Abstract
The initial transient phase of an emerging epidemic is of critical importance for data-driven model building, model-based prediction of the epidemic trend, and articulation of control/prevention strategies. Quantitative models for real-world epidemics need to be memory-dependent or non-Markovian, but this presents difficulties for data collection, parameter estimation, computation, and analyses. In contrast, such difficulties do not arise in the traditional Markovian models. To uncover the conditions under which Markovian and non-Markovian models are equivalent, we develop a comprehensive computational and analytic framework. We show that the transient-state equivalence holds when the average generation time matches the average removal time, resulting in minimal Markovian estimation errors in the basic reproduction number, epidemic forecasting, and evaluation of control strategy. The errors depend primarily on the generation-to-removal time ratio, while rarely on the specific values and distributions of these times. Overall, our study provides a general criterion for modeling memory-dependent processes using Markovian frameworks.
Similar content being viewed by others
Introduction
When an epidemic emerges, the initial transient phase of the disease spreading dynamics before a steady state is reached is of paramount importance, for two reasons1,2,3,4,5,6,7,8,9,10,11. First, estimating key indicators or parameters, e.g., the generation time, serial intervals, and basic reproduction number, is crucial for predicting and formulating control strategies when the underlying dynamical process has not reached a steady state. Second, it is during the transient phase that control and mitigation strategies can be effectively applied to prevent a large-scale outbreak. Prediction and control depend, of course, on a quantitative model of the epidemic process, which can be constructed based on the key parameters estimated from data collected during the transient phase. In principle, since the dynamical processes underlying real-world epidemics are generally memory-dependent in the sense that the state evolution depends on the history, a rigorous modeling framework needs to be of the non-Markovian type, but this presents great challenges in terms of data collection, parameter estimation, computation and analyses12,13,14. The difficulties can be alleviated by adopting the traditional simplified memoryless Markovian framework, which, however, may potentially result in deviations15,16,17,18,19,20,21,22,23,24,25,26. An outstanding question is, are there specific conditions under which a Markovian epidemic outbreak could be equivalent to the non-Markovian counterpart during the transient phase, enabling an accurate description of memory-dependent transmission within the Markovian framework? Additionally, another important issue remains, how are the errors of Markovian estimation determined? The purpose of this paper is to provide a comprehensive answer to these questions.
The COVID-19 pandemic has highlighted the need and importance of understanding disease spreading and transmission to accurately predict, control, and manage future outbreaks through non-pharmacological interventions and vaccine allocation strategies1,2,3,4,5,6,7,8,9,10. To accomplish these goals, accurate mathematical modeling of the disease-spreading dynamics is key. In a general population, epidemic transmission occurs via some kind of point process, where individuals become infected at different points in time. It has been known that point processes in the real world are typically non-Markovian with a memory effect in which the distribution of the interevent times is not exponential2,23,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41. For example, the interevent time distribution arising from the virus transmission with COVID-19 is not of the memoryless exponential type but typically exhibits memory-dependent features characterized by the Weibull distribution2. Strictly speaking, from a modeling perspective, disease spreading should be described by a non-Markovian process. A non-Markovian approach takes into account historical memory of disease progression, mathematically resulting in a complex set of integro-differential equations in the form of convolution. (Note that besides the transmission process, where the infection capacity is highly dependent on the elapsed time from the infection, there are other types of memory-dependent factors. For instance, in certain meta-population network models, the infection capacity of a node is influenced by the transmission path leading to that node, rather than solely relying on the time of infection42. Thus, it is important to mention that we only take the elapsed time into consideration in this work.)
There are difficulties with non-Markovian modeling of memory-dependent disease spreading. The foremost is data availability. In particular, while standard epidemic spreading models are available, the model parameters need to be estimated through data. A non-Markovian model often requires detailed and granular data that can be difficult to get, especially during the early stage of the epidemic where accurate modeling is most needed12. From a theoretical point of view, it is desired to obtain certain closed-form solutions for key quantities such as the onset and size of the epidemic outbreak, but this is generally impossible for non-Markovian models15,43. Computationally, accommodating memory effects in principle makes the underlying dynamical system infinitely dimensional, practically requiring solving an unusually large number of dynamical variables through a large number of complex integro-differential equations13,14. In contrast, in an idealized Markovian point process, events occur at a fixed rate, leading to an exponential distribution for the interevent time intervals and consequently a memoryless process. If the spreading dynamics were of the Markovian type, the aforementioned difficulties associated with non-Markovian dynamics no longer exist. In particular, a Markovian spreading process can be described by a small number of ordinary differential equations with a few parameters that can be estimated even from sparse data, and the numerical simulations can be carried out in a computationally extremely efficient manner15,16,17,18,19,20,21,22,23,24,25,26. For these reasons, many recent studies of the COVID-19 pandemic assumed Markovian behaviors to avoid the difficulties associated with non-Markovian modeling3,4,5,6,7,8,9,10. The issue is whether such a simplified approach can be justified. Addressing this issue requires a comprehensive understanding of the extent to which the Markovian approach represents a good approximation to model non-Markovian type of memory-dependent spreading dynamics, and specifically of the conditions under which the Markovian theory can produce accurate results that match those from the non-Markovian model. This highlights the importance of studying the equivalence between non-Markovian and Markovian dynamics.
There were previous studies of the so-called steady-state equivalence between Markovian and non-Markovian modeling for epidemic spreading. In particular, when the system has reached a steady state, such an equivalence can be established through a modified definition of the effective infection rate18,21,22. From a realistic point of view, the equivalence limited only to steady states may not be critical as the transient phase of the spreading process before any steady state is reached is more relevant and important. For example, when an epidemic occurs, it is of fundamental interest to estimate the key indicators such as the generation time (the time interval between the infections of the infector and infectee in a transmission chain), the serial interval (the time from illness onset in the primary case to illness onset in the secondary case), and the basic reproduction number (the average number of secondary transmissions from one infected person), but they are often needed to be estimated when the dynamics have not reached a steady state2,3,4,5,6,7,8,9,10,11. It is the equivalence in the transient dynamics rather than the steady state that determines whether the transmission features in the early stages of a memory-dependent disease outbreak can be properly measured through Markovian modeling. Moreover, it is only during the transient phase that control and mitigation strategies can be effective in preventing a large-scale outbreak. Discovering when and how a non-Markovian process can be approximated by a Markovian process during the transient state is thus of paramount importance. To our knowledge, such a “transient-state equivalence”, where the Markovian and non-Markovian transmission models produce similar behaviors over the entire transient transmission period, has not been established. In fact, the conditions under which the transient equivalence may hold are completely unknown at present.
In this paper, we present results from a comprehensive study of how memory effects impact the Markovian estimations in terms of the errors that arise from the Markovian hypothesis. We consider both the steady-state and transient-state equivalences between non-Markovian and Markovian models. We first rigorously show that, in the steady state, a memory-dependent non-Markovian spreading process is always equivalent to certain Markovian (memoryless) ones. We then turn to the transient states and find that an approximate equivalence can still be achieved but only if the average generation time matches the average removal time in the memory-dependent non-Markovian spreading dynamics. Qualitatively, the equality of the two times gives rise to a memoryless correlation between the infection and removal processes, thereby minimizing the impact of any memory effects. We establish that equality gives the condition under which Markovian theory accurately describes memory-dependent transmission.
One fundamental quantity underlying an epidemic process is the basic reproduction number R0. Our theoretical analysis indicates that, when the average generation and removal times are equal, the transient-state equivalence between memory-dependent and memoryless transmissions will minimize the error of the Markovian approach in estimating R0 and lead to its accurate epidemic forecasting and prevention evaluation. Another finding is that the generation-to-removal time ratio plays a decisive role in the accuracy of the Markovian approximation. Specifically, if the average generation time is smaller (greater) than the average removal time, the Markovian approximate will lead to an overestimation (underestimation) of R0 and epidemic forecasting as well as the errors of the prevention evaluation, which can also be verified based on readily accessible clinical data of four types of real diseases.
The estimation accuracy is largely determined by the time ratio but rarely depends on the particular forms of time distributions or the specific values of the average generation and removal times. This property is of great practical significance because it is in general challenging to obtain the detailed distributions of the generation and removal times in the early stages of the epidemic44, but their average values can be reliably estimated even during the transient phase45,46,47,48,49. Moreover, based on this property, we have developed a semi-empirical mathematical relationship that connects the errors in estimating R0 with the generation-to-removal time ratio. This relationship holds practical value as it can be utilized to rectify errors in real-world scenarios. The rectification of R0 and epidemic forecasting can be accomplished through our web-based application50.
Overall, our study establishes a general criterion for modeling memory-dependent processes within the context of Markovian frameworks. Once the condition for the existence of a transient-state equivalence between Markovian and non-Markovian dynamics is fulfilled, epidemic forecasting and prevention evaluation can be carried out using the Markovian model, again based solely on the data collected from the transient phase.
Results
The overall structure of this work is depicted in Fig. 1. The section titled “Model” presents the Model building of the age-stratified Susceptible-Infected-Removed (SIR) spreading (Fig. 1a), highlighting the difference between the Markovian (memoryless) and non-Markovian (memory-dependent) dynamics (Fig. 1b). In the “Dynamical equivalence” section, we demonstrate the equivalence between Markovian (memoryless) and non-Markovian (memory-dependent) dynamics for steady state and transient dynamics, which will further lead to the accurate description of memory-dependent dynamics by the Markovian theory (Fig. 1c). The section “Markovian approximation of memory-dependent spreading dynamics” analyzes the errors of the Markovian approach in estimating R0, epidemic forecasting, and prevention evaluation (Fig. 1d). (Note that this study involves numerous parameters and variables, all of which have been comprehensively detailed and listed for reference in Supplementary Table S1 of Supplementary Note 1.)
a SIR Model. Each individual belongs to one of the three states: susceptible (S), infected (I), or removed (R). When infected (i), a susceptible individual will switch into the I state (ii) and gain the ability to infect others (iii). An infected individual is removed (through recovery or death) with a probability (iv). b Non-Markovian versus Markovian process. The infection capacity of an infected individual is characterized by the infection time distribution \({\psi }_{\inf }(\tau )\) and its removal can be described by the removal time distribution ψrem(τ). For the non-Markovian process, the distributions can assume quite general forms, while the distributions are exponential for a Markovian process. c Equivalence between non-Markovian and Markovian processes: (i) steady-state equivalence holds under all conditions; (ii) transient-state equivalence only holds when Tgen is equal to Trem. d Markovian estimation of memory-dependent process. (i) The initial phase of the Monte Carlo simulation is used to fit the parameters according to the Markovian theory. (ii) Important issues such as the estimation of R0, epidemic forecasting, and the evaluation of vaccination strategies can be addressed by the theory. (iii) The remaining data generated by the Monte Carlo simulation is used to test the accuracy of the estimated R0, epidemic forecasting, and prevention evaluation.
Model
We articulate an age-stratified SIR spreading dynamics model, in which the entire population is partitioned into various age groups with intricate age-specific contact rates among them. The distribution of population across different age groups is represented by an age distribution vector (p), with the age-dependent contact matrix (A) quantifying the transmission rates between different age groups. Both p and A can be constructed from empirical data51,52. For convenience, to distinguish between the actual dynamical process and its theoretical treatment, throughout this paper we use the terms “memory-dependent” and “memoryless” to describe actual spreading processes and Monte Carlo simulations, while in various theoretical analyses, the corresponding terms are “non-Markovian” and “Markovian.”
The mechanism of disease transmission across different age groups and the recovery or death of infected individuals can be described by the SIR compartmental model, as illustrated in Fig. 1a, where the individuals possess three types of states: susceptible (S), infected (I), and removed (R). Susceptible individuals (S) have not contracted the disease and are at risk of being infected. Infected individuals (I) have contracted the disease and can infect others. Removed individuals (R) who have recovered or died from the disease. There are two dynamical processes: (1) infection during which susceptible individuals become infected by others and transition to the I state so as to become capable of infecting others, as shown in Fig. 1a(i–iii), and (2) removal during which infected individuals recover or die from the disease transmission and transition to the R state, as shown in Fig. 1a(iv). The ability to infect others of an infected individual can be characterized by the infection time distribution, \({\psi }_{\inf }(\tau )\) where τ denotes the time elapsed between the time the individual is infected and the current time, and the probability of the infection process occurring during the time interval [τ,τ + dτ) is given by \({\psi }_{\inf }(\tau )d\tau\), as shown in Fig. 1b(i). Likewise, the removal process is described by the removal time distribution, ψrem(τ), where the probability of a removal occurring within the time interval [τ,τ + dτ) is given by ψrem(τ)dτ, as shown in Fig. 1b(ii). The time distributions of the infection and removal processes with memory effects are general, with the exponential distributions associated with the memoryless process being a special case of the memory-dependent process.
The generic memory-dependent SIR spreading dynamics can be described by a set of deterministic integro-differential equations:
where sl(t), il(t), and rl(t), respectively, denote the fractions of the susceptible, infected, and removed individuals in age group l. ŕl = rl(0) represents the initial fraction of the removed individuals in age group l. The term cl(t) = 1 − sl(t) = il(t) + rl(t) represents the fraction of cumulative infections (including both infections and removals; note that the physical meaning of dcl(t) is the new infected fraction in age group l at time t, which can further spread the infection to others, so the value of dcl(0) is considered as the fraction of infection seeds, where the initial removed fraction is not taken into account) in age group l, while pm denotes the m-th element of the vector p, Alm denotes the lm-th element of matrix A, k is a parameter to adjust the overall contacts and n is the total number of age groups. The quantity \({\omega }_{\inf }(\tau )\) represents the hazard function of \({\psi }_{\inf }(\tau )\), meaning the rate at which infection happens at τ, given that the infection has not occurred before τ. Ψrem(τ) is the survival function of ψrem(τ), meaning the probability that the removal has not occurred by τ (see Method for detailed calculations for hazard and survival functions). When the infection and removal time distributions are known, Eqs. ((1)–(3)) provide an accurate description of generic (including memory-dependent and memoryless) SIR spreading Monte Carlo simulations in an age-stratified population-based system. As shown in Fig. 2a–c, the theory is validated by the agreement between the numerical solutions of Eqs. ((1)–(3)) and the results from direct Monte Carlo simulations (see Method for a detailed description of the Monte Carlo simulation procedure).
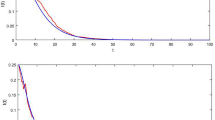
a–c The solid brown, blue, and green curves represent the theoretical results of the susceptible, infected, and removed fractions, while the solid orange, red, and purple curves show the corresponding results of 100 independent Monte Carlo simulations (\({\alpha }_{\inf }=3\), \({\beta }_{\inf }=4\), αrem = 2.5, βrem = 3.6, where α and β respectively represent the shape and scale parameters of time distributions of Weibull types, and subscripts “\(\inf\)” and “rem” indicate the time distributions of infection and removal, respectively; R0 = 1.86). d The orange and green curves, respectively, depict the removed fractions from the memory-dependent and memoryless Monte Carlo simulations of 100 independent realizations with steady-state equivalence (\({\alpha }_{\inf }=1.5\), \({\beta }_{\inf }=2.63\), αrem = 2, βrem = 2.4 for memory-dependent simulations; γ = 0.11 and μ = 0.14 for memoryless simulations; identical basic reproduction number as R0 = 1.86 for the two types simulations). e Red + and blue × markers, respectively, represent the steady-state removed fractions of memory-dependent and memoryless Monte Carlo simulations for different values of R0, where each marker is the result of averaging 100 independent simulations. The orange curve is the numerical calculations from Eq. (7), and the vertical dashed line denotes the critical point R0 = 1. f–g For Tgen = Trem in the non-Markovian theory (\({\alpha }_{\inf }=1.57\), \({\beta }_{\inf }=5.57\), αrem = 1.57, βrem = 7.79), the blue and green curves in (f) denote the susceptible and removed fractions, while the black × markers represent the inferred susceptible fractions calculated by substituting removed fractions in Eq. (11), which agrees with the susceptible curve calculated from Eq. (1). The red and green curves in (g) denote the non-Markovian infected and removed fractions, while the orange and purple dashed curves are the corresponding curves of the Markovian transmission (γ = 0.20, μ = 0.12) obtained from Eqs. (13, 14), which agree with the non-Markovian results. They all have the same basic reproduction number, i.e., R0 = 1.87. (The Euler-Lotka equation assumes exponential growth of a disease outbreak during the initial stage. As a result, the Markovian curves in (g) slightly deviate from the non-Markovian ones as the cumulative infections increase.) h–i For Tgen ≠ Trem in the non-Markovian theory (\({\alpha }_{\inf }=0.74\), \({\beta }_{\inf }=4.16\), αrem = 3.32, βrem = 7.80), the inferred susceptible curve in (h) does not match the numerical result, and the infected and removed curves of the Markovian transmission in (i) (γ = 0.41, μ = 0.28) obtained from Eqs. (13, 14) do not match the corresponding non-Markovian results. They all have the same basic reproduction number, i.e., R0 = 1.87. j Five scenarios for the non-Markovian time-distribution setting (within each scenario, \({T}_{\inf }\) and Trem are fixed): Weibull, \({T}_{\inf }=5\), Trem = 7 (blue+); Weibull, \({T}_{\inf }=5\), Trem = 5 (red × ); Weibull, \({T}_{\inf }=7\), Trem = 7 (purple □); log-normal, \({T}_{\inf }=5\), Trem = 7 (green Δ); gamma, \({T}_{\inf }=5\), Trem = 7 (orange ♢). The value of Tgen is modified to adjust \(\ln \eta\) for better visualization.
Equations (1–3) provide a general framework encompassing both non-Markovian and Markovian descriptions. If the infection and removal time distributions are exponential: \({\psi }_{\inf }(\tau )=\gamma {e}^{-\gamma \tau }\) and ψrem(τ) = μe−μτ, Eqs. (1–3) can be reformulated into a Markovian theory and further simplified into a set of ordinary differential equations with the constant infection and removal rates γ and μ (a detailed derivation of these equations is presented in Supplementary Note 2) :
While the non-Markovian and Markovian theories [Eqs. (1–3) and Eqs. (4–6), respectively] accurately describe the memory-dependent and memoryless Monte Carlo simulations, we focus on whether the Markovian theory can accurately capture memory-dependent dynamics and how memory effects influence its accuracy. For this purpose, we seek to establish the equivalence between Markovian and non-Markovian approaches for describing spreading dynamics.
Dynamical equivalence
Equations (1–6) provide a base to study the steady-state equivalence and transient-state equivalence between non-Markovian and Markovian theories, where a steady state characterizes the long-term dynamics of disease spreading and a transient state is referred to as the short-term behavior prior to system’s having reached the steady state. As illustrated in Fig. 1c, note that steady-state equivalence means that the two types of spreading dynamics attain identical steady states18,21,22, whereas transient-state equivalence implies that two types of dynamics are consistent throughout the entire transmission period. Transient-state equivalence thus implies steady-state equivalence, but not vice versa. Since Eqs. (1–6) also provides a numerical framework for Monte Carlo simulations, the terms “steady-state equivalence” and “transient-state equivalence” not only describe the connection between non-Markovian and Markovian theories, but also illustrate the relationship between memory-dependent and memoryless processes. Therefore, the equivalence between the two theories implies the equivalence between the two corresponding processes, and vice versa.
Steady-state equivalence
Equations (1)–(3) give the following transcendental equation for determining the steady state (see Supplementary Note 3 for detailed derivation):
where \({\tilde{s}}_{l}=\mathop{\lim }\nolimits_{t\to +\infty }{s}_{l}(t)\) and \({\tilde{r}}_{l}=\mathop{\lim }\nolimits_{t\to +\infty }{r}_{l}(t)\) denote the fractions of the susceptible and removed individuals in age group l at the steady state (note that \({\tilde{s}}_{l}=1-{\tilde{r}}_{l}\), because at steady state, no infection exists), while śl = sl(0) and ŕl = rl(0) represent the initial fractions of the susceptible and removed individuals in this age group. For non-Markovian dynamics, basic reproduction number R0 can be determined by:
For Markovian dynamics, R0 is given by:
where \({{{\Lambda }}}_{\max }\) is the maximum eigenvalue of the matrix kA ∘ p, and ∘ denotes a row-wise Hadamard product between a matrix and a vector (see Supplementary Note 3 for a detailed description). Since Eq. (7) applies to both non-Markovian and Markovian dynamics, an identical R0 value in the two cases will result in equivalent steady states from the same initial conditions. Consequently, for a given non-Markovian spreading process, there exists an infinite number of Markovian models with the same steady state, as the R0 value is only determined by the ratio of γ to μ, but not by either value.
As shown in Fig. 2d, memory-dependent and memoryless spreading dynamics that reach the same steady state with the identical R0 value confirm the steady-state equivalence. Figure 2e demonstrates that, even for R0 ranging from 0.023 to 4.63, the equivalent memory-dependent and memoryless spreading dynamics still produce highly consistent steady states that can be calculated from Eq. (7), which share the same critical point of phase transition at R0 = 1.
Transient-state equivalence
From the preceding section, we used the basic reproduction number R0, a fundamental metric quantifying the number of secondary infections generated by a single individual, to characterize the steady-state equivalence. Here, we propose to quantify the transient-state equivalence through the average generation time Tgen that measures the “velocity” at which secondary infections occur. This time can be calculated as2,53:
where
is the generation time distribution. Effectively, Tgen measures the average duration of disease transmission from an infected individual to the next generation of individuals. Likewise, the average infection time \({T}_{\inf }\) and the average removal time Trem are defined as the mean values of the infection and removal time distributions:
In calculating Tgen, the individual’s removal is taken into account, while \({T}_{\inf }\) measures the average time of the first disease transmission of an infectious individual without factoring in removal. In the classical memoryless transmission with exponential distributions \({\psi }_{\inf }(\tau )\) and ψrem(τ), the equality Tgen = Trem holds. However, for memory-dependent spreading, the possible scenarios are: Tgen = Trem, Tgen < Trem, or Tgen > Trem. Specifically, because Tgen, \({T}_{\inf }\) and Trem all represent the mean values of distributions, it is possible for Trem to be shorter than Tgen or \({T}_{\inf }\) in some situations. And our web-based application demonstrates the impact of parameters on the time distributions (infection, removal, and generation) as well as their average times50.
For a non-Markovian spreading process, if the equality Tgen = Trem holds, it can be approximately equivalent to a Markovian one in the transient state, because it satisfies the following equation:
which exhibits a memoryless transmission pattern similar to Markovian dynamics (see Supplementary Note 4 for a detailed analysis). Intuitively, the equality Tgen = Trem signifies that the infection and removal processes occur concurrently, which in turn leads to a memoryless relationship between the two processes, thereby minimizing the memory effects. Furthermore, to determine the corresponding Markovian parameters γ and μ of the Markovian transmission which is equivalent to the non-Markovian dynamics in the transient state, we need to utilize the Euler-Lotka equation45,53,54:
where g denotes the growth rate of the non-Markovian dynamics and is another measure of how quickly the epidemic is spreading within a population. Therefore, we can calculate the values of the basic reproduction number, R0, and growth rate, g, in the non-Markovian dynamic by using Eqs. (8), (10), and (12). Additionally, the Markovian form of ψgen(τ) according to Eq. (10) is μe−μτ, and the equivalent Markovian and non-Markovian dynamics in the transient state have the same values of R0 and the equal values of g. By substituting ψgen(τ) = μe−μτ and the calculated R0 and g into Eq. (12), we can determine the value of μ. Furthermore, using Eq. (9), we can find the value of γ based on μ. Hence, the Markovian parameters γ and μ are determined as follows:
And we provide visualizations that illustrate how the values of γ and μ are influenced by the distribution parameters in our web-based application50.
As illustrated in Fig. 2f–g, when the equality Tgen = Trem holds for the non-Markovian dynamics, Eq. (11) holds, which can be seen by comparing the susceptible curve calculated from Eq. (1) to that inferred from Eq. (11), as shown in Fig. 2f. In this case, the Markovian spreading curves deduced from Eqs. ((13) and (14)) closely align with the non-Markovian transient curves, as shown in Fig. 2g. However, as shown in Fig. 2h–i, if the equality does not hold, the equivalence in transient states breaks down. It is important to note that the Euler-Lotka equation assumes an exponential growth of a disease outbreak and is only reasonable at the initial stage. Consequently, as the cumulative infections increase (Fig. 2g), the Markovian curves will exhibit slight deviations from the non-Markovian counterparts. Meanwhile, because the equivalent dynamics share the same R0, they will ultimately reach the same steady state, ensuring that deviations will diminish while they approach the steady state.
To evaluate, under different values of the generation-to-removal time ratio η ≡ Tgen/Trem for non-Markovian dynamics we introduce a metric, ε, to quantify the difference from the corresponding Markovian results calculated from Eqs. ((13) and (14)) (see “Method” for detailed definition of ε). Figure 2j shows, for non-Markovian numerical calculations, five scenarios under various forms of time distributions \({\psi }_{\inf }(\tau )\) and ψrem(τ) constrained by certain average infection and removal times: Weibull, \({T}_{\inf }=5\), Trem = 7; Weibull, \({T}_{\inf }=7\), Trem = 7; Weibull, \({T}_{\inf }=5\), Trem = 5; log-normal, \({T}_{\inf }=5\), Trem = 7; and gamma, \({T}_{\inf }=5\), Trem = 7 (see “Method” for detailed definitions of Weibull, log-normal, and gamma distributions). The Tgen value is adjusted to obtain different values of \(\ln \eta\). For \(\ln \eta =0\), i.e., Tgen = Trem, the “distance” ε between the transient states of non-Markovian and Markovian dynamics with parameters determined from Eqs. ((13)–(14)) is minimal. Otherwise, ε increases as \(\ln \eta\) deviates from zero. Meanwhile, Fig. 2j shows that ε depends primarily on the ratio of Tgen to Trem, but rarely on the values of Tgen, \({T}_{\inf }\), or Trem. Meanwhile, the specific form of the time distributions also has limited influence on it.
Furthermore, it is important to note that the condition where Tgen = Trem in a non-Markovian dynamic ensures transient-state equivalence between this non-Markovian transmission and a Markovian one, but according to Eqs. (13) and (14), it does not imply that the average generation and removal times of the non-Markovian dynamic must be equal to those of the equivalent Markovian one. For instance, if a non-Markovian dynamic satisfies the condition of transient-state equivalence and we keep its average generation and removal times fixed, altering the shape of the corresponding time distributions will change the transmission speed53. This change, in turn, affects the infection and removal rates of the equivalent Markovian dynamic, leading to different average generation and removal times for the Markovian equivalent dynamic (see Supplementary Fig. S1 and Supplementary Notes 5 and 6 for a detailed analysis).
Markovian approximation of memory-dependent spreading dynamics
As illustrated in Fig. 1d, testing the applicability of Markovian theory for memory-dependent spreading dynamics requires three steps. The first step is fitting, where the memory-dependent Monte Carlo simulation data are divided into two parts: (a) a short early stage used as the training data for fitting the Markovian parameters in Eqs. ((4)–(6)), and (b) the remaining testing data for evaluating the performance of the Markovian model (see “Method” for details of the fitting procedure). The second step is to employ the Markovian model, equipped with fitted Markovian parameters, to accomplish various tasks, such as estimating R0, predicting outbreaks and assessing the prevention effects of different vaccination strategies. The third step is testing, i.e., evaluating the accuracy of the Markovian model, e.g., by comparing the estimated and actual R0 values, disease outbreaks and prevention effects. As real-world disease spreading is subject to environmental, social and political disturbing factors, for the fitting and testing steps, we conduct Monte Carlo simulations of stochastic memory-dependent disease outbreaks to generate the training and testing data.
Here, we first analyze the influence of η on the estimation of R0 using the Markovian theory, and design two tasks to evaluate the applicability of the theory in epidemic forecasting and prevention evaluation of memory-dependent spreading. For comparison, we also generate the corresponding results from the non-Markovian theory in the two tasks.
Estimation of basic reproduction number
Estimating basic reproduction number R0 is crucial for determining the ultimate prevalence of disease spreading and for assessing the effectiveness of various disease containment measures45,53,54. When using the Markovian theory to fit the early-stage transmission of a memory-dependent process, a key parameter that can affect the estimation of R0 is the ratio η. To develop an analysis, recall the basic principle for estimating R0: disease spreading dynamics can be viewed as a combination of two parallel processes: infection and removal. In particular, the infection process is the reproduction of the disease within each generation, where each infected individual generates an average of R0 newly infected individuals in the subsequent generation after a mean time period Tgen. In the removal process, infected individuals are removed from the spreading chain, where each generation takes an average time Trem to be removed. For a Markovian type of dynamics with constant γ and μ, the equality Tgen = Trem holds. Consequently, during the Markovian fitting step, the average number of new infections upon the removal of a single infected individual is taken as the value of R0. For memory-dependent spreading, if the equality Tgen = Trem holds, the memory-dependent spreading curves will possess an approximate memoryless feature so that R0 can be still be estimated by counting the number of new infections at the time when the current generation of infections is removed, as shown in Fig. 3a. However, for Tgen < Trem, more than one generation is produced while the current generation is removed, R0 estimated by the Markovian theory will represent an overestimate, as shown in Fig. 3b. For Tgen > Trem, less than one generation is created during Trem, the Markovian theory will give an underestimate of R0, as shown in Fig. 3c.
a–c Mechanism of the R0 estimation. The red arrows represent the infection process of the next generation by the current generation, while the dashed arrows denote the removal of the current generation. The relationship between Tgen and Trem influences the number of new infections when the current generation of infections is removed. For Tgen = Trem, the number of new infections is exactly R0. For Tgen < Trem, the number of new infections is greater than R0. For Tgen > Trem, the number of new infections is smaller than R0. d There are three distinct categories of memory-dependent disease spreading, all sharing the same value of the basic reproduction number R0 as 1.86: Tgen < Trem (blue curves; \({\alpha }_{\inf }=1.5\), \({\beta }_{\inf }=4.64\), αrem = 2.5, βrem = 5), Tgen = Trem (red curves; \({\alpha }_{\inf }=2.5\), \({\beta }_{\inf }=5\), αrem = 2.5, βrem = 5), and Tgen > Trem (green curves; \({\alpha }_{\inf }=3.5\), \({\beta }_{\inf }=5.32\), αrem = 2.5, βrem = 5), where the fractions of cumulative infected individuals (i.e., sum of infected and removed fractions) are simulated using 100 independent realizations. The predicted future evolution of the spreading dynamics by the Markovian theory with the fitted parameters are also shown: Tgen < Trem (blue + symbols), Tgen = Trem (red × symbols), and Tgen > Trem (green Δ symbols). The gray area marks the training data (see Method for detailed approach of training data selection). e The relationship between \(\ln (\ln {R}_{0}/\ln {\hat{R}}_{0})\) (R0 represents the real basic reproduction number, while \(\hat{{R}_{0}}\) denoted the estimated one) and \(\ln \eta\). The horizontal and vertical dotted lines show that the equality between Tgen and Trem results in an accurate estimation of R0 and the dashed line represents a linear fitting with the slope 1.59. Inset: the relation between \(\ln {\hat{R}}_{0}\) and η with the asymptotic behaviors: for η → 0, \({\hat{R}}_{0}\to +\infty\) (i.e., \(\ln {\hat{R}}_{0}\to +\infty\)), and for η → + ∞, \({\hat{R}}_{0}\to 1\) (i.e., \(\ln {\hat{R}}_{0}\to 0\)).
Figure 3d shows the Markovian fitting for memory-dependent spreading simulation curves, which exhibit identical values of R0 under Tgen = Trem (red curves), Tgen < Trem (blue curves), and Tgen > Trem (green curves). When the equality Tgen = Trem holds, the Markovian theory with fitted parameters generates accurate predictions of the future evolution (red × symbols). For Tgen < Trem, the outbreak in the initial stage is accelerated, resulting in an overestimation by the Markovian theory (blue + symbols). For Tgen > Trem, the initial outbreaks are decelerated, leading to an underestimation by the Markovian approach (green Δ symbols).
The above qualitative insights lead to a semi-empirical relationship between the Markovian-estimated basic reproduction number \({\hat{R}}_{0}\) and its actual value R0 as:
where a is a positive coefficient (see “Method” for a detailed derivation). The value of a is a crucial and constant parameter in Eq. (15), and it needs to be determined by fitting it to the data. Once this constant a is obtained, the actual value of R0 can be derived by adjusting the estimated \({\hat{R}}_{0}\) based on Eq. (15), and more accurate steady state can be calculated by using Eq. (7).
Equation (15) implies the relationship \(\ln (\ln {R}_{0}/\ln {\hat{R}}_{0})=a\ln \eta\). We use Weibull time distributions \({\psi }_{\inf }(\tau )\) and ψrem(τ) constrained by certain average infection and removal times: \({T}_{\inf }=5\), Trem = 7; \({T}_{\inf }=7\), Trem = 7; \({T}_{\inf }=5\), Trem = 5; \({T}_{\inf }=7\), Trem = 5; and \({T}_{\inf }=6\), Trem = 6 for memory-dependent Monte Carlo simulations. Figure 3e shows the linear relationship between \(\ln (\ln {R}_{0}/\ln {\hat{R}}_{0})\) and \(\ln \eta\), providing support for our qualitative analysis of the Markovian estimation. The estimation of R0 also depends on the ratio η and is relatively insensitive to the specific values of Tgen, \({T}_{\inf }\), or Trem for the same form of time distribution. The results in the inset of Fig. 3e further confirm that the estimated \({\hat{R}}_{0}\) approaches 1 when Tgen is much larger than Trem and tends to +∞ when Tgen is much smaller than Trem. By fitting the available data, we have determined the value of a to be 1.59. After obtaining the value of a, we can develop our web-based application for rectifying R0 and epidemic forecasting50. We also plot additional experimental results for log-normal and gamma distributions (log-normal, \({T}_{\inf }=5\), Trem = 7; and gamma, \({T}_{\inf }=5\), Trem = 7) in Supplementary Note 7. Referring to Supplementary Fig. S2, various forms of time distributions may affect the Markovian estimation of R0. However, their influence appears less compared to the impact of the value of η. Hence, it can be concluded that the distribution form seldom affects the estimation of R0.
Epidemic forecasting
As suggested in Fig. 1d, we evaluate the efficacy of Markovian theory for epidemic forecasting. We use the early stage of Monte Carlo simulation data to fit parameters under both Markovian and non-Markovian hypotheses and then to predict future disease outbreaks. The remaining simulation data are leveraged to evaluate the accuracy of the Markovian and non-Markovian forecasting results. Regardless of the forms of time distributions in the memory-dependent Monte Carlo simulations (Weibull, log-normal, or gamma), the non-Markovian model fits the training data in a consistent manner, i.e., by assuming the actual time distributions as unknown and treating them as Weibull time distributions.
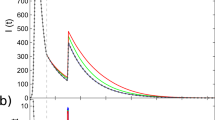
Figure 4a–c shows the evolution of the spreading dynamics from three types of memory-dependent Monte Carlo simulations with Weibull infection and removal distributions, where the shape parameters \({\alpha }_{\inf }\) and αrem are selected according to \(\ln {\alpha }_{\inf }=-0.3,\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}=1.2\) (Fig. 4a), \(\ln {\alpha }_{\inf }=0.45,\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}=0.45\) (Fig. 4b), and \(\ln {\alpha }_{\inf }=1.2,\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}=-0.3\) (Fig. 4c), for \({T}_{\inf }=5\) and Trem = 7. For the Weibull distributions, we have \({\alpha }_{\inf }\, < \,{\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\), \({\alpha }_{\inf }={\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\) and \({\alpha }_{\inf }\, > \,{\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\), corresponding to Tgen < Trem, Tgen = Trem, and Tgen > Trem, respectively. We compare the simulated cumulative infected fractions to those predicted by the Markovian and non-Markovian theories. In general, the non-Markovian theory provides more accurate predictions than the Markovian theory. For the specific parameter setting \(\ln {\alpha }_{\inf }=0.45,\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}=0.45\) (i.e., Tgen = Trem), both theories yield a high accuracy.
a–c Predicted evolution of the cumulative infected fraction (i.e., sum of infected and removed fractions) by the Markovian (orange dotted curves) and non-Markovian (red dashed curves) theories, in comparison with the Monte Carlo simulations with Weibull time distributions (blue solid curves), for three sets of simulation parameters, respectively. The results are the averages of 100 independent realizations with the standard deviations indicated by the shaded regions. And all the memory-dependent Monte Carlo simulations exhibit a similar R0 value of 1.83. The gray area marks the training data (see “Method” for detailed approach of training data selection). d, e The forecasting errors ε+ of Markovian and non-Markovian theories with respect to the memory-dependent Monte Carlo simulations in the parameter plane of \(\ln {\alpha }_{\inf }\) and \(\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\) in the range [−0.3, 1.2]. The green, blue and red squares mark the parameters of Monte Carlo simulations in (a–c), respectively. f The forecasting errors ε+ from the Markovian and non-Markovian theories for different values of \(\ln \eta\) under five scenarios of time distribution setting for Monte Carlo simulations. The corresponding estimations for a number of real-world diseases (COVID-19, SARS, H1N1 and Smallpox) are also included.
The accuracy can be assessed through the forecasting error ε+ that evaluates whether a theory overestimates or underestimates the steady-state cumulative infection, i.e., quantifying the extent of deviation between the results obtained from Markovian or non-Markovian theories and those derived from Monte Carlo simulations (see “Method” for detailed definition of ε+). A plus value of ε+ means overestimation while minus value indicates underestimation. We evaluate the accuracy measure ε+ in the parameter plane of \(\ln {\alpha }_{\inf }\) and \(\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\), ranging from −0.3 to 1.2. Figure 4d, e shows that the Markovian accuracy is sensitive to parameter changes: underestimated if \({\alpha }_{\inf }\) is greater than αrem (Tgen > Trem), overestimated when \({\alpha }_{\inf }\) is smaller than αrem (Tgen < Trem), and a high forecasting accuracy is achieved only for \({\alpha }_{\inf }={\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\) (Tgen = Trem). In contrast, the non-Markovian theory yields highly accurate results in the whole parameter plane, with only a slight underestimation for \({\alpha }_{\inf }\gg {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\). This can be primarily attributed to the growing challenge of fitting simulation data, where the absolute derivatives of R0 with respect to parameters become extremely high, making the fitting process much sensitive to the parameters55,56. This occurs when \({\alpha }_{\inf }\) is greater than αrem or Tgen is much larger than Trem, causing the Monte Carlo curves to become more sensitive to the parameters and potentially deviate from the accurate theoretical curves (see Supplementary Fig. S3 and Supplementary Note 8 for details).
Using the five scenarios specified in Fig. 2j for memory-dependent Monte Carlo simulations, we obtain the relationship between ε+ and \(\ln \eta\), as shown in Fig. 4f. It can be seen that, in the Markovian framework, an overestimation arises for Tgen < Trem, and an underestimation occurs for Tgen > Trem. Only when Tgen = Trem is an accurate estimate achieved. In general, the non-Markovian theory provides much more accurate forecasting than the Markovian theory, especially when Tgen and Trem are not equal. The results further illustrate that the specific values of Tgen, \({T}_{\inf }\), or Trem have little impact on forecasting accuracy, and the impact of the time distribution forms is much lower compared to the influence exerted by the η value.
To establish the relevance of these results to real-world diseases, we obtain the distributions of \({\psi }_{\inf }(\tau )\) and ψrem(τ) for four known infectious diseases, including COVID-19, SARS, H1N1 influenza, and smallpox, using the information in refs. 2,33,34,35,36,37,38,39,40. We then calculate the corresponding values of ε+ and \(\ln \eta\) based on the Markovian and non-Markovian approaches. As demonstrated in Fig. 4f, the positions of the four diseases in the (\(\ln \eta\), ε+) plane are consistent with the results of our estimations. Because the data were from the reports of laboratory-confirmed cases incorporating the effects of the quarantine and distancing from susceptible individuals after the confirmation of the diagnosis, Tgen of the four diseases are all smaller than the corresponding values of Trem, leading to some overestimation for the Markovian forecasting results.
Evaluation of vaccination strategies
In the development and application of a theory for disease spreading, assessing the effects of different vaccination strategies is an important task. Here we consider five prioritization strategies for vaccine distribution5: individuals under 20 years (denoted as m = 1), adults between 20 and 49 years (m = 2), adults above 20 years (m = 3), adults above 60 years (m = 4), and all age groups (m = 5), and implement these strategies in Monte Carlo simulations (see “Method” for the detailed procedure of vaccination in Monte Carlo simulations). Figure 5a shows the results of epidemic evolution with these vaccination strategies, where the shape parameters are chosen according to \(\ln {\alpha }_{\inf }=0.45\) and \(\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}=0.45\) (Tgen = Trem). Figure 5b–c shows the results from the Markovian and non-Markovian theories, respectively, with the corresponding fitted parameters for the vaccination strategies (see “Method” for the detailed procedure of vaccination in theoretical calculations). These results indicate that the Markovian and non-Markovian theories yield the correct epidemic evolution and future outbreaks under different vaccination scenarios, when Tgen = Trem.
a–c For simulation parameters chosen according to \(\ln {\alpha }_{\inf }=0.45\) and \(\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}}=0.45\), the cumulative infected fraction (i.e., sum of infected and removed fractions) curves from Monte Carlo simulations and the corresponding Markovian and non-Markovian theories with fitting parameters for five vaccination strategies. The average results are obtained from 100 independent realizations with the shaded regions representing the standard deviations. And all the memory-dependent Monte Carlo simulations exhibit a similar R0 value of 1.83. The gray area marks the training data (see “Method” for detailed approach of training data selection). d Vector δ calculated from the results in Fig. 5a–c. e, f The prevention evaluation errors ε* of Markovian and non-Markovian theories for evaluating the effects of vaccination prevention in the parameter plane \((\ln {\alpha }_{\inf },\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}})\). The green squares mark the selected parameters in (a–c). g, h The optimization failure probabilities \(\hat{\varepsilon }\) arising from the Markovian and non-Markovian within the parameter plane \((\ln {\alpha }_{\inf },\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}})\). The green squares mark the selected parameters in (a–c). i The prevention evaluation errors ε* from the Markovian and non-Markovian theories versus \(\ln \eta\) under five scenarios of time distribution setting for Monte Carlo simulations. The estimated errors for four real diseases (COVID-19, SARS, H1N1, and Smallpox) are also shown. j The optimization failure probabilities \(\hat{\varepsilon }\) from the Markovian and non-Markovian theories against \(\ln \eta\) in five different time distribution scenarios for Monte Carlo simulations. The optimization failure probabilities for four real diseases (COVID-19, SARS, H1N1, and Smallpox) are also presented. The color and symbol theme is the same as (i).
To characterize the effectiveness of different vaccination strategies in blocking disease transmission, we introduce a vaccination effectiveness vector, δ, whose m-th element quantifies the cumulative infected fraction with the m-th vaccination strategy in the steady state: \({\delta }_{m}={\tilde{c}}_{m}\), for m = 0, …, 5 (subscript m = 0 indicates the results without vaccination). Figure 5d shows that the δ vectors from the Monte Carlo simulation, Markovian and non-Markovian theories from Fig. 5a–c, respectively.
We further introduce a metric, the so-called prevention evaluation error ε*, that gauges the ability of the Markovian and non-Markovian theories to estimate the total effectiveness of vaccination, i.e., measuring the disparity between the results calculated by the Markovian or non-Markovian theories and those obtained through Monte Carlo simulations considering various vaccination strategies (see “Method” for the detailed definition of ε*). Figure 5e, f shows the average values of ε* of the two theories in the simulation parameter plane using 100 independent realizations, which are similar to those in Fig. 4d–e, indicating that the error mainly comes from the R0 estimation. In general, the Markovian theory performs well only in the diagonal area of the parameter plane where \({\alpha }_{\inf }={\alpha }_{{{{{{{{\rm{rem}}}}}}}}}\), as shown in Fig. 5e, and the non-Markovian theory outperforms the Markovian counterpart in most cases, as shown in Fig. 5f.
Meanwhile, we assess the ability of both the Markovian and non-Markovian theories to detect the optimal vaccination strategy. We also define a quantity, optimization failure probability \(\hat{\varepsilon }\), to quantify the probability of a theory failing to identify the optimal strategy, i.e., that leads to the lowest cumulative infection among these strategies (see “Method” for the detailed definition of \(\hat{\varepsilon }\)). Figure 5g–h illustrates the results of \(\hat{\varepsilon }\) for the two theories within the parameter plane \((\ln {\alpha }_{\inf },\ln {\alpha }_{{{{{{{{\rm{rem}}}}}}}}})\). While the non-Markovian theory still demonstrates superior performance, the Markovian approach proves capable of identifying the optimal strategy across a larger parameter space compared to the Markovian results depicted in Fig. 5e.
We obtain the relationships between ε* and \(\ln \eta\), as well as between \(\hat{\varepsilon }\) and \(\ln \eta\), as shown in Fig. 5i–j with the same five time-distribution scenarios as in Fig. 2j. In all cases of Fig. 5i, ε* reaches a minimum for \(\ln \eta =0\) and increases as \(\ln \eta\) deviates from zero. The results from the five scenarios further illustrates that the specific values of Tgen, \({T}_{\inf }\), or Trem play little role in the errors in vaccination evaluation, and the effect of time distribution forms is also limited. Figure 5i also includes the values of ε* for the real-world infectious diseases COVID-19, SARS, H1N1 influenza, and smallpox, which are consistent with those from the non-Markovian and Markovian theories. Regarding the results depicted in Fig. 5j, it is observed that the non-Markovian theories consistently outperform the Markovian counterparts. On the other hand, within a wide range of \(\ln \eta\) values around 0, the Markovian theories successfully identify the optimal vaccination strategy among various commonly employed ones. When the value of \(\ln \eta\) deviates from 0, Markovian theories become ineffective in determining the optimal strategy. (Note that on the left side of Fig. 5j, we only present the failures of Markovian theories to identify the optimal strategy in the Monte Carlo simulations with log-normal distribution. This is primarily due to the fact that the parameters associated with the Weibull and gamma distributions fall outside the acceptable range when we keep \({T}_{\inf }\) and Trem fixed to modify \(\ln \eta\) to a very low value.) Furthermore, we demonstrate that even when employing Markovian approaches, the optimal vaccination strategy can still be determined among these strategies considered for the four distinct real diseases.
Conducting accurate evaluations in prevention serves as the sufficient condition of the successful identification of the optimal strategy. In comparison to the prevention evaluation errors ε* of Markovian theories, the optimization failure probability \(\hat{\varepsilon }\) exhibits a wider range of \(\ln \eta\) values that result in the lowest value. The lack of mathematical continuity among these strategies is the primary reason for this. It indicates that there is no smooth transition or mathematical relationship connecting these strategies, resulting in the rank of the strategies not changing promptly when the value of \(\ln \eta\) deviates from 0. Therefore, only large errors from the Markovian theories can result in the failure to detect the optimal strategy. Based on this analysis, the extent to which \(\ln \eta\) deviates from 0, leading to the failure of Markovian theories, as well as whether such failure will occur, depends on the selection of the tested strategies.
Discussion
The COVID-19 pandemic has emphasized the importance of investigating disease transmission in human society through modeling. Empirical observations have consistently demonstrated strong memory effects in real-world transmission phenomena. The initial transient stage of an epidemic is critical for data collection, prediction, and articulation of control strategies, but an accurate non-Markovian model presents difficulties. In contrast, a Markovian model offers great advantages in parameter estimation, computation, and analyses. Uncovering the conditions under which Markovian modeling is suitable for transient epidemic dynamics is necessary.
We have developed a comprehensive mathematical framework for both Markovian and non-Markovian compartmentalized SIR disease transmissions in an age-stratified population, which allows us to identify two types of equivalence between Markovian and non-Markovian dynamics: in the steady state and transient phase of the epidemic. Our theoretical analysis reveals that, in the steady state, non-Markovian (memory-dependent) transmissions are always equivalent to the Markovian (memoryless) dynamics. However, transient-state equivalence is approximate and holds when the average generation and removal times match each other. In particular, when the average generation time is approximately equal to the average removal time, the disease transmission and removal of an infected individual exhibit a memoryless correlation, thereby minimizing the memory effects of the dynamic process. This results in highly accurate results from the Markovian theory that captures the characteristics of memory-dependent transmission based solely on the early epidemic curves. Our analysis also suggests that the Markovian accuracy is mainly determined by the value of generation-to-removal time ratio in disease transmission, where a larger-than-one (smaller-than-one) ratio can lead to underestimation (overestimation) of the basic reproduction number and epidemic forecasting, as well as the errors in the evaluation of control or prevention measures. The estimation accuracy primarily depends on this ratio, but not greatly affected by the specific values at the various times associated with the epidemic; although distribution forms might affect accuracy to some extent, their influence is much less compared to the impact of the ratio. This property exhibits substantial practical importance, because the average generation and removal times can be readily assessed based on sparse data collected from the transient phase of the epidemic, but to estimate their distributions with only sparse data is infeasible. These results provide deeper quantitative insights into the influence of memory effects on epidemic transmissions, leading to a better understanding of the connection and interplay between Markovian and non-Markovian dynamics.
There were previous studies of the equivalence between Markovian and non-Markovian transmission in the SIS model18,21,22. However, these studies addressed the steady-state equivalence rather than the transient-state equivalence. To our knowledge, our work is the first to investigate the transient-state equivalence of the SIR model. In addition, previous studies mainly examined the impact of the average generation time on the transmission dynamics, such as how the shape of the generation time distribution affects the estimation of R053 or the use of serial time distributions in estimating R0 during an epidemic45. There was a gap in the literature regarding how generation times affect the accuracy of different models. Our paper fills this gap by providing a criterion for using Markovian frameworks to model memory-dependent transmission based on the relationship between the average generation and removal times.
From an application perspective, our study suggests that the impact of the time distribution forms on Markovian estimation accuracy is limited, making it easier to select models between Markovian and non-Markovian dynamics in the initial outbreak of an epidemic based primarily on the generation-to-removal time ratio. This insight is especially useful since detailed time distribution forms are often harder to detect than their corresponding mean values. In addition, we note that in previous studies, it was observed that in various scenarios, serial intervals, albeit with larger variances, are anticipated to possess a consistent mean value with the average generation time and are more straightforward to measure2,45,46,47,48,49. Given the practical difficulties in observing the generation time, our finding of minimal impact from the distribution forms suggests that the average serial interval can be utilized as a substitute of the average generation time to determine the applicability of the Markovian theories for modeling purposes without compromising accuracy, although numerous studies have indicated that replacing the generation time distribution with the serial interval distribution may affect the analysis of transmission dynamics44,45,57. Meanwhile, based on Eq. (15), if we determine the ratio of generation-to-removal time, the estimated R0 obtained through the Markovian approach can be adjusted to approximate the true value. And our web-based application showcases the demonstration of rectifying R0 and epidemic forecasting50.
Our study highlights the critical importance of accurately quantifying R0 for achieving precise epidemic forecasting and prevention evaluation. A previous work54 revealed that the value of R0 depends on three key components: the duration of the infectious period (e.g., ψrem(τ)), the probability of infection resulting from a single contact between an infected individual and a susceptible one (e.g., \({\psi }_{\inf }(\tau )\)), and the number of new susceptible individuals contacted per unit of time. However, given the practical limitations inherent in obtaining all three components, numerous methods have been developed for estimating R0. Although our work presents a specific approach, which fits the parameters of exponential or non-exponential time distribution by using the initial outbreak curves, it is not the only one available. For example, when contact patterns are unknown, R0 can be estimated by fitting the growth rate g and the generation time distribution ψgen(τ), and then applying them in the Euler-Lotka equation45,53,54. However, since the focus of our work is on epidemic forecasting and evaluation of prevention measure where the data of contact patterns are given, R0 can be directly calculated once \({\psi }_{\inf }(\tau )\) and ψrem(τ) are fitted, without requiring the fitting of any additional quantity such as growth rate g. The estimation of R0 can also be achieved by using data in the steady state, such as the final size of an epidemic or equilibrium conditions54. However, this method is not suitable for the transient phase where only early-stage curves are available. Utilizing the approach delineated in this paper is practically more appropriate for estimating R0.
While our study focused on transmission within the SIR framework, extension to SEIR or SIS models is feasible. While we emphasized the significance of the transient-state equivalence in disease transmission, transient dynamics are more relevant or even more crucial than the steady state in nonlinear dynamical systems58. For example, in ecological systems, transient dynamics play a vital role in empirical observations and are therefore a key force driving natural evolution59,60,61,62,63,64. In neural dynamics, transient changes in neural activity can mediate synaptic plasticity, a crucial mechanism for learning and memory65,66,67. Therefore, the identification of suitable conditions for choosing between Markovian and non-Markovian dynamics may not be limited to epidemic dynamics alone and may serve as a valuable reference for other fields as well.
Taken together, our study establishes an approximate equivalence between Markovian and non-Markovian dynamics in the transient state, assuming that time distributions follow Weibull forms (see Supplementary Note 4 for details). While the applicability of our findings to most synthetic and empirical distributions has been analyzed qualitatively, a quantitative analysis requires further studies. For extreme cases with non-Weibull distributions, the transmission should be evaluated using other specific methods. While we have provided a qualitative analysis of the mechanism underlying why time distribution forms have minimal impacts on the errors of Markovian estimations, a more rigorous theoretical analysis is needed and requires further exploration. Meanwhile, in the spreading process, there are other memory effects beyond the “temporal” memory considered in our study, for example, the transmission capacity can be influenced by the previous transmission path42, which is referred to as “spatial” memory effect. However, our study specifically focuses on analyzing the impact of “temporal” memory effects. To understand how “spatial” memory effects affect the accuracy of the Markovian approach, further research is needed. In addition, due to the complexity of the nonlinear transmission, our study has produced a semi-empirical relationship to estimate the overestimation and underestimation of Markovian methods. Further research is required to develop a rigorous formula that can accurately predict these effects. Additionally, the non-Markovian SIR model utilized in this study shares similarities with the Hawkes or HawkesN process41,68 (see Supplementary Note 9 for details). These processes are commonly employed for the statistical modeling of events in diverse fields, where random events exhibit self-exciting behavior. We anticipate that our research will offer fresh perspectives and valuable insights to advance the understanding of related studies in this area.
Method
Monte Carlo simulation
In the simulation, we classify N individuals into n subgroups based on the age distribution p. The index of the subgroup to which an individual belongs is denoted by l (where 1 ≤ l ≤ n), and the index of the individual within the subgroup is denoted by u (where 1 ≤ u ≤ plN). The state of the individual u in the age group l is represented by Xlu, which includes the states S (susceptible), I (infected), W (recovered), and D (dead), where W and D both represent R (removed), while I and R both represent C (cumulative infected). For each individual, we also record the absolute time of infection and removal using two variables: \({t}_{lu}^{\inf }\) and \({t}_{lu}^{{{{{{{{\rm{rem}}}}}}}}}\), respectively. The absolute time of the system is denoted by t, and we implement the total spreading simulation step by step using a finite time step Δt as follows:
-
i.
Initialization: set t = 0, Xlu for every individual is set to S.
-
ii.
Set infection seeds: choose a set of individuals as the infection seeds and the corresponding Xlu are set to I, the corresponding \({t}_{lu}^{\inf }\) are set to 0, and \({t}_{lu}^{{{{{{{{\rm{rem}}}}}}}}}\) are set to a random value following the removal time distribution ψrem(τ).
-
iii.
Infection in one time step: calculate the infection rate, \({\hat{\omega }}_{lu}^{\inf }(t)\), of infected individual u in age group l during the current time step by
$${\hat{\omega }}_{lu}^{\inf }(t)=1-{{{\Psi }}}_{\inf }(t-{t}_{lu}^{\inf }+{{\Delta }}t)/{{{\Psi }}}_{\inf }(t-{t}_{lu}^{\inf }).$$
The probability \({\bar{\omega }}_{l}^{\inf }(t)\) of each susceptible individual in age group l being infected can be calculated by
where \({{{{{{{{\mathcal{I}}}}}}}}}_{m}(t)\) is the index set of the infected individuals in age group m at time t. The number of the susceptible individuals being infected in age group l follows a binomial distribution \(B({s}_{l}^{\star }(t){{{{{p}}}}}_{l}N,{\bar{\omega }}_{l}^{\inf }(t))\), where \({s}_{l}^{\star }(t)\) denotes the fraction of susceptible individuals in age group l at time t. Then generate a random number Nl(t) following this binomial distribution and randomly select Nl(t) susceptible individuals in age group l to set them as I state. The corresponding \({t}_{lu}^{\inf }\) of the new infected individuals are set to the current t and \({t}_{lu}^{{{{{{{{\rm{rem}}}}}}}}}\) are set to τrem + t, where the random τrem follow the removal time distribution ψrem(τ).
-
iv.
Removal in one time step: check if \({t}_{lu}^{{{{{{{{\rm{rem}}}}}}}}}\) of each infected individual is during the current time step. If this condition is satisfied, set their state to D with the probability σl and to W otherwise, where σl is the infection fatality rate of age group l69. Then let t ← t + Δt.
-
v.
Repeat the process iii) and iv), until no individual with I index exists.
Time distributions
In the numerical calculations or Monte Carlo simulations, we employ three types of time distributions, i.e., Weibull, log-normal, and gamma, to describe the non-Markovian or memory-dependent transmission process.
For Weibull time distribution, it follows:
where α and β denote the shape and scale parameters, respectively.
The log-normal time distribution is defined as follows:
The gamma time distribution is expressed as follows:
where Γ(⋅) denotes gamma function, while α and β represent the shape and scale parameters, respectively.
Additionally, the survival function could be calculated by:
while the hazard function could be derived from:
For time distribution, survival function, hazard function and the corresponding parameters, incorporating subscripts “\(\inf\)” and “rem” signifies those related to infection and removal, respectively. For example, \({\psi }_{\inf }(\tau )\) represents the time distribution of infection, while \({\alpha }_{\inf }\) denotes its shape parameter.
Derivation of semi-empirical estimation of basic reproduction number
Intuitively, the period of Trem can accommodate Trem/Tgen = 1/η time intervals of length Tgen, corresponding to the result of 1/η generations of infections. This can lead to an exponential increase in the number of infections during Trem. This intuition suggests a relationship between the fitted basic reproduction number \({\hat{R}}_{0}\) and the actual R0, which can be expressed as an exponential function:
where f(⋅) is a monotonically increasing function that satisfies three conditions. First, f(1) = 1, indicating that \({\hat{R}}_{0}\) can be accurately estimated when Tgen = Trem. Second, f(0) = 0, meaning that if Trem is an extremely small fraction of Tgen, the transmission will take a long time to reach the steady state, causing the curve to be flat in the initial stage and potentially causing the Markovian fitting to produce the estimate \({\hat{R}}_{0}=1\). Third, f( + ∞) = +∞, implying that if Trem is extremely large compared to Tgen, the transmission will quickly reach the steady state, causing the Markovian fitting to give an extremely large estimate of \({\hat{R}}_{0}\). Because the actual transmission process involves many complicated nonlinear relationships, identifying the specific form of the function f(⋅) is a challenging task. We thus assume
where a is an unknown positive coefficient. This leads to Eq. (15).
Definition of errors
The difference ε between non-Markovian and the corresponding Markovian results calculated from Eqs. ((13), (14)) is defined as:
where the pairs (s‡, s†), (i‡, i†) and (r‡, r†) correspond to the non-Markovian and Markovian susceptible, infected and removed curves, respectively. The 2-norm \(\parallel \!\!\cdot {\parallel }_{2,{{\mathbb{T}}}_{\theta }^{{{{\ddagger}}} }}\) on time duration \({{\mathbb{T}}}_{\theta }^{{{{\ddagger}}} }\) ensures that ε measures the “distance” between non-Markovian and the Markovian transient states. It is not appropriate to set time duration as the total transmission period because the cumulative infected fraction approaches the steady-state value asymptotically, making it difficult to determine the exact time point of the steady state. To address this issue, we choose \({{\mathbb{T}}}_{\theta }^{{{{\ddagger}}} }\) as \([0,{t}_{\theta }^{{{{\ddagger}}} }]\), where \({t}_{\theta }^{{{{\ddagger}}} }\) is the time when the non-Markovian cumulative infected fraction c‡(t) reaches the θ percentile point within the range that spans from its initial value to its steady-state value. For instance, let ć‡ denote the cumulative infected fraction in non-Markovian calculation at time 0, i.e., ć‡ = c‡(0), while \({\tilde{c}}^{{{{\ddagger}}} }\) represents this fraction at the steady state, i.e., \({\tilde{c}}^{{{{\ddagger}}} }=\mathop{\lim }\nolimits_{t\to +\infty }{c}^{{{{\ddagger}}} }(t)\). \({t}_{\theta }^{{{{\ddagger}}} }\) is determined to satisfy the following equation:
Therefore, θ determines the time period during which we measure the “distance” ε between non-Markovian and the Markovian transient states. The value of θ in Fig. 2j is selected as 50 (see Supplementary Figs. S4, S5 and Supplementary Note 10 for more selection of θ and detailed analysis).
The forecasting error ε+ that evaluates whether a theory overestimates or underestimates the steady-state cumulative infected fraction is defined as:
where \({\tilde{t}}^{\star }\) denotes the time when the Monte Carlo stochastic simulation reaches the steady state when no infection occurs in the population, \({c}^{\lozenge}({\tilde{t}}^{\star })\) and \({c}^{\star }({\tilde{t}}^{\star })\) are the cumulative infected fractions from theory and simulation, respectively. A positive value of ε+ indicates overestimation, whereas a negative value indicates underestimation.
The prevention evaluation error ε*, which gauges the ability of the Markovian and non-Markovian theories to estimate the total effectiveness of vaccination, is defined as:
where δ⋆ is the result from Monte Carlo simulation, δ◇ represents the result from theoretical calculation and ∥ ⋅ ∥2 is the 2-norm of a vector.
The optimization failure probability \(\hat{\varepsilon }\), which measures the probability that a theory fails to identify the optimal vaccination strategy, is defined as:
where
\({\delta }_{(l)}^{\star }\) and \({\delta }_{(l)}^{\lozenge}\) represent the vectors, δ⋆ and δ◇, for the l-th experiment, respectively, and z denotes the total number of experiments (in this paper, z is set to 100). Consequently, \(\hat{\varepsilon }\) quantifies the fraction of experiments in which a theory fails to identify the optimal vaccination strategy, and serves as a measure of the probability of failure in optimizing the vaccination strategy.
Selection of training data
In our study, we selected the curves of all states that occurred prior to the time point at which the cumulative infected fraction reached a specific percentile ζ situated between the initial and steady-state cumulative infected fractions, as the training data. For example, let ć⋆ denote the cumulative infected fraction at time 0, while \({\tilde{c}}^{\star }\) represents this fraction at the steady state. The selection of the maximum time point of the time duration \({{\mathbb{T}}}_{\zeta }^{\star }\) for training data, denoted as \({t}_{\zeta }^{\star }\), is determined to meet the following equation:
Therefore, ζ determines the time duration, i.e., \({{\mathbb{T}}}_{\zeta }^{\star }=[0,{t}_{\zeta }^{\star }]\), of the training data, which could vary over different Monte Carlo simulations. In our study, we consistently choose ζ to be 20. Note that choosing a specific constant time period as the training data may not be appropriate, as it can result in an overabundance of data points for fitting due to some instances of fast transmission already having reached the steady state, while some instances of slow transmission may not have spread out yet, leading to invalid training data.
Fitting method
Because the removal process is independent of the infection one, we divide the fitting method into two parts: removal parameter fitting and infection parameter fitting. Specifically, we use c⋆(t) and r⋆(t) to denote the cumulative infected and removed fractions of a Monte Carlo simulation at time t. These two types of data are substituted into the Eq. (3) to fit the parameters of ψrem(τ). To explain further, we can define a loss function:
where \({t}_{\zeta }^{\star }\) represents the maximum time point of the training data, \({\hat{r}}^{\star }(t)\) denotes the removed fraction calculated by cumulative infected fraction, i.e.,
In this equation, Ψrem(τ) is the survival function of a specific removal time distribution ψrem(τ), such as Weibull, log-normal, or gamma distributions for the non-Markovian framework, and exponential distribution for the Markovian framework. By minimizing the loss function Lrem, we can determine the optimal parameters for removal time distribution ψrem(τ). This can be accomplished using the L-BFGS-B optimization algorithm, which is well-suited for minimizing the loss function70.
Likewise, we use \({c}_{l}^{\star }(t)\) to denotes the cumulative infected fraction of age group l of a Monte Carlo simulation at time t and \({s}_{l}^{\star }(t)=1-{c}_{l}^{\star }(t)\) represents the corresponding susceptible fraction (note that cumulative infected fraction is precisely defined as the sum of the infected and removed fractions, which is accurately expressed as \({c}_{l}^{\star }(t)={i}_{l}^{\star }(t)+{r}_{l}^{\star }(t)=1-{s}_{l}^{\star }(t)\), leading to \({s}_{l}^{\star }(t)=1-{c}_{l}^{\star }(t)\)). After obtaining the removal parameters, these two types of data are put into Eq. (1) to fit the infection time distribution parameters. In details, we define a loss function \({L}_{\inf }\), which is given by:
where \({\hat{c}}_{l}^{\star }(t)\) represents the cumulative infected fraction of age group l at time t calculated by the cumulative infected fractions of simulation data by using the equation:
In this equation, \({\omega }_{\inf }(\tau )\) is the hazard function of a specific infection time distribution \({\psi }_{\inf }(\tau )\), such as Weibull, log-normal, or gamma distributions for the non-Markovian framework, and exponential distribution for the Markovian framework. And Ψrem(τ) is the survival function of the removal time distribution ψrem(τ), which is already obtained by fitting. To determine the optimal parameters for the infection time distribution \({\psi }_{\inf }(\tau )\), we minimize the loss function \({L}_{\inf }\) using the L-BFGS-B optimization algorithm70.
Vaccination method
We assume that the individuals will build enough immune protection from the disease κ days after vaccination with the probability ρ, where ρ is the vaccine efficacy. In Monte Carlo simulations, if a susceptible individual gets vaccinated at time tvac, he/she will be marked with the probability ρ. When the absolute time reaches tvac + κ, if this individual has not been infected, he/she will be set to a state called protected state, indicating that this individual is protected from the disease and will never be infected.
In theoretical calculation, when a fraction of individuals in age group l get vaccinated with the detailed vaccination fraction υl, vaccination time tvac and the fraction of susceptible individuals, which have not been vaccinated by time t, is recorded (denoted as sl,*(tvac)). When the absolute time reaches tvac + κ, the corresponding value of sl(tvac + κ) will be set as \({s}_{l}({t}_{{{{{{{{\rm{vac}}}}}}}}}+\kappa )[1-\frac{\rho {\upsilon }_{l}}{{s}_{l,*}({t}_{{{{{{{{\rm{vac}}}}}}}}})}]\).
Data availability
All relevant data are available at https://github.com/fengmi9312/Validity-of-Markovian-for-Memory/tree/main/FigureData.
Code availability
The web-based application can be visited at https://cns.hkbu.edu.hk/toolbox/Validity-of-Markovian-for-Memory/main.html. The GitHub repository, which includes the source code for all the figure results, the web-based application, and an additional Python application, can be accessed at https://github.com/fengmi9312/Validity-of-Markovian-for-Memory.git.
References
Dong, E., Du, H. & Gardner, L. An interactive web-based dashboard to track covid-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020).
Ferretti, L. et al. Quantifying SARS-COV-2 transmission suggests epidemic control with digital contact tracing. Science 368, eabb6936 (2020).
Giordano, G. et al. Modeling vaccination rollouts, SARS-COV-2 variants and the requirement for non-pharmaceutical interventions in Italy. Nat. Med. 27, 993–998 (2021).
Jentsch, P. C., Anand, M. & Bauch, C. T. Prioritising COVID-19 vaccination in changing social and epidemiological landscapes: a mathematical modelling study. Lancet Infect. Dis. 21, 1097–1106 (2021).
Bubar, K. M. et al. Model-informed COVID-19 vaccine prioritization strategies by age and serostatus. Science 371, 916–921 (2021).
Buckner, J. H., Chowell, G. & Springborn, M. R. Dynamic prioritization of COVID-19 vaccines when social distancing is limited for essential workers. Proc. Natl Acad. Sci. 118, e2025786118 (2021).
Goldstein, J. R., Cassidy, T. & Wachter, K. W. Vaccinating the oldest against COVID-19 saves both the most lives and most years of life. Proc. Natl Acad. Sci. 118, e2026322118 (2021).
Viana, J. et al. Controlling the pandemic during the sars-cov-2 vaccination rollout. Nat. Commun. 12, 1–15 (2021).
Matrajt, L., Eaton, J., Leung, T. & Brown, E. R. Vaccine optimization for COVID-19: who to vaccinate first? Sci. Adv. 7, eabf1374 (2021).
Matrajt, L. et al. Optimizing vaccine allocation for COVID-19 vaccines shows the potential role of single-dose vaccination. Nat. Commun. 12, 1–18 (2021).
Zhao, S. et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-NCOV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 92, 214–217 (2020).
Metcalf, C. J. E. & Lessler, J. Opportunities and challenges in modeling emerging infectious diseases. Science 357, 149–152 (2017).
Riley, S. Large-scale spatial-transmission models of infectious disease. Science 316, 1298–1301 (2007).
Levin, S. A., Grenfell, B., Hastings, A. & Perelson, A. S. Mathematical and computational challenges in population biology and ecosystems science. Science 275, 334–343 (1997).
Pastor-Satorras, R., Castellano, C., Van Mieghem, P. & Vespignani, A. Epidemic processes in complex networks. Rev. Mod. Phys. 87, 925 (2015).
Lambiotte, R., Tabourier, L. & Delvenne, J.-C. Burstiness and spreading on temporal networks. Eur. Phys. J. B 86, 1–4 (2013).
Lin, Z.-H. et al. Non-markovian recovery makes complex networks more resilient against large-scale failures. Nat. Commun. 11, 1–10 (2020).
Cator, E., Van de Bovenkamp, R. & Van Mieghem, P. Susceptible-infected-susceptible epidemics on networks with general infection and cure times. Phys. Rev. E 87, 062816 (2013).
Boguná, M., Lafuerza, L. F., Toral, R. & Serrano, M. Á. Simulating non-markovian stochastic processes. Phys. Rev. E 90, 042108 (2014).
Van Mieghem, P. & Van de Bovenkamp, R. Non-markovian infection spread dramatically alters the susceptible-infected-susceptible epidemic threshold in networks. Phys. Rev. Lett. 110, 108701 (2013).
Starnini, M., Gleeson, J. P. & Boguñá, M. Equivalence between non-markovian and Markovian dynamics in epidemic spreading processes. Phys. Rev. Lett. 118, 128301 (2017).
Feng, M., Cai, S.-M., Tang, M. & Lai, Y.-C. Equivalence and its invalidation between non-markovian and Markovian spreading dynamics on complex networks. Nat. Commun. 10, 1–10 (2019).
Min, B., Goh, K.-I. & Kim, I.-M. Suppression of epidemic outbreaks with heavy-tailed contact dynamics. EPL 103, 50002 (2013).
Karrer, B. & Newman, M. E. Message passing approach for general epidemic models. Phys. Rev. E 82, 016101 (2010).
Wang, Y., Chakrabarti, D., Wang, C. & Faloutsos, C. Epidemic spreading in real networks: an eigenvalue viewpoint. In Proc. 22nd International Symposium on Reliable Distributed Systems, 25–34 (IEEE, 2003).
Chakrabarti, D., Wang, Y., Wang, C., Leskovec, J. & Faloutsos, C. Epidemic thresholds in real networks. ACM Trans. Inf. Syst. Secur. 10, 1–26 (2008).
Keeling, M. J. & Grenfell, B. T. Disease extinction and community size: modeling the persistence of measles. Science 275, 65–67 (1997).
Barabasi, A.-L. The origin of bursts and heavy tails in human dynamics. Nature 435, 207–211 (2005).
Pappalardo, L. et al. Returners and explorers dichotomy in human mobility. Nat. Commun. 6, 1–8 (2015).
Yan, X.-Y., Wang, W.-X., Gao, Z.-Y. & Lai, Y.-C. Universal model of individual and population mobility on diverse spatial scales. Nat. Commun. 8, 1–9 (2017).
Bratsun, D., Volfson, D., Tsimring, L. S. & Hasty, J. Delay-induced stochastic oscillations in gene regulation. Proc. Natl Acad. Sci. 102, 14593–14598 (2005).
Vazquez, A., Racz, B., Lukacs, A. & Barabasi, A.-L. Impact of non-poissonian activity patterns on spreading processes. Phys. Rev. Lett. 98, 158702 (2007).
Moghadas, S. M. et al. The implications of silent transmission for the control of COVID-19 outbreaks. Proc. Natl Acad. Sci. 117, 17513–17515 (2020).
Lauer, S. A. et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 172, 577–582 (2020).
Gatto, M. et al. Spread and dynamics of the COVID-19 epidemic in Italy: effects of emergency containment measures. Proc. Natl Acad. Sci. 117, 10484–10491 (2020).
Li, R. et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-COV-2). Science 368, 489–493 (2020).
Buitrago-Garcia, D. et al. Occurrence and transmission potential of asymptomatic and presymptomatic sars-cov-2 infections: a living systematic review and meta-analysis. PLoS Med. 17, e1003346 (2020).
World Health Organization, et al. Consensus document on the epidemiology of severe acute respiratory syndrome (sars). Technical Report, (World Health Organization, 2003).
Chowell, G. et al. Model parameters and outbreak control for sars. Emerg. Infect. Dis. 10, 1258 (2004).
Eichner, M. & Dietz, K. Transmission potential of smallpox: estimates based on detailed data from an outbreak. Am. J. Epidemiol. 158, 110–117 (2003).
Rizoiu, M.-A., Mishra, S., Kong, Q., Carman, M. & Xie, L. Sir-Hawkes: linking epidemic models and Hawkes processes to model diffusions in finite populations. In Proc. World Wide Web Conference, 419–428 (2018).
Rosvall, M., Esquivel, A. V., Lancichinetti, A., West, J. D. & Lambiotte, R. Memory in network flows and its effects on spreading dynamics and community detection. Nat. Commun. 5, 4630 (2014).
Grassly, N. C. & Fraser, C. Mathematical models of infectious disease transmission. Nat. Rev. Microbiol. 6, 477–487 (2008).
Ganyani, T. et al. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, march 2020. Eurosurveillance 25, 2000257 (2020).
Park, S. W. et al. Forward-looking serial intervals correctly link epidemic growth to reproduction numbers. Proc. Natl. Acad. Sci. U.S.A. 118, e2011548118 (2021).
Svensson, Å A note on generation times in epidemic models. Math. Biosci. 208, 300–311 (2007).
Klinkenberg, D. & Nishiura, H. The correlation between infectivity and incubation period of measles, estimated from households with two cases. J. Theor. Biol. 284, 52–60 (2011).
te Beest, D. E., Wallinga, J., Donker, T. & van Boven, M. Estimating the generation interval of influenza a (h1n1) in a range of social settings. Epidemiology 24, 244–250 (2013).
Champredon, D., Dushoff, J. & Earn, D. J. Equivalence of the erlang-distributed seir epidemic model and the renewal equation. SIAM J. Appl. Math. 78, 3258–3278 (2018).
Feng, M., Tian, L., Lai, Y.-C. & Zhou, C. Web-based application for generation time distribution and error rectification of Markovian modeling. https://cns.hkbu.edu.hk/toolbox/Validity-of-Markovian-for-Memory/main.html (2023).
United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects 2019 https://population.un.org/wpp/. Accessed on 29 June 2021 (2021).
Prem, K., Cook, A. R. & Jit, M. Projecting social contact matrices in 152 countries using contact surveys and demographic data. PLoS Comput. Biol. 13, e1005697 (2017).
Wallinga, J. & Lipsitch, M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc. R. Soc. B 274, 599–604 (2007).
Dietz, K. The estimation of the basic reproduction number for infectious diseases. Stat. Methods Med. Res. 2, 23–41 (1993).
Castro, M., Ares, S., Cuesta, J. A. & Manrubia, S. The turning point and end of an expanding epidemic cannot be precisely forecast. Proc. Natl Acad. Sci. 117, 26190–26196 (2020).
Case, B., Young, J.-G. & Hébert-Dufresne, L. Accurately summarizing an outbreak using epidemiological models takes time. arXiv preprint arXiv:2301.08799 (2023).
Britton, T. & Scalia Tomba, G. Estimation in emerging epidemics: biases and remedies. J. R.Soc. Interface 16, 20180670 (2019).
Lai, Y.-C. & Tél, T.Transient Chaos—Complex Dynamics on Finite Time Scales (Springer, New York, 2011).
Hastings, A. & Higgins, K. Persistence of transients in spatially structured ecological models. Science 263, 1133–1136 (1994).
Hastings, A. Transients: the key to long-term ecological understanding? Trends Ecol. Evol. 19, 39–45 (2004).
Hastings, A. Timescales and the management of ecological systems. Proc. Natl Acad. Sci. 113, 14568–14573 (2016).
Hastings, A. et al. Transient phenomena in ecology. Science 361, eaat6412 (2018).
Morozov, A. et al. Long transients in ecology: theory and applications. Phys. Life Rev. 32, 1–40 (2020).
Meng, Y., Lai, Y.-C. & Grebogi, C. Tipping point and noise-induced transients in ecological networks. J. R. Soc. Interface 17, 20200645 (2020).
Sjöström, P. J., Turrigiano, G. G. & Nelson, S. B. Rate, timing, and cooperativity jointly determine cortical synaptic plasticity. Neuron 32, 1149–1164 (2001).
Martin, S. J., Grimwood, P. D. & Morris, R. G. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu. Rev. Neurosci. 23, 649–711 (2000).
Kandel, E. R. The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030–1038 (2001).
Hawkes, A. G. Spectra of some self-exciting and mutually exciting point processes. Biometrika 58, 83–90 (1971).
Centro de coordinación de alertas y emergencias sanitarias. Actualización nº 103. Enfermedad por el coronavirus (COVID-19). 12.05.2020 (datos consolidados a las 21:00 horas del 11.05.2020) SITUACIÓN EN ESPAÑA. https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Actualizacion_103_COVID-19.pdf. (Accessed on 24 Dec 2023).
Byrd, R. H., Lu, P., Nocedal, J. & Zhu, C. A limited memory algorithm for bound-constrained optimization. SIAM J. Sci. Comput. 16, 1190–1208 (1995).
Acknowledgements
This work was partially supported by the Hong Kong Baptist University (HKBU) Strategic Development Fund; the Research Grants Council of Hong Kong (Grant No. C2005-22Y), the National Natural Science Foundation of China (Grant No. 12275229), and the Hong Kong Chinese Medicine Development Fund (Grant No. 22B2/049A) to L.T.; the Research Grants Council of Hong Kong (Grant No. GRF12201421) to C.-S.Z. This research was conducted using the resources of the High-Performance Computing Cluster Centre at HKBU, which receives funding from the Hong Kong Research Grant Council and the HKBU. Y.-C.L. was supported by the Office of Naval Research through Grant No. N00014-21-1-2323.
Author information
Authors and Affiliations
Contributions
M.F., L.T. and C.-S.Z. designed research; M.F. performed research; L.T. and C.-S.Z. contributed analytic tools; M.F., L.T. and C.-S.Z. analyzed data; M.F., L.T., Y.-C.L. and C.-S.Z. discussed the results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Physics thanks David Soriano-Paños and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, M., Tian, L., Lai, YC. et al. Validity of Markovian modeling for transient memory-dependent epidemic dynamics. Commun Phys 7, 86 (2024). https://doi.org/10.1038/s42005-024-01578-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42005-024-01578-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.