Abstract
Type 1 diabetes (T1D) is an autoimmune disease in which immune cells destroy insulin-producing beta cells. The aetiology of this complex disease is dependent on the interplay of multiple heterogeneous cell types in the pancreatic environment. Here, we provide a single-cell atlas of pancreatic islets of 24 T1D, autoantibody-positive and nondiabetic organ donors across multiple quantitative modalities including ~80,000 cells using single-cell transcriptomics, ~7,000,000 cells using cytometry by time of flight and ~1,000,000 cells using in situ imaging mass cytometry. We develop an advanced integrative analytical strategy to assess pancreatic islets and identify canonical cell types. We show that a subset of exocrine ductal cells acquires a signature of tolerogenic dendritic cells in an apparent attempt at immune suppression in T1D donors. Our multimodal analyses delineate cell types and processes that may contribute to T1D immunopathogenesis and provide an integrative procedure for exploration and discovery of human pancreatic function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The Gene Expression Omnibus accession number associated with this paper is GSE148073. Additional data are publicly available at https://hpap.pmacs.upenn.edu/. Furthermore, a user-friendly web portal for exploration of the scRNA-seq data is available at https://cellxgene.cziscience.com/e/37b21763-7f0f-41ae-9001-60bad6e2841d.cxg/.
Code availability
Where applicable, scripts used for data processing and analysis are available in the Supplemental Information and Methods and provided on GitHub at https://github.com/GregorySchwartz/multiomics-single-cell-t1d/. TooManyCells is a publicly available suite of tools, algorithms and visualizations (https://github.com/GregorySchwartz/too-many-cells/) that was extensively used in this study, and where applicable, the flags used in TooManyCells to generate specific figures are included in the Methods.
References
Powers, A. C. Type 1 diabetes mellitus: much progress, many opportunities. J. Clin. Invest. https://doi.org/10.1172/JCI142242 (2021).
Michels, A. W., Redondo, M. J. & Atkinson, M. A. The pathogenesis, natural history, and treatment of type 1 diabetes: time (thankfully) does not stand still. Lancet Diabetes Endocrinol. https://doi.org/10.1016/S2213-8587(21)00344-2 (2021).
Boldison, J. & Wong, F. S. Immune and pancreatic beta cell interactions in type 1 diabetes. Trends Endocrinol. Metab. 27, 856–867 (2016).
Unanue, E. R. & Wan, X. The immunoreactive platform of the pancreatic islets influences the development of autoreactivity. Diabetes 68, 1544–1551 (2019).
Barrett, J. C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 (2009).
Campbell-Thompson, M. et al. Network for Pancreatic Organ Donors with Diabetes: developing a tissue biobank for type 1 diabetes. Diabetes Metab. Res. Rev. 28, 608–617 (2012).
Kaestner, K. H., Powers, A. C., Naji, A., Consortium, H. & Atkinson, M. A. NIH initiative to improve understanding of the pancreas, islet and autoimmunity in type 1 diabetes: the Human Pancreas Analysis Program. Diabetes 68, 1394–1402 (2019).
Pliner, H. A., Shendure, J. & Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat. Methods 16, 983–986 (2019).
Schwartz, G. W. et al. TooManyCells identifies and visualizes relationships of single-cell clades. Nat. Methods, https://doi.org/10.1038/s41592-020-0748-5 (2020).
Tosti, L. et al. Single-nucleus and in situ RNA-sequencing reveal cell topographies in the human pancreas. Gastroenterology 160, 1330–1344 (2021).
Campbell-Thompson, M., Rodriguez-Calvo, T. & Battaglia, M. Abnormalities of the exocrine pancreas in type 1 diabetes. Curr. Diab. Rep. 15, 79 (2015).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Tersey, S. A. et al. Islet beta-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes 61, 818–827 (2012).
Marhfour, I. et al. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia 55, 2417–2420 (2012).
Engin, F. et al. Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci. Transl. Med. 5, 211ra156 (2013).
Nakayasu, E. S. et al. Comprehensive proteomics analysis of stressed human islets identifies GDF15 as a target for type 1 diabetes intervention. Cell Metab. 31, 363–374 (2020).
Unsicker, K., Spittau, B. & Krieglstein, K. The multiple facets of the TGF-β family cytokine growth/differentiation factor-15/macrophage inhibitory cytokine-1. Cytokine Growth Factor Rev. 24, 373–384 (2013).
Carstensen, M. et al. Macrophage inhibitory cytokine-1 is increased in individuals before type 2 diabetes diagnosis but is not an independent predictor of type 2 diabetes: the Whitehall II study. Eur. J. Endocrinol. 162, 913–917 (2010).
Avrahami, D. et al. Single-cell transcriptomics of human islet ontogeny defines the molecular basis of beta cell dedifferentiation in T2D. Mol. Metab. 42, 101057 (2020).
Wang, Y. J. et al. Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65, 3028–3038 (2016).
Teo, A. K. K. et al. Single-cell analyses of human islet cells reveal de-differentiation signatures. Cell Death Discov. 4, 14 (2018).
Chiang, M. K. & Melton, D. A. Single-cell transcript analysis of pancreas development. Dev. Cell 4, 383–393 (2003).
Riedel, M. J. et al. Immunohistochemical characterisation of cells co-producing insulin and glucagon in the developing human pancreas. Diabetologia 55, 372–381 (2012).
Heng, T. S. & Painter, M. W., Immunological Genome Project Consortium.The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015).
Timshel, P. N., Thompson, J. J. & Pers, T. H. Genetic mapping of etiologic brain cell types for obesity. Elife https://doi.org/10.7554/eLife.55851 (2020).
Ludwig, M. Q. et al. A genetic map of the mouse dorsal vagal complex and its role in obesity. Nat. Metab. 3, 530–545 (2021).
Loh, P. R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed-model association for biobank-scale datasets. Nat. Genet. 50, 906–908 (2018).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Rai, V. et al. Single-cell ATAC–seq in human pancreatic islets and deep learning upscaling of rare cells reveals cell-specific type 2 diabetes regulatory signatures. Mol. Metab. 32, 109–121 (2020).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies and species. Nat. Biotechnol. 36, 411–420 (2018).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394 (2016).
Segerstolpe, A. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Wang, Y. J. et al. Multiplexed in situ imaging mass cytometry analysis of the human endocrine pancreas and immune system in type 1 diabetes. Cell Metab. 29, 769–783 (2019).
Zimmerman, K. D., Espeland, M. A. & Langefeld, C. D. A practical solution to pseudoreplication bias in single-cell studies. Nat. Commun. 12, 738 (2021).
Jailwala, P. et al. Apoptosis of CD4+CD25high T cells in type 1 diabetes may be partially mediated by IL-2 deprivation. PLoS ONE 4, e6527 (2009).
Slominski, B., Skrzypkowska, M., Ryba-Stanislawowska, M., Mysliwiec, M. & Trzonkowski, P. Associations of TP53 codon 72 polymorphism with complications and comorbidities in patients with type 1 diabetes. J. Mol. Med. 99, 675–683 (2021).
Christen, U. et al. A dual role for TNF in type 1 diabetes: islet-specific expression abrogates the ongoing autoimmune process when induced late but not early during pathogenesis. J. Immunol. 166, 7023–7032 (2001).
Lombardi, A., Tsomos, E., Hammerstad, S. S. & Tomer, Y. Interferon-α: the key trigger of type 1 diabetes. J. Autoimmun. 94, 7–15 (2018).
Marro, B. S. et al. Progression of type 1 diabetes from the prediabetic stage is controlled by interferon-α signaling. Proc. Natl Acad. Sci. USA 114, 3708–3713 (2017).
Osum, K. C. et al. Interferon-γ drives programmed death-ligand 1 expression on islet beta cells to limit T cell function during autoimmune diabetes. Sci. Rep. 8, 8295 (2018).
Ziegler, A. G. et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 309, 2473–2479 (2013).
Noble, J. A. & Valdes, A. M. Genetics of the HLA region in the prediction of type 1 diabetes. Curr. Diab. Rep. 11, 533–542 (2011).
Nejentsev, S. et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450, 887–892 (2007).
Wculek, S. K. et al. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 20, 7–24 (2020).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes and progenitors. Science https://doi.org/10.1126/science.aah4573 (2017).
Appleman, L. J. & Boussiotis, V. A. T cell anergy and co-stimulation. Immunol. Rev. 192, 161–180 (2003).
Wang, L. et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 208, 577–592 (2011).
Wang, Y. J. et al. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metab. 24, 616–626 (2016).
Rodriguez-Calvo, T., Ekwall, O., Amirian, N., Zapardiel-Gonzalo, J. & von Herrath, M. G. Increased immune cell infiltration of the exocrine pancreas: a possible contribution to the pathogenesis of type 1 diabetes. Diabetes 63, 3880–3890 (2014).
Steimle, V., Siegrist, C. A., Mottet, A., Lisowska-Grospierre, B. & Mach, B. Regulation of MHC class II expression by interferon-γ mediated by the transactivator gene CIITA. Science 265, 106–109 (1994).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Wosen, J. E., Mukhopadhyay, D., Macaubas, C. & Mellins, E. D. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front Immunol. 9, 2144 (2018).
Kondo, A. et al. Highly multiplexed image analysis of intestinal tissue sections in patients with inflammatory bowel disease. Gastroenterology 161, 1940–1952 (2021).
Russell, M. A. et al. HLA class II antigen processing and presentation pathway components demonstrated by transcriptome and protein analyses of islet beta cells from donors with type 1 diabetes. Diabetes 68, 988–1001 (2019).
Squair, J. W. et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 12, 5692 (2021).
Bonifacio, E. et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for national institute of diabetes and digestive and kidney diseases consortia. J. Clin. Endocrinol. Metab. 95, 3360–3367 (2010).
Yu, L. et al. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc. Natl Acad. Sci. USA 97, 1701–1706 (2000).
Wenzlau, J. M. et al. The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc. Natl Acad. Sci. USA 104, 17040–17045 (2007).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA-sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Crowell, H. L. et al. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 11, 6077 (2020).
Howson, J. M. et al. Evidence of gene–gene interaction and age-at-diagnosis effects in type 1 diabetes. Diabetes 61, 3012–3017 (2012).
Howson, J. M., Walker, N. M., Clayton, D. & Todd, J. A., Type 1 Diabetes Genetics Consortium. Confirmation of HLA class II independent type 1 diabetes associations in the major histocompatibility complex including HLA-B and HLA-A. Diabetes Obes. Metab. 11, 31–45 (2009).
Acknowledgements
We thank our colleagues for helpful discussions, particularly: A. Chandra, E. L. Prak, B. Stanger, M. Silverman, G. Beatty, K. Z., M. Lazar, R. Vonderheide, A. Minn and E. J. Wherry. Some schematics throughout the paper were created with Biorender.com. We thank A. Georgescu for confocal microscopy and the University of Pennsylvania Diabetes Research Center for the use of the Functional Genomics Core (P30-DK19525). This work was supported by National Institute of Health grants UC4 DK112217 and U01DK112217 (to A.N., K.K., M.B., J.M., M.F., R.B.F. and G.V.), R01CA230800 and Susan G. Komen CCR185472448 (to R.B.F.) and R01HL145754, U01DK127768, U01DA052715, the Burroughs Wellcome Fund, the Chan Zuckerberg Initiative, W. W. Smith Charitable Trust, the Penn Epigenetics Institute and the Sloan Foundation awards to G.V.
Author information
Authors and Affiliations
Consortia
Contributions
C.L. and A.N. procured human pancreatic tissues. A.N. and K.H.K. acquired funding. M.G., Y.J.W. and A.M. generated scRNA-seq libraries. J.S. performed sequencing of scRNA-seq libraries. J. L. performed CyTOF experiments. M.W. and D. T. performed IMC experiments. A.K. assisted in analysis of IMC measurements. C.L.M. performed immunohistochemistry experiments. N.G. assisted in analysis of CyTOF data. W.W. assisted in analysis of scRNA-seq data. A.M. performed annotation of IMC and CyTOF data. M. Feldman supervised the HPAP tissue bank. J.H.M., A.S.J. and M.R.B. are members of the HPAP consortium. K.H.K. supervised M.G., Y.J.W., A.M., J. L., M.W., D.T., A.K. and C.L.M. for generation of scRNA-seq, CyTOF and IMC data. R.B.F. supervised A.M. and G.W.S. in analysis of scRNA-seq, CyTOF and IMC data. G.V. supervised M. Fasolino, A.R.P., N.G. and W.W. M. Fasolino., G.W.S., A.R.P. and G.V. performed computational analysis of scRNA-seq, IMC and CyTOF data. M. Fasolino and G.V. wrote the original draft and revised it with comments from all authors. K.H.K. and R.B.F. edited the original and revised manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.R.B. has a consulting arrangement with Interius BioTherapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Tune Pers, Raghavendra Mirmira and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: Isabella Samuelson.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cell numbers and clustering before complete filtering.
a) Pie chart displaying the cell numbers and proportions of each individual donor per donor type. b) Box plot displaying the average gene number per cell per donor type. c) UMAP visualization of cell clusters for all cells. d) Doublets and singlets, as identified using DoubletFinder, across cell clusters visualized by UMAP. e) UMAP visualization of the normalized gene expression counts of each canonical gene marker of each major cell type.
Extended Data Fig. 2 Doublet removal and UMI counts.
a) Doublets and singlets, as identified using Scrublet, across cell clusters visualized by UMAP per individual. b) Venn diagram indicating the number of cells deemed doublets by DoubletFinder and Scrublet, as well as cells that were commonly identified by both approaches. c) Table indicating the number of cells removed and the resulting total cell number for each step of filtering. d) Unique molecular identifier (UMI) counts per cell projected across the dendrogram visualization and clustering of all cells from Fig. 1c. Pie charts at the end of the branches display the breakdown of UMI counts per cell within that terminal cluster. Cells begin at the start pin symbol, and from there are partitioned based on similarities and differences in gene expression. e) UMI counts per cell projected across the dendrogram visualization and clustering of ductal and endocrine cells from Fig. 1d. Pie charts at the end of the branches display the breakdown of UMI counts per cell within that terminal cluster. Cells begin at the start pin symbol, and from there are partitioned based on similarities and differences in gene expression. f) Expression of genes associated with mitochondrial function projected across the dendrogram visualization and clustering of all cells from Fig. 1c. g) Expression of genes associated with mitochondrial function projected across the dendrogram visualization and clustering of ductal and endocrine cells from Fig. 1d.
Extended Data Fig. 3 Cell numbers and clustering after complete filtering.
a) Pie chart displaying the cell numbers/proportions of each individual donor per donor type. b) UMAP visualization of cell clusters for all cells. c) UMAP visualization donor groups across clusters for all cells. d) UMAP visualization of Garnett cellular classifications across clusters for all cells. e) UMAP visualization of the normalized gene expression counts of each canonical gene marker of each major cell type.
Extended Data Fig. 4 Marker gene expression confirms canonical cell types.
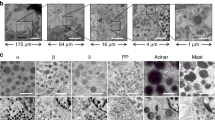
a) Dendrograms highlighting the expression of each canonical gene marker of each major cell type across the dendrogram of all cells in Fig. 1c. b) The classification of our scRNA-seq data was confirmed by a label transfer strategy using a previous single-nucleus RNA-seq data set in pancreatic islets10. c) Bar plot demonstrates percentages of agreement between previous annotation and our strategy using a label-transfer strategy. d) Dendrograms highlighting the expression of each canonical gene marker of each major cell type across the dendrogram of ductal and endocrine cells in Fig. 1d. e) To further validate the most closely related cell types to Hybrid cells, we used a label transfer strategy to a previous pancreatic islet scRNA-seq data set19. In concordance with Garnett and canonical gene markers, we corroborated the assignment of beta, alpha, and PP cells to these Hybrid cells. f) Bar plot demonstrates annotation results of label transfer for cells grouped as Hybrid cells. g) Pie chart displaying the cell numbers/proportions of each cell type defined in Fig. 1, c and d. h) Schematic of the human pancreatic islet anatomy and major cell types.
Extended Data Fig. 5 Gene and gene ontology pathways that are shared and different across disease states in Epsilon-1, Epsilon-2, and Immune cells.
(a-c) (Left) For each cell type, Venn diagrams indicate the numbers of upregulated and downregulated genes, as well as overlapping genes, across the two disease states. Circles indicate the numbers of genes that are ‘T1D enriched’ or ‘AAb enriched’. p-values presented are the results of hypergeometric CDF tests (one-tailed test for overrepresentation). (Middle) For each cell type, displayed are gene ontology pathways that are shared across T1D and AAb+ cells when compared to Control cells (top) or pathways that are differently enriched in T1D cells vs AAb+ cells (bottom). The top 20 clusters are displayed and a stringent cut-off of 1e-6 was applied to determine significant gene ontology pathways. (Right) Heatmaps displaying the degree of gene expression changes of genes (rows) that are shared (top) or differential (bottom) across AAb+ and T1D disease states. (d) GSEA analysis plots of FDR q-value vs Normalized Enrichment Score. For both ductal populations, Ductal-1 and Ductal-2, T1D cells were compared to AAb+ or Control cells to determine differentially enriched gene sets. Demarcated in red and labeled are signatures of interest.
Extended Data Fig. 6 Corroboration of HLA-DR+ Ductal cells.
(a-b) Dendrograms highlighting the expression of the MHC class II complex (a) or MHC class II activity (b) across the dendrogram of all cells in Fig. 1C. Scale bars represent normalized transcript numbers (mean across all MHC class II complex genes (a) or MHC class II activity genes (b)). (c-d) Dendrograms highlighting the expression of the MHC class II complex (c) or MHC class II activity (d) across the dendrogram of ductal and endocrine cells in Fig. 1D. Scale bars represent normalized transcript numbers (mean across all MHC class II complex genes (c) or MHC class II activity genes (d)). (e-f) Dendrograms highlighting the expression of the HLA-DPB1 (E) or KRT19 (f) across the dendrogram of ductal and endocrine cells in Fig. 1D. Scale bars represent normalized transcript numbers. (g) Dendrograms highlighting the expression of the immune-related genes across the dendrogram of ductal and endocrine cells in Fig. 1D. Scale bars represent normalized transcript numbers. (h) Dendrograms highlighting the expression of the BMPR1A across the dendrogram of ductal and endocrine cells in Fig. 1D. Scale bars represent normalized transcript numbers.
Extended Data Fig. 7 GSEA analysis across annotated cells types for dendritic cells gene sets.
a) DC1 gene signature is significantly enriched within Ductal-2 cells of T1D donors. Integrated GSEA analysis for dendritic cells gene sets from Villani et al47 across ranked lists of differentially expressed genes between T1D and control donors. b) Expression analysis of the inhibitory marker VSIR in dendritic cells demonstrates the high level of this gene in T1D ductal cells compared with control ductal cells.
Extended Data Fig. 8 CyTOF validation of canonical cell types.
a) Bar graph displaying the proportion of cells for all major pancreatic cell types from each donor group where cell annotations were obtained by our new machine-learning based strategy using CyTOF measurements across 12 donors. b) Dendrogram visualization of the immune cell cluster, CD45 positive (+) cells, as determined by the analysis of the flow cytometry by time-of-flight (CyTOF) data. c) Dendrogram visualization of the beta cell cluster, C-peptide positive (+) cells, as determined by the analysis of the CyTOF data. d) Dendrogram visualization of the alpha cell cluster, Glucagon positive (+) cells, as determined by the analysis of the CyTOF data. e) Major cell types projected on TooManyCells tree based on our machine-learning based annotation using CyTOF data (n=6,945,575 cells). f) Two-parameter CyTOF analysis of HLA-DR and cytokeratin protein expression in single cells from T1D donor #3 (HPAP023). g) Two parameter CyTOF analysis of HLA-DR and cytokeratin protein expression in single cells from Control donor #3 (HPAP034), a donor with a very low percentage of HLA-DR+ ductal cells as determined by unbiased analysis of CyTOF data with TooManyCells.
Extended Data Fig. 9 IMC validation of HLA-DR+ ductal cells.
a) Bar graph displaying the proportion of cells for all major pancreatic cell types from each donor group where cell annotations were obtained by our machine-learning-based strategy using IMC measurements. Further manual inspection of CD19 and FOXP3 staining used for annotating B and Tregs indicated low quality of these markers across tissue slides. b) Dendrogram visualization of the immune cell cluster, CD45 positive (+) cells, as determined by the analysis of the imaging mass cytometry (IMC) data analysis. c) Dendrogram visualization of the beta cell cluster, C-peptide positive (+) cells, as determined by the analysis of the IMC data analysis. d) Dendrogram visualization of the alpha cell cluster, Glucagon positive (+) cells, as determined by the analysis of the IMC data analysis. e) Major cell types projected on TooManyCells tree as they were annotated by our machine-learning based strategy using IMC data (n=1,170,001 cells).
Extended Data Fig. 10 Cellular neighborhood analysis in IMC data demonstrates the enrichment of CD4+ T cells surrounding HLA-DR+ ductal cells.
a) Bar plot displaying the proportion of HLA-DR+ cytokeratin+ cells from each pancreatic region determined by IMC. b-c) HLA-DR+ cytokeratin+ cells versus percentage of myeloid cells. For each donor group, the median of percentage of each annotated immune subtype and the median HLA-DR+ ductal cell percentage of total cells across all individual donors per donor group was computed. Only myeloid cells demonstrated significant correlation with respect to the number of HLA-DR+ cytokeratin+ cells across donor groups. d) Dendrogram visualization of the clusters of HLA-DR+ cytokeratin+ cells (red), cells neighboring HLA-DR+ cytokeratin+ (blue), and cells distant from HLA-DR+ cytokeratin+ cells (grey) as determined by leveraging the spatial architecture provided by IMC data. e) Boxplots showing the normalized protein expression of different canonical markers in cells neighboring HLA-DR+ cytokeratin+ cells (blue) versus cells neighboring random cells (grey). The number of random cells evaluated was equal to the number of HLA-DR+ cytokeratin+ cells. Differential marker expression significance for neighbors in the IMC analysis was determined using permutation tests. For each marker, the distribution of that marker value for each of the designated n neighbors was compared against 100 distributions derived from n random cells across the entire IMC tree. * indicates p-value < 0.01. Total number of cells in both blue and gray groups is 195,633. Box-and-whisker plots (centre, median; box limits, upper (75th) and lower (25th) percentiles; whiskers, 1.5 × interquartile range; points, outliers). f) CD4+ T cells are the number one immune subtypes enriched at the neighborhood of HLA-DR+ cytokeratin+ cells. Annotation of neighbors of HLA-DR+ cytokeratin+ cells was performed our machine-learning based strategy.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and legends and Supplementary Table legends
Supplementary Table 1
Donor clinical information.
Supplementary Table 2
Donor HLA information.
Supplementary Table 3
Summary statistics of scRNA-seq data.
Supplementary Table 4
Differential gene list between Beta-1 cells (positive FC) and Beta-2 cells (negative FC).
Supplementary Table 5
Differential gene list between Ductal-1 cells (positive FC) and Ductal-2 cells (negative FC).
Supplementary Table 6
Differential gene list between Ductal-2 cells (positive FC) and Acinar-2 cells (negative FC).
Supplementary Table 7
Differential gene list between Epsilon-1 cells (positive FC) and Epsilon-2 cells (negative FC).
Supplementary Table 8
Differential gene list between AAb+ cells (positive FC)) and control cells (negative FC) using individual cells strategy.
Supplementary Table 9
Differential gene list between T1D cells (positive FC) and AAb+ cells (negative FC) using individual cells strategy.
Supplementary Table 10
Differential gene list between T1D cells (positive FC) control cells (negative FC) using individual cells strategy.
Supplementary Table 11
Differential gene list between AAb+ cells (positive FC) and control cells (negative FC) using a pseudo-bulk strategy. Column gene represents the NCBI official gene symbol. P value represents the P-value uncorrected estimate. FDR is Benjamini–Hochberg FDR (that is, P-value corrected).
Supplementary Table 12
Differential gene list between T1D cells (positive FC) and AAb+ cells (negative FC) using a pseudo-bulk strategy. CColumn gene represents the NCBI official gene symbol. P value represents the P-value uncorrected estimate. FDR is Benjamini–Hochberg FDR (that is, P-value corrected).
Supplementary Table 13
Differential gene list between T1D cells (positive FC) and control cells (negative FC) using a pseudo-bulk strategy. Column gene represents the NCBI official gene symbol. P value represents the P-value uncorrected estimate. FDR is Benjamini–Hochberg FDR (that is, P-value corrected).
Supplementary Table 14
1,473 genes whose expression significantly correlates with the GAD titre in single AAb+ GAD+ donors. Column gene represents the NCBI official gene symbol. P value represents the P-value uncorrected estimate. FDR is Benjamini–Hochberg FDR (that is, P-value corrected).
Supplementary Table 15
CyTOF panel.
Supplementary Table 16
IMC panel.
Supplementary Table 17
Garnett cell-type marker file.
Rights and permissions
About this article
Cite this article
Fasolino, M., Schwartz, G.W., Patil, A.R. et al. Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes. Nat Metab 4, 284–299 (2022). https://doi.org/10.1038/s42255-022-00531-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-022-00531-x
This article is cited by
-
Systematic immune cell dysregulation and molecular subtypes revealed by single-cell RNA-seq of subjects with type 1 diabetes
Genome Medicine (2024)
-
Pancreatic draining lymph nodes (PLNs) serve as a pathogenic hub contributing to the development of type 1 diabetes
Cell & Bioscience (2023)
-
Why does the immune system destroy pancreatic β-cells but not α-cells in type 1 diabetes?
Nature Reviews Endocrinology (2023)
-
Talin-1 inhibits Smurf1-mediated Stat3 degradation to modulate β-cell proliferation and mass in mice
Cell Death & Disease (2023)
-
IFNɣ but not IFNα increases recognition of insulin defective ribosomal product-derived antigen to amplify islet autoimmunity
Diabetologia (2023)