Abstract
Germline de novo mutations (DNMs) provide the raw material for evolution. The DNM rate varies considerably between species, sexes and chromosomes. Here, we identify DNMs in the zebra finch (Taeniopygia guttata) across 16 parent-offspring trios using two genome assemblies of different quality. Using an independent genotyping assay, we validate 82% of the 150 candidate DNMs. DNM rates are consistent between both assemblies, with estimates of 6.14 × 10–9 and 6.36 × 10–9 per site per generation. We observe a strong paternal bias in DNM rates (male-to-female ratio ɑ ≈ 4), but this bias is in transition mutations only, leading to a transition-to-transversion ratio of 3.18 and 3.57. Finally, we find that DNMs tend to be randomly distributed across chromosomes, not associated with recombination hotspots or genic regions. However, the sex chromosome chrZ shows a roughly fourfold increased DNM rate compared to autosomes, which is more than the expected increase due to chrZ spending two-thirds of its time in males. Overall, our results further enhance our understanding of DNMs in passerine songbirds.
Similar content being viewed by others
Introduction
Germline de novo mutations (DNMs) introduce novel genetic variants into a population and thereby provide the raw material for deleterious, neutral and adaptive evolutionary change1. DNMs come in different forms and sizes, as they can alter single nucleotides (point mutations) to large chromosomal regions that affect genome structure (deletions, duplications, inversions and translocations). Single nucleotide DNMs are at least an order of magnitude more abundant than larger structural mutations2,3,4,5,6 and are of primary interest to such diverse fields as phylogenetics7,8, ecology and evolution9,10,11, population genetics12,13 and human medicine14.
Thus, considerable efforts have been made to quantify DNM rates. Initially, this was achieved indirectly by estimating the frequency of occurrence of monogenic dominant diseases2,15,16. Similarly, molecular evolutionary analyses used gene sequence alignments between species to estimate the DNM rate at fourfold degenerate sites17, assuming that mutations at these sites are selectively neutral. By focusing on specific genes, both these methods lacked genome-wide coverage. With the advent of high-throughput sequencing, direct genome-wide estimates of the DNM rate were obtained through sequencing of pedigree trios18,19. These studies also allowed to precisely locate DNMs in the genome and relate them to genomic features like CpG islands, recombination rate, replication time, chromatin state, distance to telomeres, expression levels, functionally constrained regions (e.g., genes and promoters) and autosomal vs sex chromosomal sites20,21,22,23. The use of trio sequencing to estimate DNM rates was pioneered in humans19, but became more common in non-model organisms in recent years9,24,25, which allowed inferences on the molecular mechanisms causing DNMs24,26, the degree of variation4 and of the male bias in DNM rates across vertebrates9.
DNMs arise spontaneously during DNA replication (“replication-driven”) or DNA mismatch repair (“damage-induced”)27,28. It has long been thought that DNMs occur predominantly during cell division in mitosis and meiosis, but recent evidence challenges this interpretation and suggests that unrepaired DNA damage is responsible for most DNMs28, as discussed in detail below. DNMs can happen anywhere in the genome, but they are not entirely randomly distributed22,29,30. Across vertebrates, CpG sites consistently have higher DNM rates than non-CpG sites, and the most common mutation is the deamination of a methylated C to a T because the methyl group lowers the energy required for deamination (summarized in Gorelick31). Because of this, and because the exchange of a pyrimidine (C, T) for another pyrimidine and of a purine (A, G) for another purine base is conformationally more likely17, the transition-to-transversion ratio (transitions: A > G, C > T, transversions: A > C, A > T, C > A, C > G) is shifted to a value larger than the expected 2/49,23. In addition, the high rate of C to T transitions and the reduced efficiency of exonucleases in regions of high GC content shift the mutational spectrum from stronger (G or C) to weaker (A or T) base pairings17,19. In humans, DNMs are more likely to appear near recombination locations13,32, possibly because the DNA break and repair machinery is prone to introducing DNMs22. Recent studies suggested that functionally constrained regions harbor fewer DNMs33,34, but these results have been questioned repeatedly and not been supported across model organisms21,35,36,37.
From the above, it can be concluded that the DNM rate varies within a genome38. However, it also varies considerably between individuals and between species9,39,40. Across eukaryotes, DNM rates vary 5000-fold and are positively correlated with generation time, genome size and nucleotide diversity between species4. And even within the avian clade (18 species compared by Bergeron et al. 9), DNM rates estimated by trio sequencing vary 40-fold and range from 1.0 × 10–9 to 39.8 × 10–9 mutations per site per generation9, with most of this variation explained by phylogeny rather than, for example, between-individual variation (phylogenetic heritability H2 = 0.99 (95% CI 0.96–1.00) estimated using the evolvability R package (v2.0.0)41).
DNM rates differ between sexes. Males tend to have higher DNM rates than females9, and inherit more DNMs with increasing age at conception17,42,43. Across the great apes, males contribute 2–4 times as many mutations as females (human: male-to-female ratio α = 3.23 (95% CI 2.69–3.87) (ratio and confidence interval calculated from data in Gao et al. 27), chimpanzee: α = 4.37 (95% CI 0.96–1.00), gorilla: α = 2.00 (95% CI 1.11–3.74), orangutan: α = 4.13 (95% CI 2.00–3.05) (ratios and confidence intervals calculated from data in Besenbacher et al. 7)), and this range of α seems to be relatively constant across mammals9,24. Birds also show a high male bias, particularly passerine birds, with a male-to-female ratio of α = 7.6 (95% CI 4.3–13.5)9. This male-biased mutation rate has long been attributed to the fact that spermatogenesis continues throughout reproductive life, while oogenesis is arrested at the embryonic stage. Because of that, the number of germline cell divisions prior to reproduction increases with paternal age44,45,46, and if the occurrence of DNMs is mainly replication driven, this increases the likelihood of DNMs in males relative to females. However, this view has been challenged in recent years, because also females inherit more DNMs with increasing age at conception13,24,47,48. While the effect is less pronounced than in males, it leads to a relatively stable male-to-female DNM transmission ratio α across paternal ages at conception, which does not align with the number of cell divisions in the female and male germline27,28. In fact, the male bias in DNM transmissions is already present at the beginning of reproductive maturity (puberty), that is at a developmental stage when males and females had equal numbers of cell divisions in their germlines. Taken together, this rather suggests that most DNMs are damaged induced and that males have less effective DNA mismatch repair mechanisms28.
Sex chromosomes appear in unequal numbers in females and males. In female heterogametic systems (birds), females have chromosomes Z and W and males have two copies of chromosome Z. In male heterogametic systems (mammals), males have chromosomes X and Y and females two copies of chromosome X. Thus, in contrast to the autosomes, sex chromosomes spend different proportions of time in males. Chromosome Z resides two-thirds of its time in males, while chromosome W none of its time. Accordingly, chromosome X spends only one-third of its time in males and chromosome Y all of its time. Given that the DNM rate is higher in males, when analyzing females and males combined, we expect to see more DNMs on chromosomes Z and Y and fewer DNMs on chromosomes W and X in comparison to the autosomes23,49. Empirical estimates from trio sequencing in humans50 and from molecular evolutionary analyses of autosomal and sex chromosomal genes in avian species51 support this hypothesis. On the other hand, the hypothesis of adaptive mutation-rate evolution, which poses that the DNM rate on chromosome X (or chromosome Z) is reduced to avoid recessive deleterious mutations to be exposed in the hemizygous state, would counteract this male mutation bias, but has only limited support20,51.
The zebra finch (Taeniopygia guttata) is a passerine songbird that serves as a model organism for studying vertebrate neurology, behavior, physiology, ecology, genetics and evolution52,53,54,55,56,57,58,59. A draft genome using Sanger sequencing of BAC clones and next-generation sequencing technology is available since the year 2010 (NCBI reference number: GCF_000151805.1)58. In the year 2021, a high-quality third-generation genome assembly with reduced false gene duplications and increased assembly continuity was published (GCF_003957565.2)60. The challenge of identifying DNMs through trio sequencing is formidable4 and may also depend on the quality of the available reference genome. The use of different human reference genomes led to discordant single nucleotide variant calls61 and any such difference would also perpetuate DNM rate estimations. Thus, here we use 16 parent-offspring trios to detect and compare the overlap of DNMs called on the two zebra finch genome assemblies. We validate DNMs with an individual genotyping assay, estimate DNM rates separately for autosomes and sex chromosomes and characterize DNMs according to parental origin, mutational spectrum and their associations with genomic features.
Results
De novo mutations (DNMs) across individuals and genome assemblies
The 16 offspring carried 2–15 candidate DNMs each (Fig. 1). In total, we detected 150 candidate DNMs across the two genome versions. Out of these, 111 (74%) were present in both the first- and third-generation genome assemblies, whereas 11 and 28 DNMs were unique to the first and third-generation genome assemblies, respectively (Fig. S13B). Thus, a combined 80% and 91% of all candidate DNMs were shared between the two assemblies, serving as an initial means of validation for these DNMs. Using the number of candidate DNMs and the size of the callable genome (see Methods), we calculated an average (uncorrected) DNM rate of μu1,g = 7.09 × 10–9 (95% CI 5.94 × 10–9–8.47 × 10–9) and μu3,g = 7.36 × 10–9 (95% CI 6.23 × 10–9–8.69 × 10–9) per site per generation in the first- and third-generation genomes, respectively (Table 1).
In the first breeding season, pairs F07051 x F07052 (family 1) and F07053 x F07054 (family 2) produced eight offspring. In the second breeding season, pairs F07053 x F07052 (family 3) and F07051 x F07054 (family 4) produced another eight offspring. Circles represent females and squares males. Numbers within each offspring are the number of candidate DNMs in the first- and third-generation genome assemblies.
We attempted to verify all candidate DNMs using an independent genotyping method and received genotypes for 134 candidate DNMs with a call rate of 96.8%. N = 111 of these were bona fide DNMs (false discovery rate [FDR] first-generation genome = 17/(122–13) = 15.60% and FDR third-generation genome = 19/(139–16) = 15.45%). Among the 23 candidate DNMs that failed verification, 11 were homozygous for the alternative allele in the focal individual and homozygous for the reference allele in all other individuals, which means that they were likely DNMs but their genotypes did not match between genotyping methods and we thus considered them false-positives (Table S9). Using the FDR and the estimated false negative rate (FNR, see Methods), we calculated an average (corrected) DNM rate of μc1,g = 6.14 × 10–9 (95% CI 5.07 × 10–9–7.43 × 10–9) and μc3,g = 6.36 × 10–9 (95% CI 5.32 × 10–9–7.60 × 10–9) per site per generation in the first- and third-generation genomes, respectively (Table 1).
Considering that the age at reproduction in the parental individuals was rather high (mean = 3.31) (cf. Bird et al. 62 for an overview of generation lengths across avian species), we also calculated the per year DNM rates, which were μc1,y = 1.85 × 10–9 (95% CI 1.53 × 10–9–2.24 × 10–9) and μc3,y = 1.92 × 10–9 (95% CI 1.61 × 10–9–2.30 × 10–9) in the two genome assemblies (Table 1).
In all following analyses, we used the bona fide DNMs, excluding false-positives and those variants that we were not able to verify. Thus, we used 92 and 105 bona fide DNMs in the two genome assemblies (Table S8).
Parental origin of DNMs
We inferred the parental origin of 71 and 79 (77.2% and 75.2%) bona fide DNMs in the first- and third-generation genomes, respectively. Four DNMs were located on chrZ and were removed for all analyses concerning parental DNM origin to avoid compositional bias resulting from the hemizygous chrZ in females. The majority of DNMs were of paternal origin (80.6% and 78.7%, P = 4 × 10–7 and P = 6 × 10–7, respectively; Fig. 2, Table S8), which corresponds to an autosomal male-to-female ratio α = 4.15 (95% CI 2.34–7.94) and α = 3.69 (95% CI 2.18–6.62).
We did not observe a significant association between parental age and the number of DNMs inherited (Table S19), but this might have been expected given our small sample size and low variation in parental age. However, we did find more DNMs of paternal origin in the second round of breeding after the partners had changed and the parents were therefore older (P = 0.020 and P = 0.0086 in the first- and third-generation genome, respectively; Table S20).
DNM types
The majority (76.1% and 78.1%) of bona fide DNMs were transition mutations (i.e., A > G or C > T) in the first- and third-generation genomes (all P < 2.2 × 10–16; Fig. 3A, B). Accordingly, the transition-to-transversion ratios (ti/tv) were 3.18 and 3.57, respectively. Most of the transitions were of paternal origin (male-to-female ratios α = 6.57 and α = 5.00, P = 4 × 10–8 and P = 2 × 10–7 for the first- and third-generation genomes, respectively; Table S21), and fathers tentatively transmitted relatively more C > T transitions than mothers (P = 0.027 and P = 0.16; Fig. 3C, D). On the other hand, transversions were almost equally likely to occur in fathers and mothers (male-to-female ratios α = 1.33 and α = 1.50, P = 0.79 and P = 0.61; Table S21), and there was a tendency for mothers to transmit relatively more C > A (P = 0.082 and P = 0.11) and C > G (P = 0.094 and P = 0.11) transversions than fathers (Fig. 3C, D). The zebra finch mutation spectrum did not significantly differ from the human DNM dataset (with N = 32,595 paternal and N = 8426 maternal DNMs)48 (Fig. S15A), but showed a tendency to deviate in paternal DNMs (Ptotal = 0.36 and 0.26, Ppaternal = 0.064 and 0.055, Pmaternal = 0.086 and 0.31; Fig. S15A–F), and this tendency was driven by a relatively higher C > T transition rate in zebra finches compared to humans (P = 3.4 × 10–3 and P = 5.2 × 10–3; Fig. S15C, D).
Absolut (A, B) and relative (C, D) numbers in the first-generation genome and in the third-generation genome assembly, respectively. Asterisks highlight significant differences in the relative frequencies of DNM types between males and females (*P < 0.05). DNMs at CpG sites are highlighted and occur only in C > T transitions.
Distribution and association of DNMs with genomic features
Genomic distribution
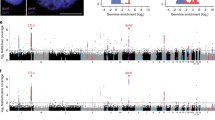
DNMs appeared to be uniformly distributed across the genome without any clustering (Fig. 4A, B). We found a significant positive effect of the callable chromosome size on the number of DNMs per chromosome (β ± SE = 1.00 ± 0.12, P < 2 × 10–16 and β ± SE = 1.04 ± 0.11, P < 2 × 10–16 for the first- and third-generation genomes, respectively; Table S22, Fig. 4C, D). The estimates were close to 1 for both genome assemblies, meaning that the number of DNMs scaled linearly with the callable chromosome size63. At the same time, we found a significant positive effect of the sex chromosome chrZ (β ± SE = 1.32 ± 0.49, P = 7.5 × 10–3 and β ± SE = 1.41 ± 0.44, P = 1.3 × 10–3 for the first- and third-generation genomes, respectively). The DNM rate was 3.73× (95% CI 0.76–8.25) and 4.11× (95% CI 1.15–8.82) higher on chrZ than on the autosomes, which was higher than the expected values of 1.20 and 1.19, respectively, although 95% CIs overlap these values slightly. In the third-generation genome assembly, chrW was present and we observed a single DNM across the entire pedigree, which translated to a strongly increased DNM rate on chrW (β ± SE = 5.24 ± 1.19, P = 1 × 10–16). However, this estimate should be treated with caution because of the low sample size (chrW-to-autosome ratio = 189.1× (95% CI 0.0–1439.7)). The offsprings’ sex had no effect on the number of DNMs (P = 0.50 and P = 0.75 for the first- and third-generation genomes, respectively; Table S22).
A Chromosomal distribution of DNMs in the first-generation genome and (B) in the third-generation genome assembly. Bars to the left show the number of DNMs on each chromosome. Bars to the right represent chromosomes and positions of DNMs are marked by individual and family. In both (C) the first-generation genome and (D) the third-generation genome assembly, the association between callable chromosome size and the number of DNMs on each chromosome is highly significant. chrZ is highlighted in yellow.
Genomic features
C > T transition DNMs were enriched at CpG sites in the genome (odds ratio [OR] = 13.97, 95% CI 7.91–24.67 and OR = 17.33, 95% CI: 10.65–28.21 in the first- and third-generation genomes; Table S23, Fig. 3). None of the other transition or transversion DNMs occurred at CpG sites. There was no difference in the proportion of DNMs located in coding, intronic or intergenic regions (P = 0.34; Table S24).
Recombination hotspots
Recombination hotspots had been mapped on the first-generation genome64. We tested whether DNMs were enriched in these hotspots using a randomization procedure (see Methods for details). The number of DNMs in windows of different sizes around hotspots did not significantly differ from the number of DNMs in randomly placed windows across the genome (all P > 0.18; Table S25, Fig S14).
Discussion
Here we estimate the DNM rate in two zebra finch genome assemblies. The estimates from the two assemblies deviate by less than 5%, not least because most DNMs were found in both assemblies. DNMs were predominantly of paternal origin and because of that more often found on the sex chromosome chrZ than on the autosomes. DNMs were enriched in transitions and these were preferentially located in CpG islands. Transversions did not show a bias in parental origin.
Offspring inherited 2–15 DNMs from their parents (mean = 8.19, SD = 3.73, coefficient of variation CV = SD/mean = 0.46). The coefficient of variation allows comparing the relative amount of variation in populations having different means65, and the CV we found for zebra finches is within the range of other avian species (CV range = 0.13–0.55)9,25,66.
We estimated a per-generation DNM rate of μc,g = 6.1 and 6.4 × 10–9 (individual range = 3.3 × 10–9–2.3 × 10–8 and 3.0 × 10–9–2.2 × 10–8). Using only two zebra finch trios, Bergeron et al. 9 estimated a mean per-generation DNM rate of μc,g = 5.8 × 10–9, with large interindividual variation (range 4.7–7.0 × 10–9), which is consistent with our estimate. On the other hand, our estimate of the per-year DNM rate (μc,y = 1.9 × 10–9) was lower than the naïve estimate of Bergeron et al. 9 (4.7 × 10–9), but similar to their modeled estimate (2.1 × 10–9). For other passerine species, naïve and modeled rates per year of Bergeron et al. 9 are broadly similar (blackbird [Turdus merula]: 2.0 × 10–9, Siberian stonechat [Saxicola maurus]: 2.2 × 10–9, blue tit [Cyanistes caeruleus]: 3.3 × 10–9 and collared flycatcher [Ficedula albicollis]: 2.3 × 10–9)9,25. Population genetic estimates of the per-year mutation rate in wild zebra finches cover similar ranges (2.1–2.8 × 10–9)64, and the substitution rate at fourfold degenerate sites is also similar (2.2 × 10–9)67. Overall, this suggests a relatively conserved mutation rate across passerines and that pedigree-based and population genetic methods yield robust and comparable results4.
We observed a transition to transversion ratio of ~3, with a predominance of mutations from strong to weak base pairings (C, G > A, T). Transitions are changes between nucleotides that maintain the same chemical class of pyrimidine or purine base and, therefore, occur more frequently. This is particularly true for the methylation and deamination of a cytosine to a thymine31, which might explain the enrichment of DNMs in CpG islands across the genome. Thus, our results align with the expected outcomes and are consistent with prior research across various vertebrates, and notably avian species (as summarized in Bergeron et al. 9).
We find that in zebra finches around 80% of all DNMs are introduced by the father (male-to-female ratio α ≈ 4), which is towards the lower end of estimates derived across multiple passerine species9. Zebra finches have relatively small seminal glomera and their sperm production rates are low in comparison to other passerine species68. They are lifetime monogamous and this might lead to reduced sperm competition and less sperm production69, which could reduce the male bias in DNM occurrence70. A recent study on great reed warblers (Acrocephalus arundinaceus) found the same low sex bias66. Great reed warblers are polygynous with low extra-pair paternity rates71,72,73, which might imply low sperm competition and thus low levels of sperm production68,74,75.
In our zebra finch pedigree, the male bias in DNM occurrence was caused by transitions, whereas transversion mutations occurred at similar rates in both sexes. Globally, the mutation spectrum did not differ between zebra finches and humans, but it appeared that male zebra finches inherited relatively more C > T transitions than human males and zebra finch females. Evidence from humans and experimental systems suggests that C > T transitions often occur spontaneously and are associated less with DNA replication17,76. Indeed, C > T transitions in humans are relatively more often transmitted by mothers than by fathers47,48, again suggesting that they arise mainly from DNA damage (although there also appears to be a non-negligible effect of DNA replication on the frequency of C > T transitions77). Since humans should have more cell divisions in their paternal germline than zebra finches78, mutation types associated with DNA replication (i.e., mutation types other than C > T) would be expected to be more common in human males than in zebra finch males. Thus, human males would inherit relatively fewer C > T transitions than zebra finch males, which is consistent with our observation. In comparison to oocytes, DNA in sperm cells is hypermethylated in vertebrates79. This could explain why the male germline is more prone to deamination at methylated CpG sites than the female germline, causing relatively more C > T transitions to be inherited via fathers than mothers, which is consistent with our observation.
We also observe a ~fourfold higher DNM rate on the sex chromosome chrZ in comparison to the autosomes. Given a male-to-female ratio α > 1, an increased chrZ-to-autosome DNM ratio is expected, because chromosome Z spends two-thirds of its time in males where it is exposed to the increased male DNM rate63. However, all else being equal between autosomes and the sex chromosome Z, this ratio has an upper bound of 4/380,81. Our 95% confidence intervals just cover this upper bound, but the point estimates of a ~fourfold higher DNM rate on chrZ suggest that other factors like differences in replication time, nucleotide composition or chromatin structure may increase the DNM rate on chrZ in comparison to the autosomes74. Gene expression, ATAC-seq and methylation studies on somatic tissue in birds have shown that, in comparison to the autosomes, chromosome Z has lower gene expression and more open chromatin in females and it is hypomethylated in both sexes82, which may influence the DNM rate22,83. As explained above, DNA in sperm cells is hypermethylated in comparison to oocytes in vertebrates77. If this was also the case in zebra finches, relatively more C > T transitions would be inherited via fathers than mothers, and it could cause a higher DNM rate on chrZ than on the autosomes, which is consistent with our observation. Methylation data to support this hypothesis is currently lacking. Alternatively or additionally, the efficiency of the DNA mismatch repair machinery may differ between the sexes, with females more likely to repair deamination errors correctly17. In any case, there is no support for the hypothesis that selection reduces the DNM rate on chromosome Z to guard against the exposure of recessive deleterious mutations in the hemizygous state20. This is consistent with findings from molecular evolutionary estimates of the mutation rate on the sex chromosome Z and autosomes in birds51.
Overall, our study provides robust estimates of the DNM rate in Australian zebra finches, which are essential for phylogenetic divergence time estimation66, demographic modeling84,85 and population genetic analyses1. Our estimates of the DNM rate using direct trio-sequencing are in good agreement with population genetic estimates of the mutation rate per year in wild zebra finches. Furthermore, we show that the DNM rate is higher on the sex chromosome chrZ, and that this increased DNM rate cannot solely be explained by the fourfold increased genome-wide mutation rate in males, which may further enhance the “fast-Z-effect” in birds86.
Materials and methods
Populations and individuals
Two breeding pairs of Australian zebra finches (Taeniopygia guttata castanotis) were set up individually in cages at the Max Planck Institute for Biological Intelligence in Seewiesen, Germany. These individuals originated from three distinct captive populations, called “Seewiesen” (study population 18 in Forstmeier et al. 87), “Cracow” (study population 11) and wild-derived “Bielefeld” (study population 4). We chose parents from different source populations to maximize the background genetic diversity. Upon pairing, birds were on average 3.14 (SD = 0.75) years old. All eggs were transferred to an incubator for four days and embryos were subsequently collected for DNA isolation (stored in EtOH). After a sufficient number of eggs had been laid, breeding partners were switched and embryos were sampled again. By then, parents were on average 3.26 (SD = 0.63) years old. Thus, there were in total four families and we selected the parents and four offspring from each family for sequencing (N = 20 individuals in total). Within each family, offspring were full-sibs and among families, offspring had eight half-sibs and four unrelated peers (Fig. 1; Table S1). DNA was isolated from the adults’ blood (stored in Queen’s lysis buffer) using the Nucleo Spin Blood Quick Pure Kit (Macherey & Nagel) and from embryonic tissue using the DNeasy Blood & Tissue Kit (Qiagen) according to manufacturers’ guidelines.
Housing, breeding, banding and blood sampling for parentage assignment of captive zebra finches do not qualify as animal experimentation according to the relevant national and regional laws and are fully covered by the housing and breeding permit of the Max Planck Institute for Biological Intelligence (# 311.4-si, Landratsamt Starnberg, Germany). We have complied with all relevant ethical regulations for animal use.
Sequencing
Paired-end libraries were prepared using the Illumina TruSeq DNA Nano LT protocol with an insert size of 400 bp. Paired-end 150 bp sequencing with an approximate coverage of 40× per sample (~55G bases per sample) was done using the Illumina HiSeq4000 at the Institute of Clinical Molecular Biology (IKMB) at Kiel University, Germany.
Quality control and mapping
For each of the 20 raw data files, we trimmed low-quality bases and adapters with TrimGalore (v0.6.7; https://github.com/FelixKrueger/TrimGalore) using parameters “–paired –retain_unpaired –quality 25 –phred33 –length 36 –stringency 3”, leaving us with 1068G bases of cleaned data (range per individual 47.5–60.0G; Table S2). Reads were mapped using BWA MEM (v0.7.17)88 with default settings to two zebra finch genome versions: the more fragmented but well-annotated genome WUSTL 3.2.4 (GCF_000151805.1)58, and the more contiguous but less well-annotated genome bTaeGut1_v1.p (GCF_003957565.2)60. In the following, we refer to these as “first-generation genome” and “third-generation genome” assemblies, respectively (see Table S3 for a comparison of summary statistics of the two genome versions). We assessed the mapping quality with QualiMap (v2.3)89 using default parameters, which yielded a mean coverage per individual of 42× (range: 37–47×) and 49× (range: 44–56×) for the first- and third-generation genomes, respectively (Tables S4 and S5). We used GATK (v4.1.9)90 MarkDuplicatesSpark to flag duplicates.
Variant calling
To generate a set of “known sites” for base quality score recalibration (BQSR), we called variants from de-duplicated BAM files with GATK HaplotypeCaller. We removed variants in low-quality regions (i.e., those in lower cases) and applied strict hard filter thresholds (SNPs: QD < 2, FS > 20, MQ < 40, MQRankSum < –2, MQRankSum > 4, ReadPosRankSum < –3 and ReadPosRankSum > 3; indels: QD < 2, FS > 200, QUAL < 30, ReadPosRankSum < –20) that were recommended by the GATK development team (2020)91 and we adjusted them based on the distributions of variant quality scores (Fig. S1). The final sets of 24,096,374 SNPs and 3,110,811 indels (first-generation genome, Table S4) and 29,147,432 SNPs and 3,146,762 indels (third-generation genome, Table S5) were used for BQSR.
Following BQSR, we employed GATK HaplotypeCaller to perform variant calling on each individual separately. Variants were called with a minimum base quality score of 15 and the following options: –output-mode EMIT_ALL_ACTIVE_SITES –emit-ref-confidence BP_RESOLUTION –bam-output. The resulting gVCF files were merged into a single VCF for all further filtering steps, and the resulting BAM files were used to manually curate the set of DNMs (see below).
Variant filtering
We summarized the quality information of all variants using GATK VariantsToTable and visually inspected the distribution of quality metrics (Fig. S2). We did not identify any outliers and applied the current best practice site-specific hard filter criteria (MQ < 40.0, QD < 2.0, FS > 60.0, SOR > 3.0, MQRankSum < –12.5, ReadPosRankSum < –8.0) for SNPs as recommended by Bergeron et al. 92. Additionally, we removed all sites in low-quality regions of the respective reference genomes (i.e., those in lower cases). This left us with a final mean of 11,955,237 SNPs per individual (first-generation genome, Table S4) and 12,636,939 SNPs per individual (third-generation genome, Table S5).
Detecting de novo mutations (DNMs)
Autosomes
Candidate DNMs were identified as Mendelian violations in each of the 16 trios consisting of parents and one offspring using bcftools (v1.12)93 with the Mendelian plugin. From these ‘Mendelian violation’ sites we selected (1) those located on autosomes and (2) those for which both parents were homozygous for the reference allele (HomRef; 0/0) and the offspring was heterozygous (Het; 1/0 or 0/1). We then applied sample-specific filters to the candidate DNMs:
-
(1)
Depth filter: Using the VCF file as input, the sequencing depth (DP) at the candidate DNM had to be within the range of 0.5× mean(DP) to 2× mean(DP), where mean(DP) is the average sequencing depth of the individual (Figs. S3 and S4). The same cut-off has been applied in other studies as well92,94.
-
(2)
Allelic balance: Using the VCF file as input, the allelic balance (AB) at the candidate DNM needed to range from 0.3 to 0.7. AB was calculated as the proportion of reads supporting the alternative allele to the total number of reads at the candidate DNM. For a true DNM, an AB of ~0.5 is expected and deviations may indicate somatic mutations, mapping errors, or sample contamination12. The thresholds were based on the AB distribution of all candidate DNMs (Fig. S5) and have also been applied in other studies12,19,92,95.
-
(3)
Genotype quality: Using the VCF file as input, the distribution of genotype qualities (GQ) was visually inspected across all candidate DNMs (Fig. S6) and a threshold was set to GQ > 40. The same threshold has been applied in Bergeron et al. 92.
-
(4)
Alternative allelic depth (AD): Using the VCF file as input, we required that parents had no reads supporting the alternative allele, which means AD = 025,43,94,96,97,98,99.
-
(5)
Alternative reads: Using the realigned BAM file (GATK) as input, we required that parents had no reads covering the alternative allele.
After these initial filtering steps, we were left with 835 (first-generation genome) and 2753 (third-generation genome) candidate DNMs (Tables S6 and S7).
Finally, we manually curated all remaining candidate DNMs by checking the read alignments for mapping errors in the raw and in the realigned BAM files using IGV (v2.15.4)100. Some of the errors were present in the raw BAM files, but some were only introduced during the realignment step. Keightley et al. 101 provided examples for mapping errors:
-
(1)
Reads carrying the candidate DNM allele also contain other SNPs or indels, and these variants are absent from other reads mapping to the same position in parents and offspring (i.e., multiple candidate DNMs are linked on a single read; (cf. Fig. S7)).
-
(2)
Reads carrying the candidate DNM allele also contain other SNPs or indels that do not violate Mendelian inheritance, but these additional variants together with the candidate DNM form more than two haplotypes in the offspring (cf. Fig. S8). In this case, it is likely that reads from a paralog were erroneously mapped.
-
(3)
Reads carrying the candidate DNM allele also contain an insertion, which is differently aligned in parents and offspring, whereby mapping errors occur (cf. Fig. S9).
Candidate DNMs that were caused by any of these mapping artifacts were removed from further analyses, leaving us with 116 (first-generation genome) and 121 (third-generation genome) candidate DNMs across all individuals. Bona fide DNMs were usually found in reads without many flanking variants.
Sex chromosomes
In birds, males are the homogametic sex (genotype Z/Z) and females are heterogametic (Z/W). Thus, for male offspring, we identified candidate DNMs as those where both parents were homozygous for the reference allele (HomRef; 0/0) and heterozygous (Het; 1/0 or 0/1) in the offspring (Fig. S10A). We filtered the candidate DNMs using the same five sample-specific filters as described above, but adapted the depth filter of mothers to 0.25× mean(DP) and 1× mean(DP) to account for the approximately halved sequencing depth of the sex chromosome chrZ in females (Fig. S11). For female offspring, we identified candidate DNMs on chrZ as those where both parents were homozygous for the reference allele (HomRef; 0/0) and offspring were homozygous for the alternative allele (HomAlt; 1/1; Fig. S10B). Candidate DNMs on chrW were identified as those homozygous for the reference allele in the mother (HomRef; 0/0) and homozygous for the alternative allele in the offspring (HomAlt; 1/1). Because candidate DNMs were hemizygous in female offspring, we could not use the allelic balance filter, but applied the remaining four sample-specific filters, again adjusting the depth filter to 0.25× mean(DP) and 1× mean(DP) in mothers and offspring. We manually curated all remaining candidate DNMs on the sex chromosomes by checking the alignments in the raw and in the realigned BAM files.
Determining the parental origin of the DNMs
Whenever there was a second variant located on the same reads as the candidate DNM and only one of the parents carried that variant in heterozygous or homozygous state, we were able to determine the parental origin of the DNM in IGV through read phasing (cf. Fig. S12). This approach makes use of the fact that two variants occurring in the same sequencing read must come from the same DNA molecule and thus be inherited from the same parent102.
Analyzing the overlap of DNMs between genome versions
Shared candidate DNMs between genome versions were identified by comparing (1) the mutation type and (2) the adjacent 100 base pairs before and after the candidate DNM. If the mutation type and the 200 adjacent base pairs were the same or reverse complementary, we considered the candidate DNM as shared between genome versions.
Those candidate DNMs that were unique to one or the other genome assembly (N = 28 and N = 33 in the first- and third-generation genome assembly, respectively; Fig S13A) were re-evaluated by aligning 2 kb before and after the candidate DNM to the genome assembly in which the DNM was missing using BLAST (v2.13.0)103 with parameters -e-value 1e-05 -max_target_seqs 100 -max_hsps 100. Successful BLAST searches yielded one unique best hit in the genome assembly in which the DNM was missing. While most DNMs were unique to a certain genome assembly because the sequence was missing from the other assembly (N = 11 in the first-generation and N = 28 in the third-generation genome; Fig. S13B), it was also the case that some DNMs were present in both assemblies but filtered out by our automated filtering pipeline because of low variant quality scores or during manual curation from one of the genome assemblies (N = 17 in the first- and N = 5 in the third-generation genome; Table S8).
DNM validation
To determine the reliability of our DNM calls and to estimate the false-positive DNM call rate (see below), we attempted to validate all candidate DNMs on the MassARRAY platform (Agena Bioscience). For that, we included all 150 candidate DNMs in six genotyping assays (25 attempted DNMs per assay) and called genotypes in all 20 individuals using the Typer Analyzer Application (v4.1.83; Agena Bioscience). Each individual was genotyped twice for each candidate DNM and all but one genotype call was consistent between the two genotyping trials (99.96% consistent calls). For 6 variants, genotyping failed in all individuals. For a further 9 variants, genotyping failed in the focal individual carrying the candidate DNM. Finally, genotyping failed in the focal parental individual for 1 variant (Table S9). These 16 variants were removed prior to FDR estimation (see below). For the remaining variants, the call rate was 96.8%. A candidate DNM was considered a bona fide DNM when both parents were homozygous for the reference allele and the offspring was heterozygous.
Estimating the de novo mutation rate
We estimated uncorrected per-site DNM rates for the entire genome and separately for the autosomes and sex chromosomes. Specifically, uncorrected (subscript u) per-site per-generation (subscript g) DNM rates of autosomes and sex chromosomes in the first- or third-generation genome assembly (subscripts 1 and 3, respectively) are given by the formulas:
N candidate DNMauto, N candidate DNMchrZ and N candidate DNMchrW are the total number of candidate DNMs on autosomes, chrZ and chrW, respectively. CGauto, CGchrZ and CGchrW are the sizes of the callable genome for each individual on autosomes, chrZ and chrW, respectively. The callable genome size of autosomes is calculated as the sum of all sites (1) that are not in low-complexity genomic regions (i.e., not lower case), (2) where both parents are homozygous for the reference allele and (3) where all individuals of a trio pass the depth filter, the genotype quality filter and the no alternative reads filter7. We calculated the callable genome size of the sex chromosomes by adjusting the depth filter to 0.25× mean(DP) and 1× mean(DP) in mothers and female offspring (as described in the methods section ‘Detecting de novo mutations’). We extracted callable sites from the gVCF files that contained all sites in the reference genome (obtained with the --emit-ref-confidence BP_ RESOLUTION in GATK HaplotypeCaller, see above). For the first-generation genome, the callable genome size ranged between 520,715,343 and 537,064,330 bp (mean = 529,796,788 bp) on autosomes and between 8,215,727 and 12,801,080 bp (mean = 10,571,665 bp) on chrZ. For the third-generation genome, it was between 571,840,269 and 588,282,136 bp (mean = 581,200,530 bp) on autosomes, between 10,836,034 and 14,135,852 bp (mean = 12,535,258 bp) on chrZ, and between 137,365 and 162,714 bp (mean = 148,686 bp) on chrW (Table S10). We also calculated the callable genome size for each chromosome (i.e., the callable chromosome size) to test for an association with the number of DNMs (see below; Tables S11 and S12) and to estimate the DNM rate of each chromosome (Tables S13 and S14).
To account for false-negative and false-positive DNM calls in the estimation of the DNM rate, we used the formulas (subscript c stands for ‘corrected’):
FDR is the false-discovery rate and FNR is the false-negative rate. The FDR was estimated from the validation genotyping as the number of candidate DNMs that were not successfully verified as bona fide DNMs relative to all successfully genotyped DNMs. The FNR is mostly influenced by the allelic balance filter7,94,104. Thus, it has been suggested to estimate the FNRauto by calculating the proportion of bona fide heterozygous sites on autosomes that are filtered out in the offspring by the allelic balance filter when one parent is homozygous for the reference allele (0/0) and the other parent is homozygous for the alternative allele (1/1). The FNRchrZ is the false-negative rate on chrZ, which was estimated by calculating the proportion of bona fide heterozygous sites on male offsprings’ chrZ (males have genotype Z/Z) that are filtered out in the male offspring by the allelic balance filter when one parent is homozygous for the reference allele (0/0) and the other parent is homozygous for the alternative allele (1/1). The FNRaverage was estimated by calculating the proportion of all bona fide heterozygous sites (on the autosomes for all offspring and on chrZ for male offspring, see above) that are filtered out in the offspring by the allelic balance filter. In the first-generation genome, 503,937 out of 20,424,004 bona fide heterozygous sites on autosomes (2.47%) and 10,148 out of 318,949 bona fide heterozygous sites on chrZ (3.18%) were filtered, resulting in an FNRaverage of 2.48%. In the third-generation genome, 549,302 out of 25,756,985 bona fide heterozygous sites on autosomes (2.13%) and 9,972 out of 381,737 bona fide heterozygous sites on chrZ (2.61%) were filtered, yielding an FNRaverage of 2.14% (Tables S10 and S15–S18).
We derived 95% binomial confidence intervals (CI) for the uncorrected and corrected DNM rate with the binconf() function and default Wilson scores from the Hmisc package (v5.1-1)105 in R (v4.2.3)106.
We calculate the per-year (subscript y) and per-site DNM rates (μc,y) by dividing the corrected (μc,g) per-site DNM rate by the average age at reproduction of the parental individuals.
Characterizing de novo mutations
Parental origin
We could determine the parental origin of 83 bona fide DNMs (74.8% out of 111 bona fide DNMs) and we tested whether there was a parental bias (either male or female) using a binomial test in R (v4.2.3). In humans, the parental origin of DNMs can be determined in only 15–30% of cases through read phasing102. The most likely explanation for this difference in read phasing efficiency is the differing nucleotide diversity52, which is around π = 0.082 in zebra finches64 and π = 0.001 in humans.
For all analyses concerning parental DNM origin, we removed four DNMs with known parental origin located on chrZ to avoid compositional bias resulting from the hemizygous chrZ in females. We derived the male-to-female ratio α using a generalized linear model with a binomial error distribution and a logit link function and fitted the number of paternally and maternally inherited DNMs as the dependent variable (using the cbind() function) and the intercept as the sole predictor. The male-to-female ratio α can be derived through back-transforming the parameter estimate (yielding x) and calculating the ratio x/(1–x).
To examine the association between bona fide DNMs and parental age, we fitted a generalized linear mixed-effects model using the lme4 package (v1.1.32)107 in R (v4.2.3). We used the number of DNMs of each individual on each chromosome as our dependent variable and fitted five predictors: paternal age (scaled) as a covariate, maternal age (scaled) as a covariate, callable chromosome size (log-transformed) as a covariate, whether a chromosome was an auto- or gonosome as a factor (two levels: autosome and chrZ or three levels: autosome, chrZ and chrW) and the individual’s sex as a factor (two levels). We fitted individual ID and chromosome ID as two random intercept effects. We assessed whether our data was overdispersed using the performance (v0.10.2)108 and DHARMa (v0.4.6)109 packages in R (v4.2.3). Because we did not observe any overdispersion, we used a Poisson error distribution and a log link function (see Knief and Forstmeier110). Because the same parents were involved in the two breeding seasons, we also tested whether there were any differences in the number of maternal and paternal bona fide DNMs between the two breeding seasons using a binomial test in R (v4.2.3).
Mutation spectrum
We assessed whether bona fide DNMs were more likely to be transitions (A > G or T > C and C > T or G > A) or transversions (A > C or T > G, A > T or T > A, C > A or G > T and C > G or G > C). Because there are two types of transitions and four of transversions, we used a binomial test with a hypothesized probability of success p = 2/6. We tested for relative differences between the sexes in transition and transversion frequencies using Fisher’s exact test for each of the six mutation types (2 transition + 4 transversion types) vs the background mutation types as described in Jónsson et al. 48. To check whether bona fide DNMs were located in CpG islands, we assessed whether the DNM had a C or G as the reference allele and was followed by a G or C, respectively. We tested whether bona fide DNMs were more likely to appear at CpG sites by comparing the number of DNMs at CpG and non-CpG (any nucleotides except CpG) sites to the total callable genome-wide numbers of CpGs and non-CpGs (sum of callable genome size of each offspring) using a chi-squared test. We calculated the odds ratio and the 95% CI of the contingency table using the chisquare package (v0.9)111 in R (v4.2.3).
Comparison of the mutation spectrum between zebra finches and humans
We downloaded the largest DNM dataset collected via trio-sequencing in humans so far48 and derived the mutation spectrum in the same way as described above. We then tested (1) whether the spectrum of the six mutation types differed globally between zebra finches and humans for either mothers, fathers or both sexes combined using Fisher’s exact test. (2) We tested whether the relative frequencies of each of the six mutation types differed between zebra finches and humans in mothers, fathers or both sexes combined using Fisher’s exact test.
Association with genes
We used the gene annotations on the third-generation genome to count how many of the bona fide DNMs were located in coding, intronic or intergenic regions. We calculated the size of the callable genome covering coding, intronic or intergenic regions and performed a chi-squared test to determine whether the occurrence of bona fide DNMs was associated with these genomic regions.
Association with recombination hotspots
Recombination hotspots have been mapped to the first-generation genome by Singhal et al. 64. We examined whether bona fide DNMs were located more often in the vicinity of recombination hotspots than expected by chance. For that, we counted the number of DNMs in windows of 1–20 kb (1, 2, 5, 10, 20 kb) centered on the hotspots. We constructed a null-distribution by placing the same number of same-sized windows randomly on the genome and counting how many DNMs were covered by these randomized windows. We repeated this procedure 10,000 times for each window size and checked whether more DNMs were located at recombination hotspots than expected by our simulated null distribution.
Association with auto- or gonosomes
To test whether bona fide DNMs occurred at a different frequency on autosomes than on the sex chromosome chrZ, we fitted the same generalized linear mixed-effects model with Poisson error distribution as described above (see section ‘Parental origin’) using the lme4 package (v1.1.32) in R (v4.2.3), but left out the two parental age covariates, because they did not turn out to be significantly associated with the number of DNMs. Thus, we included three predictors: callable chromosome size (scaled and log-transformed) as a covariate, whether a chromosome was an auto- or gonosome as a factor (two levels: autosome and chrZ or three levels: autosome, chrZ and chrW) and the individual’s sex as a factor (two levels). We again fitted individual ID and chromosome ID as two random intercept effects, and assessed model fit using the performance (v0.10.2) and DHARMa (v0.4.6) packages in R (v4.2.3). If the number of DNMs on a chromosome scaled linearly with log-transformed callable chromosome size, we expected an estimate for log(CG) of β = 163.
We used this model to also derive the chrZ-to-autosome ratio of the DNM rates. Specifically, we back-transformed the parameter estimates for auto- and gonosome and calculated their ratios. We derived 95% confidence intervals (CIs) through parametric bootstrapping with 1000 iterations. Expected chrZ-to-autosome ratios were first derived by Miyata et al. 80. They assumed an equal DNM rate on the sex chromosomes and the autosomes and an equal sex ratio, which leads to an upper bound of the chrZ-to-autosome ratio of 4/3. We adjusted their formula to incorporate different numbers of female (NF = 9) and male (NM = 7) offspring as:
where α is the male-to-female DNM ratio. In case of an equal sex ratio (NF = NM), this formula simplifies to \(2/3* \left(2* \alpha +1\right)/(1+\alpha )\), which is the formula given by Miyata et al. 80.
Statistics and reproducibility
All analysis scripts and data are publicly available (raw whole genome sequencing data of the 20 zebra finches at NCBI: SRR28223484–SRR28223503, validation genotyping data and analysis scripts at the Open Science Framework: https://doi.org/10.17605/OSF.IO/N496T)112. Sample sizes for each statistical analysis are given in the corresponding figures or supplementary material. We used two-tailed statistical tests and considered P < 0.05 as statistically significant. All analyses were performed blind to the outcome.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The supplementary Tables S1–S25 can be found in Supplementary Data. Whole genome re-sequencing data of the 20 zebra finches is available through NCBI (SRR28223484–SRR28223503). Validation genotyping data is available through the Open Science Framework (https://doi.org/10.17605/OSF.IO/N496T)112.
Code availability
Analysis scripts are available through the Open Science Framework (https://doi.org/10.17605/OSF.IO/N496T)112.
References
Yoder, A. D. & Tiley, G. P. The challenge and promise of estimating the de novo mutation rate from whole‐genome comparisons among closely related individuals. Mol. Ecol. 30, 6087–6100 (2021).
Kondrashov, A. S. Direct estimates of human per nucleotide mutation rates at 20 loci causing mendelian diseases. Hum. Mutat. 21, 12–27 (2003).
Belyeu, J. R. et al. De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2396 families. Am. J. Hum. Genet. 108, 597–607 (2021).
Wang, Y. & Obbard, D. J. Experimental estimates of germline mutation rate in eukaryotes: a phylogenetic meta-analysis. Evol. Lett. 7, 216–226 (2023).
Kloosterman, W. P. et al. Characteristics of de novo structural changes in the human genome. Genome Res. 25, 792–801 (2015).
Sung, W. et al. Evolution of the insertion-deletion mutation rate across the tree of life. G3 GenesGenomesGenetics 6, 2583–2591 (2016).
Besenbacher, S., Hvilsom, C., Marques-Bonet, T., Mailund, T. & Schierup, M. H. Direct estimation of mutations in great apes reconciles phylogenetic dating. Nat. Ecol. Evol. 3, 286–292 (2019).
Scally, A. & Durbin, R. Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet. 13, 745–753 (2012).
Bergeron, L. A. et al. Evolution of the germline mutation rate across vertebrates. Nature 615, 285–291 (2023).
Lynch, M. Evolution of the mutation rate. Trends Genet. 26, 345–352 (2010).
Zhang, M., Yang, Q., Ai, H. & Huang, L. Revisiting the evolutionary history of pigs via de novo mutation rate estimation in a three-generation pedigree. Genomics Proteom. Bioinform. 20, 1040–1052 (2022).
Besenbacher, S. et al. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat. Commun. 6, 5969 (2015).
Kessler, M. D. et al. De novo mutations across 1,465 diverse genomes reveal mutational insights and reductions in the Amish founder population. Proc. Natl Acad. Sci. 117, 2560–2569 (2020).
Acuna-Hidalgo, R., Veltman, J. A. & Hoischen, A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 17, 241 (2016).
Haldane, J. B. S. The rate of spontaneous mutation of a human gene. J. Genet. 31, 317–326 (1935).
Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl Acad. Sci. 107, 961–968 (2010).
Ségurel, L., Wyman, M. J. & Przeworski, M. Determinants of mutation rate variation in the human germline. Annu. Rev. Genomics Hum. Genet. 15, 47–70 (2014).
Roach, J. C. et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328, 636–639 (2010).
Kong, A. et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012).
McVean, G. T. & Hurst, L. D. Evidence for a selectively favourable reduction in the mutation rate of the X chromosome. Nature 386, 388–392 (1997).
Rodriguez-Galindo, M., Casillas, S., Weghorn, D. & Barbadilla, A. Germline de novo mutation rates on exons versus introns in humans. Nat. Commun. 11, 3304 (2020).
Seplyarskiy, V. B. & Sunyaev, S. The origin of human mutation in light of genomic data. Nat. Rev. Genet. 22, 672–686 (2021).
Hodgkinson, A. & Eyre-Walker, A. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 12, 756–766 (2011).
Wu, F. L. et al. A comparison of humans and baboons suggests germline mutation rates do not track cell divisions. PLoS Biol. 18, e3000838 (2020).
Smeds, L., Qvarnström, A. & Ellegren, H. Direct estimate of the rate of germline mutation in a bird. Genome Res. 26, 1211–1218 (2016).
Wang, R. J. et al. Examining the effects of hibernation on germline mutation rates in grizzly bears. Genome Biol. Evol. 14, evac148 (2022).
Gao, Z. et al. Overlooked roles of DNA damage and maternal age in generating human germline mutations. Proc. Natl Acad. Sci. 116, 9491–9500 (2019).
Hahn, M. W., Peña-Garcia, Y. & Wang, R. J. The ‘faulty male’ hypothesis for sex-biased mutation and disease. Curr. Biol. 33, R1166–R1172 (2023).
Benzer, S. On the topography of the genetic fine structure. Proc. Natl Acad. Sci. USA 47, 403–415 (1961).
Reijns, M. A. M. et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 (2015).
Gorelick, R. Evolution of dioecy and sex chromosomes via methylation driving Muller’s ratchet. Biol. J. Linn. Soc. 80, 353–368 (2003).
Halldorsson, B. V. et al. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363, eaau1043 (2019).
Frigola, J. et al. Reduced mutation rate in exons due to differential mismatch repair. Nat. Genet. 49, 1684–1692 (2017).
Monroe, J. G. et al. Mutation bias reflects natural selection in Arabidopsis thaliana. Nature 602, 101–105 (2022).
Charlesworth, B. & Jensen, J. D. Population genetic considerations regarding evidence for biased mutation rates in Arabidopsis thaliana. Mol. Biol. Evol. 40, msac275 (2023).
Liu, H. & Zhang, J. Is the mutation rate lower in genomic regions of stronger selective constraints? Mol. Biol. Evol. 39, msac169 (2022).
Majic, P. & Payne, J. L. Developmental selection and the perception of mutation bias. Mol. Biol. Evol. 40, msad179 (2023).
Hodgkinson, A., Ladoukakis, E. & Eyre-Walker, A. Cryptic variation in the human mutation rate. PLoS Biol. 7, e1000027 (2009).
Goldmann, J. M. et al. Differences in the number of de novo mutations between individuals are due to small family-specific effects and stochasticity. Genome Res. 31, 1513–1518 (2021).
Ness, R. W., Morgan, A. D., Vasanthakrishnan, R. B., Colegrave, N. & Keightley, P. D. Extensive de novo mutation rate variation between individuals and across the genome of Chlamydomonas reinhardtii. Genome Res. 25, 1739–1749 (2015).
Bolstad, G. H. et al. Genetic constraints predict evolutionary divergence in Dalechampia blossoms. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130255 (2014).
de Manuel, M., Wu, F. L. & Przeworski, M. A paternal bias in germline mutation is widespread in amniotes and can arise independently of cell division numbers. eLife 11, e80008 (2022).
Wang, R. J. et al. De novo mutations in domestic cat are consistent with an effect of reproductive longevity on both the rate and spectrum of mutations. Mol. Biol. Evol. 39, msac147 (2022).
Crow, J. F. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 1, 40–47 (2000).
Ellegren, H. & Fridolfsson, A.-K. Male–driven evolution of DNA sequences in birds. Nat. Genet. 17, 182–184 (1997).
Makova, K. D. & Li, W.-H. Strong male-driven evolution of DNA sequences in humans and apes. Nature 416, 624–626 (2002).
Goldmann, J. M. et al. Parent-of-origin-specific signatures of de novo mutations. Nat. Genet. 48, 935–939 (2016).
Jónsson, H. et al. Parental influence on human germline de novo mutations in 1548 trios from Iceland. Nature 549, 519–522 (2017).
Johnson, N. A. & Lachance, J. The genetics of sex chromosomes: evolution and implications for hybrid incompatibility. Ann. N. Y. Acad. Sci. 1256, E1–E22 (2012).
Francioli, L. C. et al. A framework for the detection of de novo mutations in family-based sequencing data. Eur. J. Hum. Genet. 25, 227–233 (2017).
Axelsson, E., Smith, N. G. C., Sundström, H., Berlin, S. & Ellegren, H. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol. Biol. Evol. 21, 1538–1547 (2004).
Knief, U. et al. Fitness consequences of polymorphic inversions in the zebra finch genome. Genome Biol. 17, 199 (2016).
Knief, U. et al. A sex-chromosome inversion causes strong overdominance for sperm traits that affect siring success. Nat. Ecol. Evol. 1, 1177–1184 (2017).
Pariser, E. C., Mariette, M. M. & Griffith, S. C. Artificial ornaments manipulate intrinsic male quality in wild-caught zebra finches (Taeniopygia guttata). Behav. Ecol. 21, 264–269 (2010).
Royle, N. J., Hartley, I. R. & Parker, G. A. Sexual conflict reduces offspring fitness in zebra finches. Nature 416, 733–736 (2002).
Wang, D., Forstmeier, W. & Kempenaers, B. No mutual mate choice for quality in zebra finches: time to question a widely held assumption. Evolution 71, 2661–2676 (2017).
Wang, D. et al. Irreproducible text-book “knowledge”: the effects of color bands on zebra finch fitness. Evolution 72, 961–976 (2018).
Warren, W. C. et al. The genome of a songbird. Nature 464, 757–762 (2010).
Zann, R. A. The Zebra Finch: A Synthesis of Field and Laboratory Studies (Oxford University Press, Oxford, 1996).
Rhie, A. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021).
Li, H. et al. Exome variant discrepancies due to reference-genome differences. Am. J. Hum. Genet. 108, 1239–1250 (2021).
Bird, J. P. et al. Generation lengths of the world’s birds and their implications for extinction risk. Conserv. Biol. 34, 1252–1261 (2020).
Gelman, A. & Hill, J. Data Analysis Using Regression and Multilevel/Hierarchical Models (Cambridge University Press, Cambridge, 2007).
Singhal, S. et al. Stable recombination hotspots in birds. Science 350, 928–932 (2015).
Sokal, R. R. & Rohlf, F. J. Biometry: The Principles and Practices of Statistics in Biological Research (W. H. Freeman, New York, 1995).
Zhang, H., Lundberg, M., Tarka, M., Hasselquist, D. & Hansson, B. Evidence of site-specific and male-biased germline mutation rate in a wild songbird. Genome Biol. Evol. 15, evad180 (2023).
Nam, K. et al. Molecular evolution of genes in avian genomes. Genome Biol. 11, R68 (2010).
Birkhead, T. R. & Møller, A. P. Sperm Competition and Sexual Selection (Academic Press, San Diego, 1998).
Birkhead, T. R., Pellatt, E. J., Brekke, P., Yeates, R. & Castillo-Juarez, H. Genetic effects on sperm design in the zebra finch. Nature 434, 383–387 (2005).
Blumenstiel, J. P. Sperm competition can drive a male-biased mutation rate. J. Theor. Biol. 249, 624–632 (2007).
Brouwer, L. & Griffith, S. C. Extra-pair paternity in birds. Mol. Ecol. 28, 4864–4882 (2019).
Hasselquist, D., Bensch, S. & von Schantz, T. Low frequency of extrapair paternity in the polygynous great reed warbler, Acrocephalus arundinaceus. Behav. Ecol. 6, 27–38 (1995).
Leisler, B., Beier, J., Staudter, H. & Wink, M. Variation in extra-pair paternity in the polygynous Great Reed Warbler (Acrocephalus arundinaceus). J. F.ür. Ornithol. 141, 77–84 (2000).
Ellegren, H. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc. Biol. Sci. 274, 1–10 (2007).
Bartosch-Härlid, A., Berlin, S., Smith, N. G. C., Møller, A. P. & Ellegren, H. Life history and the male mutation bias. Evolution 57, 2398–2406 (2003).
Taylor, J., Tyekucheva, S., Zody, M., Chiaromonte, F. & Makova, K. D. Strong and weak male mutation bias at different sites in the primate genomes: insights from the human-chimpanzee comparison. Mol. Biol. Evol. 23, 565–573 (2006).
Seplyarskiy, V. B. et al. Population sequencing data reveal a compendium of mutational processes in the human germ line. Science 373, 1030–1035 (2021).
Drost, J. B. & Lee, W. R. Biological basis of germline mutation: comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ. Mol. Mutagen 25, 48–64 (1995).
Jiang, L. et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153, 773–784 (2013).
Miyata, T., Hayashida, H., Kuma, K., Mitsuyasu, K. & Yasunaga, T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 52, 863–867 (1987).
Gillespie, J. H. The Causes of Molecular Evolution (Oxford University Press, New York, 1994).
Catalán, A., Merondun, J., Knief, U. & Wolf, J. B. W. Chromatin accessibility, not 5mC methylation covaries with partial dosage compensation in crows. PLoS Genet. 19, e1010901 (2023).
Xia, B. et al. Widespread transcriptional scanning in the testis modulates gene evolution rates. Cell 180, 248–262.e21 (2020).
Lapierre, M., Lambert, A. & Achaz, G. Accuracy of demographic inferences from the site frequency spectrum: the case of the Yoruba population. Genetics 206, 439–449 (2017).
Zeng, K., Jackson, B. C. & Barton, H. J. Methods for estimating demography and detecting between-locus differences in the effective population size and mutation rate. Mol. Biol. Evol. 36, 423–433 (2019).
Mank, J. E., Axelsson, E. & Ellegren, H. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 17, 618–624 (2007).
Forstmeier, W., Segelbacher, G., Mueller, J. C. & Kempenaers, B. Genetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata). Mol. Ecol. 16, 4039–4050 (2007).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Okonechnikov, K., Conesa, A. & García-Alcalde, F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294 (2016).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
GATK team. I am unable to use VQSR (recalibration) to filter variants. https://gatk.broadinstitute.org/hc/en-us/articles/360037499012-I-am-unable-to-use-VQSR-recalibration-to-filter-variants (2020).
Bergeron, L. A. et al. The Mutationathon highlights the importance of reaching standardization in estimates of pedigree-based germline mutation rates. eLife 11, e73577 (2022).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
Bergeron, L. A. et al. The germline mutational process in rhesus macaque and its implications for phylogenetic dating. GigaScience 10, giab029 (2021).
Francioli, L. C. et al. Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 47, 822–826 (2015).
Koch, E. M. et al. De novo mutation rate estimation in wolves of known pedigree. Mol. Biol. Evol. 36, 2536–2547 (2019).
Luo, X. et al. Trio deep-sequencing does not reveal unexpected off-target and on-target mutations in Cas9-edited rhesus monkeys. Nat. Commun. 10, 5525 (2019).
Pfeifer, S. P. Direct estimate of the spontaneous germ line mutation rate in African green monkeys. Evolution 71, 2858–2870 (2017).
Sasani, T. A. et al. Large, three-generation human families reveal post-zygotic mosaicism and variability in germline mutation accumulation. eLife 8, e46922 (2019).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Keightley, P. D., Ness, R. W., Halligan, D. L. & Haddrill, P. R. Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics 196, 313–320 (2014).
Goldmann, J. M., Veltman, J. A. & Gilissen, C. De novo mutations reflect development and aging of the human germline. Trends Genet. 35, 828–839 (2019).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Thomas, G. W. C. et al. Reproductive longevity predicts mutation rates in primates. Curr. Biol. 28, 3193–3197.e5 (2018).
Harrell Jr, F. E. Hmisc: Harrell Miscellaneous. https://CRAN.R-project.org/package=Hmisc (2023).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing (2022).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Lüdecke, D., Ben-Shachar, M. S., Patil, I., Waggoner, P. & Makowski, D. Performance: an R package for assessment, comparison and testing of statistical models. J. Open Source Softw. 6, 3139 (2021).
Hartig, F. Dharma: residual diagnostics for hierarchical (multi-level / mixed) regression models. https://CRAN.R-project.org/package=DHARMa (2022).
Knief, U. & Forstmeier, W. Violating the normality assumption may be the lesser of two evils. Behav. Res. Methods 53, 2576–2590 (2021).
Alberti, G. chisquare: chi-square and g-square test of independence, power and residual analysis, measures of categorical association. https://CRAN.R-project.org/package=chisquare (2024).
Yang, S. Characterization and distribution of de novo mutations in the zebra finch. OSF https://doi.org/10.17605/OSF.IO/N496T (2024).
Acknowledgements
D.W. is funded by the Chinese Academy of Sciences (CAS Pioneer Hundred Talents program), the Third Xinjiang Scientific Expedition Program (Grant No. 2022xjkk0801), and the National Natural Science Foundation of China (32270452). U.K. is funded through the German Research Foundation (DFG). We are grateful to Bart Kempenaers and Wolfgang Forstmeier for breeding zebra finches in the facilities at the Max Planck Institute for Biological Intelligence in Seewiesen. We further thank the Institute of Clinical Molecular Biology (IKMB) at Kiel University, Germany, for sequencing and genotyping. We are grateful to Xu Zhang and Xuebo Xi for help with computational problems. HPC computing was partly performed on the BioHPC hosted at Leibniz Rechenzentrum Munich funded by the German Research Foundation (grant INST 86/2050-1 FUGG).
Author information
Authors and Affiliations
Contributions
D.W. and U.K. conceived of the study. U.K. designed the study and produced the data. S.Y. and X.L. performed the analysis with help from D.W. and U.K. U.K. completed the experimental genotype verification. All authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Aylin Bircan and David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liang, X., Yang, S., Wang, D. et al. Characterization and distribution of de novo mutations in the zebra finch. Commun Biol 7, 1243 (2024). https://doi.org/10.1038/s42003-024-06945-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06945-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.