, A., Matsumoto, H., Mizutani, S. & Tsujimoto, M. Molecular cloning of adipocyte-derived leucine aminopeptidase highly related to placental leucine aminopeptidase/oxytocinase. J Biochem (Tokyo) 125, 931–8 (1999).
Schomburg, L., Kollmus, H., Friedrichsen, S. & Bauer, K. Molecular characterization of a puromycin-insensitive leucyl-specific aminopeptidase, PILS-AP. Eur J Biochem 267, 3198–207 (2000).
Saveanu, L. et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol 6, 689–97 (2005).
Chang, S. C., Momburg, F., Bhutani, N. & Goldberg, A. L. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a "molecular ruler" mechanism. Proc Natl Acad Sci U S A 102, 17107–12 (2005).
Nguyen, T. T. et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol 18, 604–13 (2011).
Kochan, G. et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci U S A 108, 7745–50 (2011).
Tsujimoto, M. & Hattori, A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta 1751, 9–18 (2005).
Rogi, T., Tsujimoto, M., Nakazato, H., Mizutani, S. & Tomoda, Y. Human placental leucine aminopeptidase/oxytocinase. A new member of type II membrane-spanning zinc metallopeptidase family. J Biol Chem 271, 56–61 (1996).
Rammensee, H. G., Friede, T. & Stevanoviic, S. MHC ligands and peptide motifs: first listing. Immunogenetics 41, 178–228 (1995).
DiBrino, M. et al. Endogenous peptides bound to HLA-A3 possess a specific combination of anchor residues that permit identification of potential antigenic peptides. Proc Natl Acad Sci U S A 90, 1508–12 (1993).
Evnouchidou, I. et al. The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS ONE 3, e3658 (2008).
Rotzschke, O. et al. Dominant aromatic/aliphatic C-terminal anchor in HLA-B*2702 and B*2705 peptide motifs. Immunogenetics 39, 74–7 (1994).
Palmer, E. & Naeher, D. Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat Rev Immunol 9, 207–13 (2009).
Collins, E. J., Garboczi, D. N. & Wiley, D. C. Three-dimensional structure of a peptide extending from one end of a class I MHC binding site. Nature 371, 626–9 (1994).
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67, 271–81 (2011).
CCP4 (Collaborative Computational Project, No. 4). The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–3 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004).
Kyrieleis, O. J., Goettig, P., Kiefersauer, R., Huber, R. & Brandstetter, H. Crystal structures of the tricorn interacting factor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations. J Mol Biol 349, 787–800 (2005).
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–23 (1997).
Acknowledgements
We thank Dr. Anand M. Saxena for assistance on data collection, Dr. Masafumi Tsujimoto for providing cDNA clone of ERAP1, Dr. Sara Kaloo for assistance on cell culture and Drs. Jessica E. Garb and Mark Hines for critical reading of the manuscript. This work was supported by Grants AI068831 and AI078134 from the NIH.
Author information
Authors and Affiliations
Contributions
All authors performed the experiment and discussed the results. A.G., D.L. and H.-C.G analysed the experimental data and contributed to manuscript preparation. H.-C.G wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Gandhi, A., Lakshminarasimhan, D., Sun, Y. et al. Structural insights into the molecular ruler mechanism of the endoplasmic reticulum aminopeptidase ERAP1. Sci Rep 1, 186 (2011). https://doi.org/10.1038/srep00186
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00186
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.