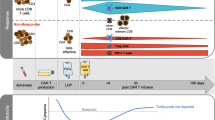
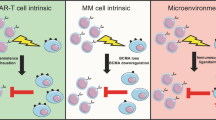
Abstract
Markers that predict response and resistance to chimeric antigen receptor (CAR) T cells in relapsed/refractory multiple myeloma are currently missing. We subjected mononuclear cells isolated from peripheral blood and bone marrow before and after the application of approved B cell maturation antigen-directed CAR T cells to single-cell multiomic analyses to identify markers associated with resistance and early relapse. Differences between responders and nonresponders were identified at the time of leukapheresis. Nonresponders showed an immunosuppressive microenvironment characterized by increased numbers of monocytes expressing the immune checkpoint molecule CD39 and suppressed CD8+ T cell and natural killer cell function. Analysis of CAR T cells showed cytotoxic and exhausted phenotypes in hyperexpanded clones compared to low/intermediate expanded clones. We identified potential immunotherapy targets on CAR T cells, like PD1, to improve their functionality and durability. Our work provides evidence that an immunosuppressive microenvironment causes resistance to CAR T cell therapies in multiple myeloma.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq and scRNA-seq, BCR sequencing and TCR sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE234261. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
Processing and analysis code related to this study is deposited in the GitHub repository at https://github.com/fraunhofer-izi/Rade_Grieb_et_al_2023.
References
Merz, M. et al. Adjusted comparison of outcomes between patients from CARTITUDE-1 versus multiple Myeloma Patients with Prior Exposure to PI, Imid and Anti-CD-38 from a German Registry. Cancers 13, 5996 (2021).
Munshi, N. C. et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N. Engl. J. Med. 384, 705–716 (2021).
Berdeja, J. G. et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): a phase 1b/2 open-label study. Lancet 398, 314–324 (2021).
Martin, T. et al. Ciltacabtagene autoleucel, an anti-B-cell maturation antigen chimeric antigen receptor T-cell therapy, for relapsed/refractory multiple myeloma: CARTITUDE-1 2-year follow-up. J. Clin. Oncol. 41, 1265–1274 (2022).
Da Vià, M. C. et al. Homozygous BCMA gene deletion in response to anti-BCMA CAR T cells in a patient with multiple myeloma. Nat. Med. 27, 616–619 (2021).
Samur, M. K. et al. Biallelic loss of BCMA as a resistance mechanism to CAR T cell therapy in a patient with multiple myeloma. Nat. Commun. 12, 868 (2021).
Gagelmann, N. et al. Access to and affordability of CAR T-cell therapy in multiple myeloma: an EBMT position paper. Lancet Haematol. 9, e786–e795 (2022).
Gazeau, N. et al. Effective anti-BCMA retreatment in multiple myeloma. Blood Adv. 5, 3016–3020 (2021).
Deng, H. et al. Efficacy of humanized anti-BCMA CAR T cell therapy in relapsed/refractory multiple myeloma patients with and without extramedullary disease. Front. Immunol. 12, 720571 (2021).
Haradhvala, N. J. et al. Distinct cellular dynamics associated with response to CAR-T therapy for refractory B cell lymphoma. Nat. Med. 28, 1848–1859 (2022).
Sheih, A. et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat. Commun. 11, 219 (2020).
Bai, Z. et al. Single-cell antigen-specific landscape of CAR T infusion product identifies determinants of CD19-positive relapse in patients with ALL. Sci. Adv. 8, eabj2820 (2022).
Boiarsky, R. et al. Single cell characterization of myeloma and its precursor conditions reveals transcriptional signatures of early tumorigenesis. Nat. Commun. 13, 7040 (2022).
Cohen, Y. C. et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat. Med. 27, 491–503 (2021).
Dutta, A. K. et al. Single-cell profiling of tumour evolution in multiple myeloma—opportunities for precision medicine. Nat. Rev. Clin. Oncol. 19, 223–236 (2022).
Ledergor, G. et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 24, 1867–1876 (2018).
Merz, M. et al. Deciphering spatial genomic heterogeneity at a single cell resolution in multiple myeloma. Nat. Commun. 13, 807 (2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Phipson, B. et al. propeller: testing for differences in cell type proportions in single cell data. Bioinformatics 38, 4720–4726 (2022).
van der Leun, A. M., Thommen, D. S. & Schumacher, T. N. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer 20, 218–232 (2020).
Cohen, A. D. et al. B cell maturation antigen-specific CAR T cells are clinically active in multiple myeloma. J. Clin. Invest. 129, 2210–2221 (2019).
Nakamura, K., Smyth, M. J. & Martinet, L. Cancer immunoediting and immune dysregulation in multiple myeloma. Blood 136, 2731–2740 (2020).
Merz, M. et al. Spatiotemporal assessment of immunogenomic heterogeneity in multiple myeloma. Blood Adv. 7, 718–733 (2022).
Zavidij, O. et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat. Cancer 1, 493–506 (2020).
Clements, A. N. & Warfel, N. A. Targeting PIM kinases to improve the efficacy of immunotherapy. Cells 11, 3700 (2022).
Keane, N. A., Reidy, M., Natoni, A., Raab, M. S. & O’Dwyer, M. Targeting the Pim kinases in multiple myeloma. Blood Cancer J. 5, e325 (2015).
Chatterjee, S. et al. Targeting PIM kinase with PD1 inhibition improves immunotherapeutic antitumor T-cell response. Clin. Cancer Res. 25, 1036–1049 (2019).
Moesta, A. K., Li, X.-Y. & Smyth, M. J. Targeting CD39 in cancer. Nat. Rev. Immunol. 20, 739–755 (2020).
Vucinic, V. et al. S287: factors influencing autologous lymphocyte collections for chimeric antigen receptor (CAR) T-cells—the role of T-cell senescence. HemaSphere 6, 188 (2022).
Chen, P.-H. et al. Activation of CAR and non-CAR T cells within the tumor microenvironment following CAR T cell therapy. JCI Insight 5, e134612 (2020).
Dhodapkar, K. M. et al. Changes in bone marrow tumor and immune cells correlate with durability of remissions following BCMA CAR T therapy in myeloma. Blood Cancer Discov. 3, 490–501 (2022).
Mathewson, N. D. et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 184, 1281–1298 (2021).
Melenhorst, J. J. et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature 602, 503–509 (2022).
Wang, B. et al. Chimeric antigen receptor T cell therapy in the relapsed or refractory multiple myeloma with extramedullary disease—a single institution observation in China. Blood 136, 6 (2020).
García-Guerrero, E. et al. All-trans retinoic acid works synergistically with the γ-secretase inhibitor crenigacestat to augment BCMA on multiple myeloma and the efficacy of BCMA-CAR T cells. Haematologica 108, 568–580 (2023).
Merz, M. et al. Cytogenetic subclone formation and evolution in progressive smoldering multiple myeloma. Leukemia 34, 1192–1196 (2020).
Merz, M. et al. Prognostic significance of cytogenetic heterogeneity in patients with newly diagnosed multiple myeloma. Blood Adv. 2, 1–9 (2017).
Kumar, S. et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 17, e328–e346 (2016).
Campbell, T. et al. Uses of anti-BCMA chimeric antigen receptors. Worldwide patent WO2021091978A1 (2021).
Schecter, J. M. & Fan, X. BCMA-targeted CAR-T cell therapy for multiple myeloma. Worldwide patent WO2022116086A1 (2022).
Germain, P.-L., Lun, A., Meixide, C. G., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 10, 979 (2022).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Koh, W. & Hoon, S. MapCell: learning a comparative cell type distance metric with Siamese neural nets with applications toward cell-type identification across experimental datasets. Front. Cell Dev. Biol. 9, 767897 (2021).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Andreatta, M. et al. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 12, 2965 (2021).
Andreatta, M., Berenstein, A. J. & Carmona, S. J. scGate: marker-based purification of cell types from heterogeneous single-cell RNA-seq datasets. Bioinformatics 38, 2642–2644 (2022).
Fu, R. et al. clustifyr: an R package for automated single-cell RNA sequencing cluster classification. F1000Res. 9, 223 (2020).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res. 9, 47 (2020).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Tickle, T., Tirosh, I., Brown, M. & Haas, B. InferCNV: inferring copy number alterations from tumor single cell RNA-seq data. GitHub https://github.com/broadinstitute/inferCNV/wiki (2023).
Wang, Y. et al. iTALK: an R package to characterize and illustrate intercellular communication. Preprint at bioRxiv https://doi.org/10.1101/507871 (2019).
Dimitrov, D. et al. Comparison of methods and resources for cell–cell communication inference from single-cell RNA-seq data. Nat. Commun. 13, 3224 (2022).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Blache, U. et al. Advanced flow cytometry assays for immune monitoring of CAR-T cell applications. Front. Immunol. 12, 658314 (2021).
Acknowledgements
We thank S. Scharf from the Leipzig Medical Biobank for handling cryopreserved samples and D. Bretschneider, C. Mueller, K. Wildenberger and C. Wilhelm for performing fluorescence in situ hybridization analyses on clinical samples. We express our gratitude toward the participants and their families who participated in this analysis. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SPP µbone) and EU HORIZON Project CERTAINTY (101136379), and M.M. received financial support from grants from the International Myeloma Society, SpringWorks and Janssen.
Author information
Authors and Affiliations
Contributions
Conception and design: M.R., N.G., R.W., V.V. and M.M. Acquisition of data (acquired and managed participants, provided facilities, fluorescence in situ hybridization, flow cytometry, biobanking, in vitro studies and so on): L.F., P.B., A.B., S.F., P.F., C.K., S. Heyn, A.S.K., R.B., S.Y.W., E.B., S. Hoffmann, J.U., B.S., S. Hell, M.J., S.S., K.H.M., G.-N.F., M.H., U.S., K.R., U.K., U.P., V.V. and M.M. Analysis and interpretation of data (for example, statistical analysis, biostatistics and computational analysis): M.R., N.G., R.W., J. Sia, S.X., L.F., J. Scolnick, L.V., K.R., V. V. and M.M. Writing, review and/or revision of the manuscript: all authors.
Corresponding author
Ethics declarations
Competing interests
S.F.: consultant and/or speaker fees from Novartis Pharma, Janssen-Cilag, Vertex Pharmaceuticals (Germany), Kite/Gilead Sciences, MSGO and Bristol-Myers Squibb. U.K.: consultant and/or speaker fees from AstraZeneca, Affimed, Glycostem, GammaDelta, Zelluna, Miltenyi Biotec and Novartis Pharma and Bristol-Myers Squibb. M.M.: advisory boards/honoraria/research support from Amgen, BMS, Celgene, Gilead, Janssen, Stemline, Springworks and Takeda. K.H.M.: BMS (consultancy and honoraria), AbbVie (honoraria, research funding), Pfizer (honoraria), Otsuka (honoraria), Janssen (honoraria) and Novartis (consultancy). U.P.: Syros (consultancy, honoraria, research funding), MDS Foundation (membership on an entity’s Board of Directors or advisory committees), Silence Therapeutics (consultancy, honoraria, research funding), Celgene (honoraria), Takeda (consultancy, honoraria, research funding), Fibrogen (research funding), Servier (consultancy, honoraria, research funding), Roche (research funding), Merck (research funding), Amgen (consultancy, research funding), Novartis (consultancy, honoraria, research funding), AbbVie (consultancy), Curis (consultancy, research funding), Janssen Biotech (consultancy, research funding), Jazz (consultancy, honoraria, research funding), BeiGene (research funding), Geron (consultancy, research funding) and Bristol-Myers Squibb (consultancy, honoraria, membership on an entity’s Board of Directors or advisory committees, other, travel support, medical writing support, research funding). M.J.: Novartis (honoraria), Amgen (honoraria), Pfizer (honoraria), Blueprint Medicine (honoraria), BMS (honoraria) and Jazz (honoraria). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks Marco Davila and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Enrichment analysis for post-infusional BMMCs and PBMCs.
GO term enrichment analysis of significantly differentially expressed genes (non-responders vs. responders) for BMMCs and PBMCs. Terms are ranked by rich factor, which is the number of DE genes in the term divided by the number of background genes in that term. Dot plot depict the highest ranked significantly enriched GO terms for biological processes (FDR < 0.05). The dot size indicates the z-score, which is the number of DE genes with logFC >0 minus the number of DE genes with logFC <0 divided by the square root of the number of term-associated genes. Grey/white dots indicate the same number of genes with a logFC >0 and <0. Cell type abbreviations: NK = natural killer; Mono = monocyte; cDC = classical dendritic cell.
Extended Data Fig. 2 Comparing post- and pre-infusional BMMCs and PBMCs in nonCR and CR.
a, Comparison between post- and pre-infusional cell types in PBMCs from responders (CR) and non-responders (nonCR). The number of biologically independent samples for the contrasts is as follows. CR: n = 4 for post- and n = 5 for pre-infusional groups. nonCR: n = 5 for post- and n = 5 for pre-infusional groups. Significant differences (unadjusted p-values) were estimated using the empirical Bayes moderated t-statistics (two-sided) implemented in the speckle package (* p = 0.1, ** p = 0,05, *** p = 0.01, **** p = 0.001, p < 0.0001 = *****). The exact p-values for CR are as follows: NK (p = 0.00210), NK_CD56bright (p = 0.01849), B memory (p = 0.08501) and pDC (p = 0,02643), B intermediate (p = 0.074792) and HSPC (p = 0,09701). The exact p-value for nonCR, is as follows: CD14 Mono (p = 0.03526). The center of the box plots indicates the median, and the upper and lower bounds of the boxes indicate first and third interquartile ranges. Whiskers extends to 1.5 x interquartile range. b, Differential gene expression analysis comparing post- with pre-infusional BMMCs and PBMCs in CR and nonCR Only cell types with significant genes are shown (FDR < 0.05 and fold change <1.5 or >1.5). A positive log fold change indicates upregulation in post-infusional samples. c, GO term enrichment analysis of significantly differentially expressed genes (post- vs. pre-infusional). Terms are ranked by rich factor, which is the number of DE genes in the term divided by the number of background genes in that term. Dot plots depict the highest ranked significantly enriched GO terms for biological processes (FDR < 0.05). The dot size indicates the z-score, which is the number of DE genes with logFC >0 minus the number of DE genes with logFC <0 divided by the square root of the number of term-associated genes. Grey/white dots indicate the same number of genes with a logFC >0 and <0. Cell type abbreviations: NK = natural killer; gdT-Cell = gamma delta T cell; Mono = monocyte; cDC = classical dendritic cell; pDC = plasmacytoid dendritic cell; Cilta cel = Ciltacabtagene autoleucel; Ide cel = Idecabtagene vicleucel; GMP = granulocyte-monocyte progenitor, HSPC = Hematopoietic stem and progenitor cell.
Extended Data Fig. 3 Comparing post- with pre-infusional T cell subtypes in nonCR and CR.
a, Differences in T cell subtype composition between post- and pre-infusional PBMCs from CR and nonCR. For each cell type, the log fold change in mean cell fraction between post- and pre-infusional samples was calculated with the R package speckle based on arcsin square root transformation. The cell fraction calculation includes all cell types (the denominator is the sum of all cells analyzed). For clarity, only cell types with a fold change >1.5 are shown. b, GO term enrichment analysis for DEGs comparing post- with pre-infusional PBMCs in CR and nonCR. GO terms are ranked by rich factor, which is the number of DE genes in the term divided by the number of background genes in that term. Dot plot depict the highest ranked significantly enriched GO terms for biological processes (FDR < 0.05). Cell type abbreviations: CM = central memory; EM = effector memory; TEMRA = effector memory cells re-expressing CD45RA; TPEX = precursor exhausted T cells; TEX = exhausted T cell; MAIT = Mucosal-associated invariant T cell; CTL_EOMES = eomesodermin expressing cytotoxic lymphocytes; CTL_GNLY = granulysin expressing cytotoxic lymphocytes; CTL_Exh = exhausted cytotoxic lymphocytes; Tfh T follicular helper cells; Treg = regulatory T cells; gdT = gamma delta T cell.
Extended Data Fig. 4 Enrichment analysis in CAR T cells (CD4 and CD8) compared to pre-infusional T cells from patients treated with Ide-cel.
GO term enrichment analysis of significantly differentially expressed genes (CAR vs. pre-infusional T cells) in participants 12 (a) and 14 (b). Terms are ranked by rich factor, which is the number of DE genes in the term divided by the number of background genes in that term. Dot plot depict the highest ranked significantly enriched GO terms for biological processes (FDR < 0.05).
Extended Data Fig. 5 Enrichment analysis in CAR T cells (T cell subtypes) compared to pre-infusional T cells from patients treated with Ide-cel.
a, GO term enrichment analysis of significantly differentially expressed genes (CAR vs. pre-infusional T cells) in PBMCs for patient 12. No significantly enriched GO terms were found for patient 14. b, Differential cell surface protein expression for CD4 and CD8 T-cells when comparing CAR+ from PBMCs with CAR+ from BMMCs for patient 12 (BMMCs CD4 n = 41; CD8 430 // PBMCs CD4 n = 214; CD8 4699). Genes with an FDR < 0.05 and fold change <1.5 or >1.5 were considered as significantly differentially expressed. Cell type abbreviations: CM = central memory; EM = effector memory; TEMRA = effector memory cells re-expressing CD45RA; TEX = exhausted T cell.
Extended Data Fig. 6 Analysis of CD38 surface protein expression in patient 01 with samples available after exposure/becoming refractory to a CD38 antibody as well as after discontinuing the antibody.
Patient 01 had progressed with a plasma cell leukemia after frontline treatment with 4 cycles bortezomib/cyclophosphamide/dexamethasone (VCD), high-dose melphalan and autologous transplantation (HDM + ASCT) and lenalidomide (Len) maintenance. Circulating malignant plasma cells (n = 7071) showed homogeneous CD38 protein expression, which was significantly decreased 4 days after starting Daratumumab/Carfilzomib/Dexamethasone (Dara/Carf/Dex). The patient relapsed 9 months later. CD38 expression on circulating malignant plasma cells significantly increased after 4 days of 3rd line treatment with Elotuzumab/Pomalidomide/Dexamethasone (Elo/Pom/Dex). CD38 expression levels increased further upon relapsed from treatment with Idecabtagene vicleucel (Ide-cel) 2 months after the last dose of Daratumumab.
Extended Data Fig. 7 Marker genes for T cell identities.
DE genes for each T cell identity were determined using the Wilcoxon rank-sum test. Only CAR negative cells were analyzed. Genes with an FDR < 0.05 and an absolute fold change >1.5 were considered statistically significant. Genes are ranked according to FDR. For each cell identity, 5 significant genes are shown. The color intensity indicates the standardized average expression level in a cell identity. The dot size indicates the percentage of expressing cells within each cell identity of the corresponding genes.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 (pseudotime trajectory analysis of preinfusional and postinfusional CD4+CD8+ T cells and CAR T cells) and Supplementary Fig. 9 (gating strategy).
Supplementary Tables
Supplementary Table 1. Summary of ten individuals treated with Ide-cel or Cilta-cel (participants P7 and P8). Except for participants P8 and P10, all individuals had at least one high-risk cytogenetic feature detected by fluorescence in situ hybridization (mostly chromosome 1 abnormalities). Bridging therapy consisted mainly of antibody-based three-drug combinations. Participant P9 was treated with high-dose chemotherapy and autologous stem cell transplantation before CAR T cell infusion. Abbreviations: m, male; f, female; FISH, fluorescence in situ hybridization; Elo/Pom/Dex, elotuzumab/pomalidomid/dexamethasone; Benda/Carf/Dex, bendamustine/carfilzomib/dexamethasone; Dara/Pom/Dex, daratumumab/pomalidomide/dexamethasone; HD-Melphalan, high-dose melphalan; ASCT, autologous stem cell transplantation; Benda/Dex, bendamustine/dexamethasone; IsaPomDex, isatuximab/pomalidomide/dexamethasone; Dara/Len/Dex, daratumumab/lenalidomide/dexamethasone; Isa/Carf/Dex, isatuximab/carfilzomib/dexamethasone. Supplementary Table 2. Summary of samples and quality control data from scRNA-seq, TCR and BCR sequencing and surface proteomics. Abbreviations: PR, partial remission; PD, progressive disease; UMI, unique molecular identifier. Supplementary Table 3. Feature reference .csv file declaring antibody capture constructs and associated barcodes. The CellRanger multipipeline requires this configuration .csv file as input. Supplementary Table 4. Summary of differential gene expression analyses. Testing for differential gene expression was performed using the function FindMarkers (Wilcoxon rank-sum test) implemented in Seurat. Genes with an FDR of <0.05 (Benjamini–Hochberg correction) and fold change of <1.5 or >1.5 were considered DEGs. Summaries include comparisons of different cell types between nonresponders and responders from PBMCs after CAR T cell therapy (post-PBMC; non-CR versus CR), BMMCs after CAR T cell therapy (post-BMMC; non-CR versus CR), PBMCs collected at apheresis (pre-PBMC; non-CR versus CR) and comparisons of PBMCs after CAR T cell therapy versus PBMCs at apheresis in responders (CR; post versus pre) and nonresponders (non-CR; post versus pre). Supplementary Table 5. Summary of differential gene expression analyses for T cell identities. Testing for differential gene expression was performed using the function FindMarkers (Wilcoxon rank-sum test) implemented in Seurat. Genes with an FDR of <0.05 (Benjamini–Hochberg correction) and fold change of <1.5 or >1.5 were considered DEGs. Summaries include comparisons of T cell identities between nonresponders and responders from PBMCs at apheresis (pre-PB; T cell; non-CR versus CR), PBMCs after CAR T cell therapy (post-PB; T cell; non-CR versus CR), BMMCs after CAR T cell therapy (post-BM; T cell; non-CR versus CR) and comparisons of PBMCs after CAR T cell therapy versus PBMCs at apheresis in responders (CR; PB; T cell; post versus pre) and nonresponders (non-CR; PB; T cell post versus pre), CAR T cells versus preinfusional T cells in participant P12 (CAR versus pre-T cell) and CAR T cells versus preinfusional T cells in participant P14 (CAR versus pre-T cell).
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rade, M., Grieb, N., Weiss, R. et al. Single-cell multiomic dissection of response and resistance to chimeric antigen receptor T cells against BCMA in relapsed multiple myeloma. Nat Cancer (2024). https://doi.org/10.1038/s43018-024-00763-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43018-024-00763-8