Abstract
Interatomic potentials (IAPs), which describe the potential energy surface of atoms, are a fundamental input for atomistic simulations. However, existing IAPs are either fitted to narrow chemistries or too inaccurate for general applications. Here we report a universal IAP for materials based on graph neural networks with three-body interactions (M3GNet). The M3GNet IAP was trained on the massive database of structural relaxations performed by the Materials Project over the past ten years and has broad applications in structural relaxation, dynamic simulations and property prediction of materials across diverse chemical spaces. About 1.8 million materials from a screening of 31 million hypothetical crystal structures were identified to be potentially stable against existing Materials Project crystals based on M3GNet energies. Of the top 2,000 materials with the lowest energies above the convex hull, 1,578 were verified to be stable using density functional theory calculations. These results demonstrate a machine learning-accelerated pathway to the discovery of synthesizable materials with exceptional properties.
Similar content being viewed by others
Main
Atomistic simulations are the bedrock of in silico materials design. The first step in most computational studies of materials is to obtain an equilibrium structure, which involves navigating the potential energy surface (PES) across all independent lattices and atomic degrees of freedom in search of a minimum. Atomistic simulations are also used to probe the dynamical evolution of materials systems, and to obtain thermodynamic averages and kinetic properties (for example, diffusion constants). Although electronic structure methods such as density functional theory (DFT) provide the most accurate description of the PES, they are computationally expensive and scale poorly with system size.
For large-scale materials studies, efficient, linear-scaling interatomic potentials (IAPs) that describe the PES in terms of many-body interactions between atoms are often necessary. However, most IAPs today are custom-fitted for a very narrow range of chemistries: often for a single element or up to no more than four to five elements. The most popular general purpose IAPs are the AMBER family of force fields1,2 and the universal force field (UFF)3. However, both were formulated primarily for molecular/organic systems and have limited support and accuracy in modeling crystal structures. More recently, machine learning of the PES has emerged as a particularly promising approach to IAP development4,5,6,7,8. These so-called ML-IAPs typically express the PES as a function of local environment descriptors such as the interatomic distances and angles, or atomic densities, and have been demonstrated to substantially outperform classical IAPs across a broad range of chemistries9. Message-passing and graph deep learning models10,11,12 have also been shown to yield highly accurate predictions of the energies and/or forces of molecules, as well as a limited number of crystals such as Li7P3S11 (ref. 13) and LixSiy (ref. 14) for lithium-ion batteries. Nevertheless, no work has demonstrated a universally applicable IAP across the periodic table and for all crystal types.
In the past decade, the advent of efficient and reliable electronic structure codes15 with high-throughput automation frameworks16,17,18,19 has led to the development of large federated databases of computed materials data, including the Materials Project20, AFLOW21, Open Quantum Mechanical Database (OQMD)22, NOMAD23 and so on. Most of the focus has been on making use of the final outputs from the electronic structure computations performed by these databases—namely, the equilibrium structures, energies, band structures and other derivative material properties—for the purposes of materials screening and design. Less attention has been paid to the huge quantities of PES data—that is, intermediate structures and their corresponding energies, forces and stresses—amassed in the process of performing structural relaxations.
In this work we develop the formalism for a graph-based deep learning IAP by combining many-body features of traditional IAPs with those of flexible graph material representations. Using the largely untapped dataset of more than 187,000 energies, 16,000,000 forces and 1,600,000 stresses from structural relaxations performed by the Materials Project since its inception in 2011, we trained a universal IAP for materials based on graph neural networks (GNN) with three-body interactions (M3GNet) for 89 elements of the periodic table with low energy, force and stress errors. We demonstrate the applications of M3GNet in phonon and elasticity calculations, structural relaxations and so on. We further relaxed ~30 million hypothetical structures for new materials discovery.
Results
Materials graphs with many-body interactions
Mathematical graphs are a natural representation for crystals and molecules, with nodes and edges representing the atoms and the bonds between them, respectively. Traditional graph neural network models for materials24,25,26,27 have proven to be exceptionally effective for general materials property predictions24,25,26,27, but are not suitable as IAPs due to the lack of physical constraints such as a continuity of energies and forces with the changes to the length and number of bonds.
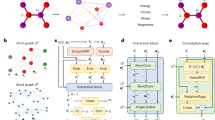
Here we develop a materials graph architecture that explicitly incorporates many-body interactions (Fig. 1). The materials graph is represented as \({{{\mathcal{G}}}}=({{{\mathcal{V}}}},{{{\mathcal{E}}}},{{{\mathcal{X}}}},\left.[{{{\textbf{M}}}},{{{\textbf{u}}}}]\right])\), where \({{{{\textbf{v}}}}}_{i}\in {{{\mathcal{V}}}}\) is atom information for atom i; \({{{{\textbf{e}}}}}_{ij}\in {{{\mathcal{E}}}}\) is the bond information for a bond connecting atoms i and j; and u is the optional global state information. A key difference with past materials graph implementations is the addition of \({{{{\textbf{x}}}}}_{i}\in {{{\mathcal{X}}}}\) (the coordinates for atom i) and M (the optional 3 × 3 lattice matrix in crystals). The graph structure is passed to a graph featurizer that embeds the pair atom distance rij—up to a certain cut-off rc—to basis functions, and the atomic number Zi to element feature spaces.
The model architecture starts from a position-included graph and then goes through a featurization process, followed by the main block and then the readout module with energy, force and stress outputs. The featurization process consists of the graph featurizer and the many-body computation module. In the graph featurizer, the atomic numbers of elements are embedded into a learnable continuous feature space and the pair bond distances are expanded into a basis set with values and derivatives of up to second order, going to zero at the boundary. The many-body computation module calculates the three- and many-body interaction atom indices and the associated angles. The main block consists of two main steps, namely, the many-body to bond module and the standard graph convolution. The many-body to bond step calculates the new bond information eij by considering the full bonding environment \({{{{\mathcal{N}}}}}_{i}\) of atom i via many-body angles such as θjik, τkijl and so on, and the bond lengths rik, rij, ril and so on. The standard graph convolution updates the bond, atom and optional state information iteratively. During the readout stage, the atom information in the graph is passed to a gated multilayer perceptron (MLP) for obtaining the atomic energy, which sums to the total energy. The derivatives of the total energy give force and stress outputs.
The model development takes inspiration from traditional IAPs such as the Tersoff bond-order potential28, where the bond interaction eij incorporates n-body interactions using all distinct combinations of n − 2 neighbors in the neighborhood \({{{{\mathcal{N}}}}}_{i}\) of atom i, excluding i and j. We will denote this materials graph with an n-body interactions neural network as MnGNet for brevity. The many-body computations of the graph produce high-order interactions such as angles θ and dihedrals τ and their interactions. The many-body interactions are then aggregated to bonds. Standard graph convolution steps subsequently update the bond, atom and state information. Such many-body calculations and graph convolutions can be repeated N times to construct models of arbitrary complexity, similar to previous materials graph network architectures25. In this work we will focus on incorporation of three-body interactions only (M3GNet).
In the case of IAP fitting, the atom information maps to atom-wise energy Ei and is summed to the total energy E, which is then used to calculate forces f and stresses σ via auto-differentiation.
M3GNet IAP
To develop an IAP using the M3GNet architecture, we used crystal structures with corresponding E, f and σ as targets as training data. The model generates trainable targets via auto-differentiation with f = − ∂E/∂x and σ = V−1∂E/∂ϵ, where x are the atomic coordinates, V is the volume and ϵ is the strain.
Benchmark on IAP datasets
As an initial benchmark, we selected a diverse DFT dataset of elemental energies and forces previously generated by Zuo and co-workers9 for face-centered cubic (fcc) nickel, fcc copper, body-centered cubic (bcc) lithium, bcc molybdenum, diamond silicon and diamond germanium. From Table 1, the M3GNet IAPs substantially outperform classical many-body potentials such as the embedded atom method (EAM) and modified EAM (MEAM); they also perform comparably with local environment-based ML-IAPs such as the Behler–Parinello neural network potential (NNP)4 and moment tensor potential (MTP)7. It should be noted that although ML-IAPs can achieve slightly smaller energy and force errors than M3GNet IAPs, it comes with a substantial loss in flexibility in handling multi-element chemistries because incorporating multiple elements in ML-IAPs usually results in a combinatorial explosion in the number of regression coefficients and the corresponding data requirements. By contrast, the M3GNet architecture represents the elemental information for each atom (node) as a learnable embedding vector. Such a framework is readily extendable to multicomponent chemistries. For instance, the M3GNet-all IAP trained on all six elements performed similarly to the M3GNet IAPs trained on individual elements. The M3GNet framework—like other GNNs—is able to capture long-range interactions without the need to increase the cut-off radius for bond construction (Supplementary Fig. 1). At the same time, unlike the previous GNN models, the M3GNet architecture still maintains a continuous variation of energy, force and stress with changes of the number of bonds (Supplementary Fig. 2), a crucial requirement for IAPs.
Universal IAP for the periodic table
To develop an IAP for the entire periodic table, we leveraged on one of the largest open databases of DFT crystal structure relaxations in the world, that is, the Materials Project20. In total, this dataset, named MPF.2021.2.8, contains 187,687 ionic steps of 62,783 compounds, with 187,687 energies, 16,875,138 force components and 1,689,183 stress components. The dataset covers an energy, force and stress range of [–28.731, 49.575] eV per atom, [–2,570.567, 2,552.991] eV Å−1 and [–5,474.488, 1,397.567] GPa, respectively (Fig. 2a,b). The majority of structures have formation energies between –5 and 3 eV per atom, as shown in Supplementary Fig. 3. Although the distribution of forces is relatively symmetric, the stress data contain a slightly higher proportion of negative (compressive) stresses than positive stresses due to the well-known tendency of the Perdew–Burke–Ernzerhof (PBE) functional to underbind. The radial distribution function g(r) (Fig. 2c) shows that the dataset also spans a broad range of interatomic distances, including small distances of less than 0.6 Å that are essential for the M3GNet model to learn the repulsive interactions at close distances. The dataset encompasses 89 elements of the periodic table with their counts shown in Fig. 2d (see the Methods and Supplementary Table 1 for more information on the MPF.2021.2.8 data).
a,b, Structural energy per atom (E) versus distributions of force (a) and stress (b) components. c, The radial distribution function g(r) (dark blue line) and pair atom distance distribution density (light blue histogram). The short distance (<1.1 Å) density is made of mostly hydrogen bonded with oxygen, carbon and nitrogen, as illustrated in the inset. d, Element counts for all atoms in the dataset, covering 89 elements across the periodic table.
In principle, an IAP can be trained on only energies, or a combination of energies and forces. In practice, the M3GNet IAP trained only on energies (M3GNet-E) was unable to achieve reasonable accuracies for predicting either forces or stresses, with mean absolute errors (MAEs) that are larger than even the mean absolute deviation of the data (Supplementary Table 2). The M3GNet models trained with energies + forces (M3GNet-EF) and energies + forces + stresses (M3GNet-EFS) achieved relatively similar energy and force MAEs, but the MAE in the stresses of the M3GNet-EFS was about half that of the M3GNet-EF model. Accurate stress predictions are necessary for applications that involve lattice changes, for example, structural relaxations or NpT molecular dynamics simulations. Our results suggest that it is critical to include all three properties (energy, force and stress) in model training to obtain a practical IAP. The final M3GNet-EFS IAP (henceforth referred to as the M3GNet model for brevity) achieved an average value of 0.035 eV per atom, 0.072 eV Å−1 and 0.41 GPa for the energy, force and stress test MAE, respectively.
On the test data, the model predictions and the DFT ground truth match well, as revealed by the high linearity and the R2 values for the linear fitting between DFT and model predictions (Fig. 3a–c). The cumulative distribution of the model errors indicate that 50% of the data have energy, force and stress errors that are smaller than 0.01 eV per atom, 0.033 eV Å−1 and 0.042 GPa, respectively (Fig. 3d–f). More stringent tests were performed using phonon and elasticity calculations, which were not part of the original training data. The M3GNet model can reproduce accurate phonon dispersion curves and density of states (DOS) of β-cristobalite, stishovite and α-quartz SiO2 (Supplementary Fig. 4) to quantitative agreements with expensive DFT computations29. The M3GNet phonon DOS centers (\(\bar{\omega }\)) from phonon calculations using predicted forces and the frozen phonon approach are also in good agreement with density functional perturbation theory-computed values with a MAE of 44.2 cm−1 (Fig. 3g)29. The systematic underestimation by the M3GNet model relative to DFT is probably due to the different choices of pseudopotentials; the DFT phonon calculations were performed using the PBEsol30 functional whereas the M3GNet training data comprised PBE/PBE + U calculations31,32. This systematic underestimation can be corrected with a constant shift of 31.6 cm−1 and the MAE reduces to 28.8 cm−1. Such errors are even smaller than a state-of-the-art phonon DOS peak position prediction model which reported MAE of 36.9 cm−1 (ref. 33). We note that the DOS peak prediction model does not exhibit a systematic shift as it was directly fitted on the data by minimizing a mean squared error. Similar to DFT, the relationship \(\bar{\omega }\propto 1/{(\overline{m})}^{2}\) (where \(\bar{\omega }\) is the average frequency and \(\overline{m}\) is the average atomic mass) is obtained (Supplementary Fig. 5). The M3GNet-calculated Debye temperatures are less accurate (Fig. 3h), which can be attributed to relatively poor M3GNet predictions of the shear moduli (R2 = 0.134; Supplementary Fig. 6); the bulk moduli predictions (R2 = 0.757), however, are reasonable.
a–c, The parity plots for the energy (a), force (b) and stress (c). The model predicted results are \(\hat{E}\), \(\hat{f}\) and \(\hat{\sigma }\). The dashed lines (y = x) are guides for the eye. d–f, The cumulative distribution of errors for the energy (d), force (e) and stress (f). The horizontal dashed lines indicate the model errors at 50%, 80% and 95% (from the bottom to the top, respectively). g, Comparisons between the model-calculated 1,521 phonon DOS centers data (\(\hat{\omega }\)) and the PBEsol DFT calculations (\(\bar{\omega }\)) by Petretto and co-workers29. h, Comparisons between the 11,848 Debye temperatures (excluding negative moduli) calculated from the M3GNet model (\({\hat{T}}_{{{{\rm{Debye}}}}}\)) and the PBE DFT elastic tensors from de Jong et al.64.
The M3GNet IAP was then applied in a simulated materials discovery workflow where the final DFT structures are not known a priori. M3GNet relaxations were performed on the initial structures from the test dataset of 3,140 materials. M3GNet relaxation yields crystals with volumes that are much closer to the DFT reference volumes (Fig. 4a). Although 50% and 5% of the initial input structures have volumes that differ from the final DFT-relaxed crystals by more than 2.4% and 22.2%, respectively, these errors are reduced to 0.6% and 6.6%, respectively, via M3GNet relaxation. Correspondingly, the errors in the predicted energies \(\hat{E}\) are also much smaller (Fig. 4b). Using the initial structures for direct model predictions, the energy differences distribute broadly, with a considerable number of structures having errors that are larger than 0.1 eV per atom. All errors here were calculated relative to the DFT energies of the final DFT-relaxed structures for each material. The overall MAE is 0.169 eV per atom with ~20% of the structures having errors that are larger than 0.071 eV per atom (Fig. 4b). These errors are far too large for reliable estimations of materials stability, given that about 90% of all inorganic crystals in the Inorganic Crystal Structure Database (ICSD) have an energy above the convex hull smaller than 0.067 eV per atom (ref. 34). By contrast, energy calculations on the M3GNet-relaxed structures yield an MAE of 0.035 eV per atom, and 80% of the materials have errors smaller than 0.028 eV per atom. The error distributions using M3GNet-relaxed structures are close to the case in which we know the DFT final structures (Fig. 4b) which suggests that the M3GNet potential can be accurate in helping obtain the correct structures. In general, relaxations with M3GNet converge rapidly, as shown in Supplementary Fig. 7. An example of M3GNet relaxation is shown in Supplementary Fig. 8 for K57Se34 (Materials Project ID mp-685089), a material with one of the largest energy changes during relaxation. Convergence is achieved after about 100 steps when the forces fall below 0.1 eV Å−1. The X-ray diffraction pattern of the M3GNet-relaxed structure also resembles the counterpart from DFT relaxation (Supplementary Fig. 8g). This relaxation can be performed on a laptop in about 22 s on a single Intel(R) Xeon(R) CPU E5-2620 v.4 2.10 GHz core, whereas the corresponding DFT relaxation took 15 h on 32 cores in the original Materials Project calculations.
a, Distribution of the absolute percentage error in volumes of M3GNet-relaxed structures relative to DFT-relaxed structures. b, The differences between M3GNet-predicted energies (\(\hat{E}\)) and ground-state energies (Egs) using the initial M3GNet- and DFT-relaxed structures; Egs is defined as the DFT energy of the DFT-relaxed crystal. The horizontal dashed lines mark the 50th, 80th and 95th percentiles of the distributions (from the bottom to the top, respectively), and the corresponding x-axis values are annotated.
New materials discovery
The ability of M3GNet to accurately and rapidly relax arbitrary crystal structures, and predict their energies, makes it ideal for large-scale materials discovery. We generated 31,664,858 candidate structures as starting points (see Methods for details), used M3GNet IAP to relax the structures and calculated the signed energy distance to the Materials Project convex hull (Ehull-m); 1,849,096 materials have a Ehull-m of less than 0.01 eV per atom.
A formation energy model based on the Matbench33 Materials Project data was developed using the same architecture as the M3GNet IAP model (see Supplementary Table 3). Materials with a difference in the signed energy distance to the Materials Project convex hull from this model (Ehull-f) and a Ehull-m of greater than 0.2 eV per atom were discarded in the subsequent DFT analysis. This extra step removes materials with higher energy prediction uncertainties, which account for 13.2% (243,820) of the predicted materials. It should be noted that this step can also be omitted to simplify the discovery workflow, although potentially with an impact on the hit rate of stable materials discovery. The top-1,000 lowest Ehull-m materials from any chemistry, and the top-1,000 metal oxides with elements from the first five rows (excluding technetium due to radioactivity and rubidium due to high dominance), were then selected for validation via DFT relaxation and energy calculations. Only the most stable polymorphs were selected for each composition. It was found that the distribution in the DFT-calculated Ehull−dft matches well with the distributions of Ehull-m (Fig. 5a). For most computational materials discovery efforts, a positive threshold—typically around 0.05–0.1 eV per atom—is applied to identify synthesizable materials. This positive threshold accounts for both errors in DFT-calculated energies and the fact that some thermodynamically meta-stable materials can be realized experimentally. Of the top-1,000 materials from any chemistry, 999 were found to have a Ehull−dft of less than 0.001 eV per atom (Fig. 5b), and none of them were in the Materials Project database. For the top-1,000 oxides, 579, 826 and 935 were found to be synthesizable on the basis of Ehull−dft thresholds of 0.001, 0.05 and 0.1 eV per atom, respectively (Fig. 5b). Out of the 579 DFT-stable oxides, only five (namely, Mg4Nb2O9, Sr3V2O8, K2SnO2, Cd(RhO2)2 and CoMnO4) were previously known and matched with the Materials Project structures. The effectiveness of the M3GNet IAP relaxations can be seen in Supplementary Fig. 9, which shows that the energy changes during subsequent DFT relaxations (of the MEG3Net-relaxed structures) are at least one order of magnitude smaller than the energy changes during M3GNet relaxation. The final M3GNet-relaxed energies are in excellent agreement with the final DFT-relaxed energies, with MAEs of 0.112 and 0.045 eV per atom for the top-1,000 materials in the any-chemistry and oxide-chemistry categories, respectively (Fig. 5c,d). Using the M3GNet IAP, we have also assessed the dynamic stability of the 1,578 materials with a Ehull−dft of less than 0.001 eV per atom using phonon calculations. A total of 328 materials do not exhibit imaginary frequencies in their M3GNet phonon dispersion curves. Four phonon dispersion curves are shown in Extended Data Fig. 1; the others are provided in ‘Data availability’ section.
a, The signed Ehull distribution for the top-1,000 lowest Ehull-m materials from any chemistry and oxides only. b, Fraction of materials below Ehull−dft among top-1,000 materials in the all and oxides categories. c,d, Plot of the final M3GNet-predicted energy against final DFT energy for the all (c) and oxides (d) categories.
As a further evaluation of the performance of M3GNet for materials discovery, we computed the discovery rate, that is, the fraction of DFT-stable materials (Ehull−dft ≤ 0) for 1,000 structures uniformly sampled from the ~1.8 million materials with a Ehull-m of less than 0.001 eV per atom. The discovery rate remains close to 1.0 up to a Ehull-m threshold of around 0.5 eV per atom, and remains at a reasonably high value of 0.31 at the strictest threshold of 0.001 eV per atom, as shown in Supplementary Fig. 10. For this material set, we also compared the DFT relaxation time cost with and without M3GNet pre-relaxation. The results show that without M3GNet pre-relaxation, the DFT relaxation time cost is about three times of that with the M3GNet relaxation, as shown in Supplementary Fig. 11.
Discussion
A universal IAP such as M3GNet has applications beyond crystal structure relaxation and stability predictions. For instance, a common application of IAPs is in molecular dynamics simulations to obtain transport properties such as diffusivity and ionic conductivity. An example of a M3GNet application is in Supplementary Fig. 12 for Li3YCl6. Training an IAP for complex multicomponent systems such as Li3YCl6 is typically a highly involved process35, whereas the M3GNet IAP can be universally applied to any material without further retraining. For example, M3GNet molecular dynamics calculations could be applied to a wide range of lithium-containing compounds to identify potential lithium superionic conductors (Supplementary Fig. 13). Furthermore, the M3GNet IAP could also serve as a surrogate model in lieu of DFT with other structural exploration techniques (for example, evolutionary algorithms such as USPEX36 and CALYPSO37, or generative models such as CDVAE38) to generate more diverse and unconstrained candidates.
It should be noted that the current M3GNet IAP reported in this work is the best that can be obtained at present with the available data. Further improvements in accuracy can be achieved through several efforts. First, the training data for the M3GNet IAP come from DFT-relaxation calculations in the Materials Project, which were performed with less stringent convergence criteria such as a lower energy cut-off and sparser k-point grids. For IAP development, the best practice is to obtain accurate energies, forces and stresses via single-point, well-converged DFT calculations for training data. Building such a database is an extensive effort that is planned for future developments in the Materials Project. Second, active learning strategies (for instance, by using the DFT relaxation data from the M3GNet-predicted stable crystals in a feedback loop) can be used to systematically improve the M3GNet IAP, especially in underexplored chemical spaces with the greatest potential for materials discoveries. Nevertheless, about 1.8 million of the 31 million candidates were predicted to be potentially stable or meta-stable by M3GNet against materials in the Materials Project, which already expands the potential exploration pool by an order of magnitude over the ~140,000 crystals in the Materials Project database today. We shall note that the potentially stable materials will need to be further verified with DFT calculations and experimental syntheses. The model uncertainty will also play a role in further decreasing the number of true discoveries. Systematic methods for quantifying uncertainty are likely to further increase model fidelity.
Finally, the M3GNet framework is not limited to crystalline IAPs or even IAPs in general. The M3GNet formalism without lattice inputs and stress outputs is naturally suited for molecular force fields. When benchmarked on MD17 and MD17-CCSD(T) molecular force-field data (Supplementary Tables 4 and 5)39,40,41, the M3GNet models were found to be more accurate than the embedded atom neural network force field42, and to perform comparably with the state-of-the-art message-passing networks and equivariant neural network models. Moreover, by changing the readout section from summed atomic energy as in Fig. 1 to intensive property readout, the M3GNet framework can be used to develop surrogate models for property prediction. We trained M3GNet models on the Matbench materials data covering nine general crystal materials properties (Supplementary Table 3)33. In all cases, the M3GNet models achieved excellent accuracies.
Methods
Data source
The Materials Project performs a sequence of two relaxation calculations19 with the PBE43 generalized gradient approximation (GGA) functional or the GGA + U method44 for every unique input crystal, typically obtained from an experimental database such as the Inorganic Crystal Structure Database45. Our initial dataset comprises a sampling of the energies, forces and stresses from the first and middle ionic steps of the first relaxation and the last step of the second relaxation for calculations in the Materials Project database that contains GGA Structure Optimization or GGA + U Structure Optimization task types as of 8 February 2021. The snapshots that have a final energy per atom greater than 50 eV per atom or atom distance less than 0.5 Å were excluded as those tend to be the result of errors in the initial input structure.
This dataset is then split into the training, validation and test data in the ratio of 90%, 5% and 5%, respectively, according to materials not data points. Three independent data splits were performed.
Materials discovery methods
To generate initial materials candidates, combinatorial isovalent ionic substitutions based on the common oxidation states of non-noble-gas element were performed on 5,283 binary, ternary and quaternary structural prototypes in the 2019 version of the ICSD45 database. Only prototypes with less than 51 atoms were selected for computational speed considerations. Further filtering was performed to exclude structures with non-integer or zero-charged atoms. A total of 31,664,858 hypothetical materials candidates were generated, more than 200 times the total number of unique crystals in the Materials Project today. The candidate space contains 294,643 chemical systems, whereas the Materials Project has only about 47,000 chemical systems. This represents a quantity and chemical diversity of materials that is inaccessible using current DFT or other IAP implementations.
All structures were relaxed using the M3GNet model and their signed energy distance to the Materials Project convex hull were calculated using the M3GNet IAP-predicted energy (Ehull-m). We acknowledge that some of the generated structures may compete with each other for stability; however, to avoid introducing additional uncertainties into the Ehull-m predictions, we have elected to compute Ehull-m relative to ground-truth DFT energies in the Materials Project as opposed to the higher uncertainty M3GNet-computed energies. A zero or negative Ehull means that the material is predicted to be potentially stable compared to known materials in MP. In total, 1,849,096 materials have Ehull-m less than 0.001 eV per atom. We then excluded materials that have non-metal ions in multiple valence states, for example, materials containing Br+ and Br− at the same time and so on. It is well-known that PBE overbinds single-element molecules such as O2, S8, Cl2 and so on, and negative anion energy corrections are applied to ionic compounds in Materials Project to offset such errors.46 However the corrections are based mostly on composition, which may artificially overstabilize materials with multivalence non-metal ions. We have developed a searchable database for the generated hypothetical structures and their corresponding M3GNet-predicted properties at http://matterverse.ai.
Model construction
Neural network definition
If we denote one layer of the perceptron model as
then the K-layer MLP can be expressed as
The K-layer gated MLP becomes
where \({{{{\mathcal{L}}}}}_{\sigma }^{K}(x)\) replaces the activation function g(x) of \({{{{\mathcal{L}}}}}_{g}^{K}(x)\) to sigmoid function σ(x), and ⊙ denotes the element-wise product. The gated MLP comprises the normal MLP before ⊙ and the gate network after ⊙.
Model architecture
The neighborhood of atom i is denoted as \({{{{\mathcal{N}}}}}_{i}\). We consider all other bonds emanating from atom i when calculating the bond interaction of eij. To incorporate n-body interactions, each eij is updated using all distinct combinations of n − 2 neighbors in \({{{{\mathcal{N}}}}}_{i}\) excluding atom j (that is, \({{{{\mathcal{N}}}}}_{i}/j\)), denoted generally as follows:
where ϕn is the update function and rik is the vector pointing from atoms i to k. In practice, this n-body information exchange involves the calculation of distances, angles, dihedral angles, improper angles and so on, which escalates combinatorially with the order n as (Mi − 1)!/(Mi − n + 1)!, where Mi is the number of neighbors in \({{{{\mathcal{N}}}}}_{i}\). We will denote this materials graph with an n-body interactions neural network as MnGNet. In this work, we will focus on the incorporation of three-body interactions only (M3GNet).
Let θjik denote the angle between bonds eij and eik. Here we expand the three-body angular interactions using an efficient complete and orthogonal spherical Bessel function and spherical harmonics basis set, as proposed by Klicpera and colleagues11. The bond update equation can then be rewritten as:
where W and b are learnable weights from the network; jl is the spherical Bessel function with the roots at zln, i.e., jl(zln) = 0; \({Y}_{l}^{0}\) is the spherical harmonics function; σ is the sigmoid activation function; \({f}_{\mathrm{c}}(r)=1-6{(r/{r}_{\mathrm{c}})}^{5}+15{(r/{r}_{\mathrm{c}})}^{4}-10{(r/{r}_{\mathrm{c}})}^{3}\) is the cut-off function ensuring that the functions vanished smoothly at the neighbor boundary47; g(x) = xσ(x) is the nonlinear activation function48; and \({\tilde{{{{\textbf{e}}}}}}_{ij}\) is a vector of length nmaxlmax, expanded by indices l = 0, 1, ... , lmax − 1 and n = 0, 1, ... , nmax − 1.
Following the n-body interaction update, several graph convolution steps are carried out sequentially to update the bond, atom and—optionally—the state information, as follows:
where ϕe(x) and \({\phi }_{e}^{\prime}(x)\) are gated MLPs, as in equation (3); ⊕ is the concatenation operator; Nv is the number of atoms; and \({{{{\boldsymbol{e}}}}}_{ij}^{0}\) represents the distance-expanded basis functions with the target values, and the first and second derivatives smoothly going towards zero at the cut-off boundary (see Methods). Such a design ensures that the target values and their derivatives up to second order change smoothly with changes in the number of bonds; u inputs and updates are optional to the models as not all structures or models have state attributes.
Materials graphs were constructed using a radial cut-off of 5 Å. For computational efficiency considerations, the three-body interactions were limited to within a cut-off of 4 Å. The graph featurizer converts the atomic number into embeddings of dimension 64. The bond distances were expanded using the continuous and smooth basis function proposed by Kocer et al.49, which ensures that the first and second derivatives vanish at the cut-off radius.
where
\({{{{\textbf{e}}}}}_{ij}^{0}\) is a vector formed by m basis functions of h(r).
In this work, we used three basis functions for the pair distance expansion.
The main blocks consist of three three-body information exchange and graph convolutions (N = 3 in Fig. 1). By default, the values of W and b in equations (5–9) give output dimensions of 64. Each gated MLP (ϕe(x) and \({\phi }_{e}^{\prime}(x)\) in equations (7) and (8)) have two layers with 64 neurons in each layer.
For the prediction of extensive properties such as total energies, a three-layer gated MLP (equation (3)) was used on the atom attributes after the graph convolution and sum the outputs as the final prediction, that is,
The gated MLP ϕ3(x) operating on node attributes vi has a layer neuron configuration of [64, 64, 1] and no activation in the last layer of the normal MLP part.
For the prediction of intensive properties, the readout step was performed as follows by including optional state information u after the main blocks.
with weights wi summing to 1 and defined as
ξ3 and \({\xi }_{3}^{\prime}\) have neuron configurations of [64, 64, 1] to ensure the output is scalar. There is no activation in the final layer of the MLP for regression targets, whereas, for classification targets, the last layer activation is chosen as the sigmoid function.
In the training of MPF.2021.2.8 data, the M3GNet model comprises three main blocks with 227,549 learnable weights.
Model training
The Adam optimizer50 was used with an initial learning rate of 0.001, with a cosine decay to 1% of the original value in 100 epochs. During optimization, the validation metric values were used to monitor the model convergence, and training was stopped if the validation metric did not improve for 200 epochs. For the elemental IAP training, the loss function was the mean squared error. For other properties, we used the Huber loss function51 with δ set to 0.01. For universal IAP training, the total loss function includes the loss for energy, forces and—in inorganic compounds—the stresses. A batch size of 32 was used in model training.
where ℓ is the Huber loss function, e is energy per atom and w are the scalar weights. The DFT subscripts indicate data from DFT.
Before M3GNet IAP fitting, we fit the elemental reference energies using linear regression of the total energies. We first featurize a composition into a vector c = [c1, c2, c3, ... , c89] where ci is the number of atoms in the composition with the atomic number i. The composition feature vector c is mapped to the total energy of the material E via E = ∑iciEi, where Ei is the reference energy for an element with atomic number i that can be obtained by linear regression of the training data. The elemental reference energies were then subtracted from the total energies to improve M3GNet model training stability. We set wf = 1 and wσ = 0.1 during training the MPF.2021.2.8 data.
Software implementation
The M3GNet framework was implemented using the TensorFlow52 package and currently runs on TensorFlow v.2.9.1. All crystal and molecular structure processing were performed using the Python Materials Genomics (pymatgen)16 v.2020.12.31. The structural optimization was performed using the FIRE53 algorithm implemented in the atomic simulation environment (ASE) v.3.22.0 (ref. 54). The molecular dynamics simulations were performed in the NVT ensemble using ASE (ref. 54). Phonon calculations were performed using the Phonopy package v.2.10.0 (ref. 55). Data analysis and visualization were performed using scikit-learn v.0.24.2 (ref. 56), statsmodels v.0.12.2 (ref. 57), matplotlib v.3.3.0 (ref. 58), seaborn v.0.11.2 (ref. 59) and pandas v.1.3.1 (ref. 60).
Data availability
The training data for the universal IAP are available at https://doi.org/10.6084/m9.figshare.19470599 (ref. 61). The phonon dispersion curves of 328 dynamically stable materials are available at https://doi.org/10.6084/m9.figshare.20217212 (ref. 62). The ICSD database used in this study is a commercial product and cannot be shared. All generated hypothetical compounds and their corresponding M3GNet predictions are provided at http://matterverse.ai. Each material can be accessed via a detail page at https://matterverse.ai/details/mv-id, where id ranges from 0 to 31,664,854. Source Data are provided with this paper.
Code availability
The source code for M3GNet is available at https://github.com/materialsvirtuallab/m3gnet and https://doi.org/10.5281/zenodo.7141391 (ref. 63).
References
Weiner, P. K. & Kollman, P. A. AMBER: assisted model building with energy refinement. A general program for modeling molecules and their interactions. J. Comput. Chem. 2, 287–303 (1981).
Case, D. A. et al. The AMBER biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Rappe, A. K., Casewit, C. J., Colwell, K. S., Goddard, W. A. & Skiff, W. M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 114, 10024–10035 (1992).
Behler, J. & Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 98, 146401 (2007).
Bartók, A. P., Payne, M. C., Kondor, R. & Csányi, G. Gaussian approximation potentials: the accuracy of quantum mechanics, without the electrons. Phys. Rev. Lett. 104, 136403 (2010).
Thompson, A. P., Swiler, L. P., Trott, C. R., Foiles, S. M. & Tucker, G. J. Spectral neighbor analysis method for automated generation of quantum-accurate interatomic potentials. J. Comput. Phys. 285, 316–330 (2015).
Shapeev, A. V. Moment tensor potentials: a class of systematically improvable interatomic potentials. Multiscale Model. Simul. 14, 1153–1173 (2016).
Zhang, L., Han, J., Wang, H., Car, R. & E, W. Deep potential molecular dynamics: a scalable model with the accuracy of quantum mechanics. Phys. Rev. Lett. 120, 143001 (2018).
Zuo, Y. et al. Performance and cost assessment of machine learning interatomic potentials. J. Phys. Chem. A 124, 731–745 (2020).
Schütt, K. et al. SchNet: a continuous-filter convolutional neural network for modeling quantum interactions. In Proc. 31st International Conference on Neural Information Processing Systems Advances in Neural Information Processing Systems 992–1002 (NIPS, 2017).
Klicpera, J., Groß, J. & Günnemann, S. Directional message passing for molecular graphs. Preprint at https://arxiv.org/abs/2003.03123 (2020).
Haghighatlari, M. et al. NewtonNet: a Newtonian message passing network for deep learning of interatomic potentials and forces. Digit. Discov. 1, 333–343 (2022).
Park, C. W. et al. Accurate and scalable graph neural network force field and molecular dynamics with direct force architecture. npj Comput. Mater. 7, 1–9 (2021).
Cheon, G., Yang, L., McCloskey, K., Reed, E. J. & Cubuk, E. D. Crystal structure search with random relaxations using graph networks. Preprint at https://arxiv.org/abs/2012.02920 (2020).
Lejaeghere, K. et al. Reproducibility in density functional theory calculations of solids. Science 351, aad3000 (2016).
Ong, S. P. et al. Python Materials Genomics (pymatgen): a robust, open-source Python library for materials analysis. Comput. Mater. Sci. 68, 314–319 (2013).
Jain, A. et al. FireWorks: a dynamic workflow system designed for high-throughput applications. Concurr. Comput. 27, 5037–5059 (2015).
Pizzi, G., Cepellotti, A., Sabatini, R., Marzari, N. & Kozinsky, B. AiiDA: automated interactive infrastructure and database for computational science. Comput. Mater. Sci. 111, 218–230 (2016).
Mathew, K. et al. Atomate: a high-level interface to generate, execute, and analyze computational materials science workflows. Comput. Mater. Sci. 139, 140–152 (2017).
Jain, A. et al. Commentary: the materials project: a materials genome approach to accelerating materials innovation. APL Mater. 1, 011002 (2013).
Curtarolo, S. et al. AFLOWLIB.ORG: a distributed materials properties repository from high-throughput ab initio calculations. Comput. Mater. Sci. 58, 227–235 (2012).
Kirklin, S. et al. The Open Quantum Materials Database (OQMD): assessing the accuracy of DFT formation energies. npj Comput. Mater. 1, 1–15 (2015).
Draxl, C. & Scheffler, M. The NOMAD laboratory: from data sharing to artificial intelligence. J. Phys. Mater. 2, 036001 (2019).
Xie, T. & Grossman, J. C. Crystal graph convolutional neural networks for an accurate and interpretable prediction of material properties. Phys. Rev. Lett. 120, 145301 (2018).
Chen, C., Ye, W., Zuo, Y., Zheng, C. & Ong, S. P. Graph networks as a universal machine learning framework for molecules and crystals. Chem. Mater. 31, 3564–3572 (2019).
Chen, C., Zuo, Y., Ye, W., Li, X. & Ong, S. P. Learning properties of ordered and disordered materials from multi-fidelity data. Nat. Comput. Sci. 1, 46–53 (2021).
Choudhary, K. & DeCost, B. Atomistic line graph neural network for improved materials property predictions. npj Comput. Mater. 7, 1–8 (2021).
Tersoff, J. New empirical approach for the structure and energy of covalent systems. Phys. Rev. B 37, 6991–7000 (1988).
Petretto, G. et al. High-throughput density-functional perturbation theory phonons for inorganic materials. Sci. Data 5, 180065 (2018).
Perdew, J. P. et al. Restoring the density-gradient expansion for exchange in solids and surfaces. Phys. Rev. Lett. 100, 136406 (2008).
Kresse, G. & Hafner, J. Ab initio molecular dynamics for liquid metals. Phys. Rev. B 47, 558–561 (1993).
Kresse, G. & Furthmüller, J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comput. Mater. Sci. 6, 15–50 (1996).
Dunn, A., Wang, Q., Ganose, A., Dopp, D. & Jain, A. Benchmarking materials property prediction methods: the Matbench test set and Automatminer reference algorithm. npj Comput. Mater. 6, 1–10 (2020).
Sun, W. et al. The thermodynamic scale of inorganic crystalline metastability. Sci. Adv. 2, e160022 (2016).
Qi, J. et al. Bridging the gap between simulated and experimental ionic conductivities in lithium superionic conductors. Mater. Today Phys. 21, 100463 (2021).
Glass, C. W., Oganov, A. R. & Hansen, N. USPEX—evolutionary crystal structure prediction. Comput. Phys. Commun. 175, 713–720 (2006).
Wang, Y., Lv, J., Zhu, L. & Ma, Y. CALYPSO: a method for crystal structure prediction. Comput. Phys. Commun. 183, 2063–2070 (2012).
Xie, T., Fu, X., Ganea, O.-E., Barzilay, R. & Jaakkola, T. Crystal diffusion variational autoencoder for periodic material generation. Preprint at https://arxiv.org/abs/2110.06197 (2021).
Chmiela, S. et al. Machine learning of accurate energy-conserving molecular force fields. Sci. Adv. 3, e1603015 (2017).
Schütt, K. T., Arbabzadah, F., Chmiela, S., Müller, K. R. & Tkatchenko, A. Quantum-chemical insights from deep tensor neural networks. Nat. Commun. 8, 13890 (2017).
Chmiela, S., Sauceda, H. E., Müller, K.-R. & Tkatchenko, A. Towards exact molecular dynamics simulations with machine-learned force fields. Nat. Commun. 9, 3887 (2018).
Zhang, Y., Hu, C. & Jiang, B. Embedded atom neural network potentials: efficient and accurate machine learning with a physically inspired representation. J. Phys. Chem. Lett. 10, 4962–4967 (2019).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 77, 3865–3868 (1996).
Anisimov, V. I., Zaanen, J. & Andersen, O. K. Band theory and Mott insulators: Hubbard U instead of stoner I. Phys. Rev. B 44, 943–954 (1991).
Hellenbrandt, M. The Inorganic Crystal Structure Database (ICSD)—present and future. Crystallogr. Rev. 10, 17–22 (2004).
Wang, L., Maxisch, T. & Ceder, G. Oxidation energies of transition metal oxides within the GGU+U framework. Phys. Rev. B 73, 195107 (2006).
Singraber, A., Behler, J. & Dellago, C. Library-based LAMMPS implementation of high-dimensional neural network potentials. J. Chem. Theory Comput. 15, 1827–1840 (2019).
Ramachandran, P., Zoph, B. & Le, Q. V. Searching for activation functions. Preprint at https://arxiv.org/abs/1710.05941 (2017).
Kocer, E., Mason, J. K. & Erturk, H. A novel approach to describe chemical environments in high-dimensional neural network potentials. J. Chem. Phys. 150, 154102 (2019).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint https://arxiv.org/abs/1412.6980 (2017).
Huber, P. J. Robust estimation of a location parameter. Ann. Math. Stat. 35, 73–101 (1964).
Abadi, M. et al. TensorFlow: a system for large-scale machine learning. In 12th Symposium on Operating Systems Design and Implementation 265–283 (OSDI, 2016).
Bitzek, E., Koskinen, P., Gähler, F., Moseler, M. & Gumbsch, P. Structural relaxation made simple. Phys. Rev. Lett. 97, 170201 (2006).
Larsen, A. H. et al. The atomic simulation environment—a Python library for working with atoms. J. Phys. 29, 273002 (2017).
Togo, A. & Tanaka, I. First principles phonon calculations in materials science. Scripta Materialia 108, 1–5 (2015).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Seabold, S. & Perktold, J. Statsmodels: econometric and statistical modeling with Python. In Proc. 9th Python in Science Conference 92–96 (SciPy, 2010).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Waskom, M. L. Seaborn: statistical data visualization. J. Open Source Softw. 6, 3021 (2021).
Pandas Development Team Pandas-Dev/Pandas: Pandas (Zenodo, 2020).
Chen, C. & Ong, S. P. MPF.2021.2.8. (FigShare, 2022); https://doi.org/10.6084/m9.figshare.19470599.v3
Chen, C. m3gnet Phonon Dispersion Curve of 328 Materials (FigShare, 2022); https://doi.org/10.6084/m9.figshare.20217212.v1
Ong, S. P. et al. materialsvirtuallab/m3gnet: v0.1.0. 2022 (FigShare, 2022); https://doi.org/10.5281/zenodo.7141391
de Jong, M. et al. Charting the complete elastic properties of inorganic crystalline compounds. Sci. Data 2, 150009 (2015).
Acknowledgements
This work was primarily supported by the Materials Project, funded by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, Materials Sciences and Engineering Division under contract no. DE-AC02-05-CH11231: Materials Project program KC23MP. The lithium superionic conductor analysis portion of the work was funded by the LG Energy Solution through the Frontier Research Laboratory (FRL) Program. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (grant no ACI-1548562). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
C.C. and S.P.O. conceived the idea and designed the work. C.C. implemented the models and performed the analysis. C.C. and S.P.O. wrote the manuscript and contributed to the discussion and revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no financial interests.
Peer review
Peer review information
Nature Computational Science thanks Ekin Cubuk and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Handling editor: Jie Pan, in collaboration with the Nature Computational Science team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 M3GNet-calculated phonon dispersion curves of four new materials predicted to be thermodynamically and dynamically stable.
a, Sr6Sc2Al4O15; b, K2Li3AlO4; c, KMN4V2O12; d, MnCd(GAO2)4.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13, Sections 1–7 and Tables 1–5.
Source data
Source Data Fig. 2
Sheet Figure_2a: 2D energy-force distribution counts Sheet Figure_2b: 2D energy-stress distribution counts Sheet Figure_2c: Radial distribution Sheet Figure_2d: Element counts in the dataset.
Source Data Fig. 3
Sheet Figure_3a: DFT energy vs M3GNet energy Sheet Figure_3b: DFT force vs M3GNet force Sheet Figure_3c: DFT stress vs M3GNet stress Sheet Figure_3g: DFT DOS center vs M3GNet DOS center Sheet Figure_3h: DFT Debye temperature vs M3GNet Debye temperature.
Source Data Fig. 4
Sheet Figure_4a: Volume change with model relax vs no model relax Sheet Figure_4b: Energy difference between M3GNet energies and DFT ground-state energies using DFT initial structures, M3GNet-relaxed structures and DFT-relaxed structures.
Source Data Fig. 5
Sheet Figure_5a: Signed Ehull of M3GNet and DFT for all cases and oxide cases Sheet Figure_5b: Stable materials ratio for all and oxide cases as a function of DFT ehull thresholds. Sheet Figure_5c: M3GNet energy vs DFT energy for the all cases. Sheet Figure_5d: M3GNet energy vs DFT energy for the oxide cases.
Source Data Extended Data Fig. 1
Sheet a-special_points: special high symmetry points of phonon dispersion curve and their high symmetry labels Sheet a-high_symmetry_line-{d}: distance and the frequency for different vibration modes. d goes from 0 to the max number of symmetry lines considered for this phonon dispersion curve. Panels b, c, d are stored in Sheet {x}-special points, Sheet {x}-high_symmetry_line-{d}, where x = b, c, or d.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, C., Ong, S.P. A universal graph deep learning interatomic potential for the periodic table. Nat Comput Sci 2, 718–728 (2022). https://doi.org/10.1038/s43588-022-00349-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-022-00349-3
This article is cited by
-
The rise of high-entropy battery materials
Nature Communications (2024)
-
Robust training of machine learning interatomic potentials with dimensionality reduction and stratified sampling
npj Computational Materials (2024)
-
Exploring the frontiers of condensed-phase chemistry with a general reactive machine learning potential
Nature Chemistry (2024)
-
MLMD: a programming-free AI platform to predict and design materials
npj Computational Materials (2024)
-
Towards atom-level understanding of metal oxide catalysts for the oxygen evolution reaction with machine learning
npj Computational Materials (2024)