Abstract
Not all MET exon 14 skipping (METex14) NSCLC patients benefited from MET inhibitors. We hypothesized an inter-tumoral heterogeneity in METex14 NSCLC. Investigations at genomic and transcriptomic level were conducted in METex14 NSCLC samples from stage I-III and recurrent/metastatic patients as discovery and validation cohort. Four molecular subtypes were discovered. MET-Driven subtype, with the worst prognosis, displayed MET overexpression, enrichment of MET-related pathways, and higher infiltration of fibroblast and regulatory T cells. Immune-Activated subtype having the most idea long-term survival, had higher tertiary lymphoid structures, spatial co-option of PD-L1+ cancer cells, and GZMK+ CD8+ T cell. FGFR- and Bypass-Activated subtypes displayed FGFR2 overexpression and enrichments of multiple oncogenic pathways respectively. In the validation cohort, patients with MET-Driven subtype had better response to MET inhibitors than those with MET overexpression. Thus, molecular subtypes of METex14 NSCLC with distinct biological and clinical significance may indicate more precise therapeutic strategies for METex14 NSCLC patients.
Similar content being viewed by others
Introduction
MET Exon14 skipping (METex14) is an oncogenic driver occurring in 1–3% of patients with lung adenocarcinoma (LUAD)1,2,3 and lung squamous carcinoma (LUSC)4. METex14 NSCLC responds to the treatment of MET tyrosine kinase inhibitors (MET-TKIs), with the reported objective response rate (ORR) ranging from 32% to 67.9% and median progression-free survival (mPFS) was from 5.4–9.7 months5,6,7,8. Thus, not all METex14 NSCLC patients benefited from the treatment of MET inhibitors. We hypothesized an inter-tumoral heterogeneity in ex14 NSCLC determining response of METex14 NSCLC to MET-TKIs. And it would be of clinical importance in influencing decision of management for MET ex14 NSCLC patients.
Regarding complex genomic locations and various types of mutations, METex14 NSCLC may putatively harbor biological heterogeneity9,10. Meanwhile, MET expression varied in METex14 NSCLC patients. As reported, METex14 NSCLC with MET over-expression got more benefit from MET inhibitor treatment6,11. On the other side, NSCLC patients with METex14 displayed higher levels of PD-L1 both at transcription and protein levels. Higher immune infiltration and IFN-γ signatures were also reported by previous studies12,13. However therapeutic efficacy of immunotherapy remained controversial in patients with METex14 NSCLC14,15,16. These recent translational and clinical studies strongly indicated a potential inter-heterogeneity in METex14 NSCLC.
While no systematic investigation by far has been conducted to disclose the inter-tumor heterogeneity in METex14 NSCLC. To address this hypothesis, comprehensive genomic and transcriptomic profiling was performed together with the immunophenotype of the tumor microenvironment. A better understanding of the biological and tumor microenvironmental diversity of METex14 NSCLC may provide insight into a more precise evaluation the risk of disease recurrence at the early disease stage. It may also indicate novel therapeutic approaches for advanced/metastatic NSCLC patients with METex14.
Results
Transcriptomic profiling reveals Intertumoral heterogeneity among METex14 NSCLC patients
Limited genomic location-specific differentially expressed genes in MET ex14 NSCLC were discovered (Supplementary Fig. 1), which indicated the limited impact of various genomic locations of MET ex14 on inter-tumoral heterogeneity.
Transcriptomics-based molecular subtypes were discovered revealing inter-tumoral heterogeneity in various malignant solid tumors17,18,19,20. In this study, targeted RNA sequencing was successfully conducted in the discovery and validation cohort with the success rate of 97.67% (126/129) and 95.52% (64/67), respectively (Fig. 1). To understand the inter-tumor heterogeneity, enrichment of pathways in KEGG, Reactome, Hallmark was evaluated in each sample utilizing RNA-seq data from discovery cohort (n = 126) followed by k-means clustering. As informed by consensus cumulative distribution function (CDF) and the relative change in area under the curve (AUC) for CDF, (Supplementary Fig. 2a, b), four well-distinguished subtypes (κ = 4) in METex14 NSCLC from the discovery cohort were discovered for the first time (Fig. 1, Fig. 2a, b), with 38 cases (30.2%) of subtype A, 19 cases (15.1%) of subtype B, 24 cases (19.0%) of subtype C and 45 cases (35.7%) of subtype D.
a The consensus matrix represented as heatmap for the chosen optimal cluster number (k = 4) for unsupervised clustering analysis on RNA-seq data. b The t-SNE plot shows the localization of each of the four tumor classes. c Comparison of clinical characteristics between molecular subtypes. d Disease-free survival of the four subtypes in the discovery cohort. Forest plots showing multivariate (e) and univariate (f) analysis of the molecular subtype and clinical risk factors associating with DFS.
Molecular subtype distinct clinic-pathological significance in METex14 NSCLC patients
Subtype A was significantly associated with clinical (P = 0.0023) and pathologic (P < 0.001) stage II/III, especially with clinical and pathological T2-3 (P < 0.001) (Fig. 2c, Table 1). Subtype A also displayed clinic-pathologically more aggressive characteristics, such as invasive subtype (P < 0.0001) and pleural invasion (P = 0.004) (Fig. 2c, Table 1). In 107 patients with long-term follow-up, subtype A and C had the most unfavored and best prognosis, respectively (Fig. 2d, P = 0.029). Of note, three-year DFS rate of subtype C reached to 100% (Fig. 2d). Multi and univariate cox regression analysis showed subtype A was independently associated with shorter DFS (hazard ratio [HR] = 6.68 [95% CI 1.80–24.88], P = 0.005) (Fig. 2e, f). These well-defined molecular subtypes with unique clinic-pathologic features suggested subtype-specific biological features.
Comprehensive genomic profiling displayed a relatively well-balanced subtype-specific genomic characteristics
To explore subtype-specific genomic characteristics, comprehensive genomic profiling was conducted and analyzed. MET amplification (~5% in 120 samples) was balanced across four subtypes (Supplementary Fig. 3a). TP53, which had a mutation frequency of 31.6% in subtype A and was rarer in the other subtypes (Supplementary Fig. 3a). No association between TP53 mutations and prognosis was observed, either across the cohort or only within subtype A (Supplementary Fig. 3b, c). FGF19, AKT2, and FGF3 amplifications were significantly higher in subtype A; FGF3, PDCD1, and FGF19 deletions were more commonly occurred in subtype B and D (Supplementary Fig. 3a). No association was found between these CNV gain/loss and prognosis, mRNA expression, tumor microenvironment features, signaling pathways enrichment (Supplementary Fig. 4a–c). No significantly distributed specific mutation signature was found across four subtypes (Supplementary Fig. 3a). No differential distribution of genomic locations of MET ex14 variants was observed across these four subtypes (Supplementary Fig. 3d, e).
Unique activated oncogenic pathways in four molecular subtypes
Variant allele frequency (VAF) of METex14 was highest in subtype A (Fig. 3a, P = 0.0022), together with MET gene and c-MET protein over expression (Fig. 3b–f, P < 0.001). Pathways mostly enriched in subtype A were associated with promoting cell-cycle, DNA replication and MET-driven cell motility and PTK2 signaling (Fig. 4a–c).
a GSVA analysis of differentially expressed genes in the four subtypes based on the curated KEGG, Reactome, and Hallmark gene sets. b ssGSEA analysis of differentially expressed genes based on the KEGG gene set. c ssGSEA result showed that COL1A1, LAMB3, MET, and FN1 were significantly enriched in three pathways “MET activates PTK2 signaling”, “MET promotes cell motility”, and “signaling by MET” in subtype A. d ssGSEA result showed that FGFR2, FGFR4, FGFR3, and FGFR1 were significantly enriched in “SIGNALING_BY_FGFR” pathway in subtype B. e ssGSEA result showed that NOTCH4, NOTCH1, CREBBP, MAML2, and NCOR2 were significantly enriched in “NOTCH_SIGNALING_PATHWAY” pathway in subtype D.
Subtype B was mostly related to FGFR pathways, such as PI3K_CASCADE_FGFR1-4, and DOWTREAM SIGNALING OF ACTIVATED FGFR 1-4 (Fig. 4a, b). And Subtype B was characterized by FGFR2 over-expression (Fig. 4d).
Subtype C was characterized by the enrichment of immune-related pathways, such as MHC_CLASS_II_ANTIGEN, INNATE IMMUNE SYSTEM, and ADAPTIVE IMMUNE SYSTEM (Fig. 4a, b).
Activation of multiple signaling pathways, such as NOTCH, FGFR, PIP2, PI3K-AKT, ERBB2, and IKBA were found in subtype D (Fig. 4a, b). Further analysis of the “NOTCH_SIGNALING_PATHWAY” pathway revealed that NOTCH4, NOTCH1, CREBBP, MAML2, and NCOR2 were highly expressed (Fig. 4e).
Distinct tumor immune microenvironment (TME) in four molecular subtypes
Part of METex14 NSCLC patients benefited from immune checkpoint inhibitors21, but others did not22,23, which suggested heterogenous tumor microenvironment in METex14 NSCLC. As discovered, each molecular subtype displayed a distinct TME phenotype (Fig. 5a, Supplementary Fig. 5a–e). Subtype A displayed highly immunosuppressive TME characterized by infiltration of regulatory T (Treg) cells and PDCD1 (PD-1) expression (Fig. 5a, b, Supplementary Fig. 5f, Supplementary Fig. 6a). Meanwhile highly enriched cancer-associated fibroblasts (CAF) signatures can also be observed in subtype A tumors, especially for pan-CAF (pCAF), desmoplastic CAF (dCAF) and LRRC15 + CAF (Fig. 5c). Histologically, condensed desmoplastic phenotype and fibrosis in H&E staining further supported the findings of enriched CAF signatures in subtype A (Fig. 5d).
a Signatures from 28 immunity, 29 immunity, AO, Danaher and MCPCounter reference sets were found to hold significant differences among four subtypes. b Expression of four cancer-associated fibroblasts (CAF) signatures, myofibroblastic CAF (myCAF), development CAF (dCAF), inflammatory CAF (iCAF) and pCAF, and c fibroblasts score in the four subtypes. d A representative H&E staining of subtype A. e Expression of STAT signature, IFN-γ related genes, and effector T cell gene signature of the four subtypes. f Expression of 18 genes related to IFN-γ signature in the four subtypes. g Representative example of multiple immunofluorescences of Treg cells of subtype A. h Representative example of multiple immunofluorescences of effector CD8 + T cell of subtype C. i Representative example of multiple immunofluorescences of CD20 + B cell of subtype C.*, **, and *** represent P < 0.05, P < 0.01, and P < 0.001, respectively.
On the other hand, anti-tumor immunity was more prominent in subtype C, with significantly elevated enrichment of Effector cells, T cells, Th1 signature, B cell, Coactivation molecules (Supplementary Fig. 5a–e, Supplementary Fig. 6b), and increased level of inflammatory cancer-associated fibroblast (iCAF) (Fig. 5c). Moreover, pro-inflammatory signatures like STAT signature, IFN-γ signature, and effector T cell signature were also significantly enriched in Subtype C compared with other subtypes (Fig. 4e). Of note, IFN-γ related genes was also highly upregulated in subtype C (Fig. 4f).
In parallel, mIF showed an aggregation of CD4+/FOXP3+ Treg cells in tumor tissues of subtype A (Fig. 5g), but almost invisible in the tumor tissue of the other three subtypes (Supplementary Fig. 7). In subtype C, a larger number of infiltrated effector CD8 + T cells (CD8+/GZMK+) and spatial co-option with PD-L1+ cancer cells (Fig. 5h) were observed compared to the other three subtypes (Supplementary Fig. 8). Higher number of tertiary lymphoid structures (TLS) characterized by accumulated CD20+ B cells were also detected in subtype C (Fig. 5i, Supplementary Fig. 9). These findings proved that in METex14 NSCLC from the discovery cohort, patients with immune enriched subtype (subtype C) potentially benefited from ICIs, while patients with subtype A, prominent fibrosis and Treg infiltration, would putatively be resistant to ICIs.
Constructing subtype classifier and predicting molecular subtypes in relapsed/metastatic MET ex14 NSCLC patients
To translate biological diversity of METex14 NSCLC into a clinically feasible tool, a subtype classifier was constructed based on the transcriptomic data in the discovery cohort (Fig. 6a). Previous studies in cancers such as SCLC17, CRC18,24, and breast cancer19,25,26 have demonstrated the feasibility of using early-stage patients for subtype identification and then validation in advanced patients. An independent cohort of relapsed/metastatic NSCLC patients was enrolled (Fig. 1) to verify the performance of the subtype classifier. To ensure the biological comparability, differences in gene expression, tumor microenvironment, and signaling pathways enrichment between the discovery cohort and validation cohort patients were analyzed by tSNE methods. Overall, a well-balanced expression profile was found between early- and late-stage NSCLC with METex14 mutations (Supplementary Fig. 3f–h).
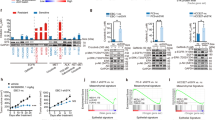
a Schematic diagram of model construction process. b The predictive performance in 5-fold cross-validation of the prediction model based on (the discovery cohort). c Leave-one-out cross-validation showing the model prediction results of the discovery cohort. d ssGSEA result showed that COL1A1, MET, FN1, and LAMB3 were significantly enriched in three pathways “MET activates PTK2 signaling”, “MET promotes cell motility”, and “signaling by MET” in subtype A in the independent validation cohort. e Expression of MET gene in the four subtypes in the independent validation cohort. f ssGSEA result showed that FGFR2 was significantly enriched in “SIGNALING_BY_FGFR2” pathway in subtype B in the independent validation cohort. g Expression of FGFR2 gene in the independent validation cohort. h Signatures from 28 immunity, 29 immunity, AO, Danaher, and MCPCounter with significant differences among four subtypes in the independent validation cohort. i ssGSEA result showed that differentially expressed genes were enriched in NOTCH, PI3K-AKT, ERBB2, and MAPK SIGNALING pathways in subtype D in the independent validation cohort. *, **, and *** represent P < 0.05, P < 0.01, and P < 0.001, respectively.
Briefly, 20 leading genes were screened for each subtype using univariant logistic regression, and a total of 80 genes were obtained (Supplementary Fig. 10a). 39 of 80 genes were selected by lasso-logistic regression for classifier construction (Supplementary Fig. 10b). Subtype-specific genes in each group were also selected (Supplementary Fig. 10c). Five-fold cross-validation and LOOV showed with more than 80% accuracy (Figs. 6b), and 90.5% of overall concordance rate in LOOV (Fig. 6c) respectively.
In the validation cohort, four subtypes were predicted and showed well-balanced clinic-pathological characteristics (Supplementary Table 1). Benchmarked MET-related signaling pathways were also enriched in predicted subtype A (MET-Driven) (68.8%) (Fig. 6d), with the expression of MET genes was significantly higher in this subtype (Fig. 6e). A tendency for FGFR-related signaling pathways was observed to be enriched in predicted subtype B (FGFR Activated) (Fig. 6f), and FGFR2 gene expression was significantly upregulated (Fig. 6g). TME phenotype of each subtype in this validation set similarly reveal that predicted subtype C (Immune-Activated) (21.9%) was immunologically activated (Fig. 6h). And activation of NOTCH, PI3K-AKT, ERBB2 and MAPK were observed in predicted subtype D (Bypass-Activated) (Fig. 6i).
Of note, one patient (a 68-year-old female, non-smoker) with Immune-Activated subtype in the validation cohort was treated with immune-chemo combined therapy (nivolumab + pemetrexed + carboplatin) and achieved a partial response with a PFS of 16 months (Supplementary Fig. 11).
MET-Driven patients displayed better responses to MET-targeting tyrosine kinase inhibitors
To understand the drug vulnerability of each subtype of METex14 NSCLC, the survival of 41 patients in the independent validation cohort with the treatment of MET inhibitors was analyzed (Fig. 7a). 26 patients were MET-Driven subtype (subtype A) and 15 patients belonged to other subtypes. Baseline characteristics were comparable between patients with MET-Driven subtype and those with the other three subtypes (Supplementary Table 2). The follow-up cut-off was June 2023. The median progression-free survival (PFS) of the 41 patients was 7.5 months (Fig. 7b). Patients with MET-Driven subtype responded better to MET inhibitors and displayed longer PFS than the other three predicted subtypes (median PFS: 9.9 months vs 5.9 months vs 6.3 months vs 4.5 months, P = 0.029, Fig. 7 a, c), (median PFS, 95%CI: 9.9 months,7-NR vs 5.9 months, 4-NR; HR, 95%CI: 0.377, 0.175–0.811, P = 0.0093, Fig. 7d). Moreover, MET-Driven subtype independently associated with the benefit of MET inhibitors (HR, 95%CI: 0.36, 0.16–0.79, P = 0.011, Fig. 7f).
a Swimmer plot showed the follow-up information of the 41 patients treated with MET-TKI in a time range of 35 months. b Kaplan-Meier plot showed progression-free survival (PFS) of all the 41 patients treated with MET-TKI. c PFS of the patients who received MET-TKI treatment with four predicted subtypes. d PFS of the predicted subtype A patients and patients with the other three predicted subtypes received MET-TKI treatment. e PFS of the patients received MET-TKI treatment with high or low MET expression. f Forest plot of showing multivariate COX regression of the molecular subtype and clinical risk factors associated with PFS.
However, PFS was not significantly different for patients with high MET expression and those with low MET expression (median PFS: 9.6 months vs 7.0 months; HR,95%CI: 0.572, 0.261–1.254, P = 0.15, Fig. 7e). These results suggested MET inhibitors may be used more precisely, especially to those with MET-Driven subtype.
Discussion
Short-term response to MET inhibitors observed in multiple clinical trials as well as dis-concordant efficacy of immunotherapy leads to the following hypothesis: there is an inter-patient/tumor heterogeneity with clinical and biological significance in patients with METex14 NSCLC21. As suggested by this study, diverse genomic locations, or types of METex14 variants are not directly associated with inter-tumoral heterogeneity. In parallel, recent clinical trials of MET-TKIs suggested there is no direct association between the locations or type of the METex14 alterations and therapeutic efficacy of the targeted MET inhibitor6,27.
In this study, four subtypes (MET-Driven, FGFR-Activated, Immune-Activated and Bypass-Activated) with differentially clinic-pathological characteristics, prognosis and drug vulnerability were discovered for the first time at transcriptional level (Fig. 8). Co-occurred genomic alterations either TP53 or copy number variations had limited impact on clinical and biological characterizations of MET ex 14 NSCLC. MET-Drive subtype (30.2% in stage I-III patients; 68.8% in relapsed/metastatic patients), showed higher VAF of METex14 together with overexpression of MET gene, c-MET protein, and enrichment of MET-related pathways. As reported, the frequency of METex14 and expression of MET correlated with the efficacies of MET-TKIs5,27. Parallelly, MET-Driven patients were significantly vulnerable to MET-TKIs and displayed longer PFS than other subtypes.
MET-Driven tumors displayed an immunosuppressive phenotype with infiltration of Tregs and LRRC15+ and desmoplastic fibroblasts. High expression of the LRRC15+ CAF and desmoplastic CAFs signature was associated with poor response to immune-checkpoint blockade in several different human tumor types28,29. As such, even in MET-Driven tumors, PD-L1, as well as other inhibitory immune checkpoint molecules21,30 were upregulated in tumor cells, this immunosuppressive and exclusive TME would attenuate the efficacy of immunotherapy31. These findings may provide a preliminary explanation of the paradox in clinics of why significant proportions of PD-L1 positive METex14 NSCLC patients fail to respond to PD-1/PD-L1 treatment.
Oppositely, Immune-Activated subtype (19.0% in stage I-III patients; 21.9% in relapsed/metastatic patients) were characterized by significant enrichment of proinflammatory signatures like IFN-γ, effector T cells and inflammatory CAFs. Immunofluorescence further suggested increased infiltrated CD8+/GZMK+ T cells and TLS in immune-activated subtype. It can be inferred that patients with immune activated phenotype may benefit from immunotherapy. Recent studies reported portions of NSCLC patients harboring METex14 alterations displayed durable response to immunotherapy, which supported this hypothesis. Particularly, partial, or complete responses over more than 18 months were seen in six patients (46.2%) with METex14 alterations32. In addition, four NSCLC patients with METex14 and MET amplification had long-lasting response to second- or late line immunotherapy12. Immune-Activated subtype may define a subset of METex14 NSCLC patients may benefit from immunotherapy or a combination of immune checkpoint inhibitors with MET-TKIs.
FGFR-Activated subtype (15.1% in stage I-III patients; 3.1% in relapsed/metastatic patients) in METex14 NSCLC was mainly characterized by FGFR1-4 overexpression, especially FGFR2 overexpression, and enrichment of FGFR signaling pathways. Previous studies have found that activation of the FGFs-FGFRs pathways contributes to NSCLC pathogenesis and progression via autocrine and paracrine loops33,34. Cancer-associated fibroblast with myofibroblast differentiation also displayed activation of FGF-FGFR pathways mediating drug resistance35. Activation of these pathways may trigger resistance of multiple therapeutic modalities like anti-MET36, anti-EGFR37, and anti-vascular endothelial growth factor (VEGF)38 therapies. As suggested by a recent investigation on FGFR1-4 over-expressed breast cancer, FGFR inhibitor can cause shrinkage of tumors with elevation of FGFR1-4 mRNA39. Thus, the potential application of FGFR inhibitors might be considered as a combinational strategy with MET inhibitors for FGFR-overexpressed patients with METex14.
Bypass subtype, featuring enrichment of multiple oncogenic pathways, including NTRK, NOTCH, ERBB2, PI3K-AKT and MAPK pathways, may confer therapeutic resistance to MET tyrosine kinase inhibitors40,41,42. Biologically, METex14 could lead to trans-activation of multiple intracellular signaling pathways including RAS-MAPK43, SMAD244 resulting tumor progression and resistant to MET-targeted therapies. And, it is scientifically rational that there is mixture of genetic and non-genetic clones in malignant tumors45. And the mixture of various pathway-dominated clones promotes cancer progression46. Clinically, among the MET-TKI treated patients in this study, Bypass-Activated patients displayed the most inferior response to MET-TKI, with significantly shorter mPFS. Thus, it can be inferred that METex14 NSCLC patients in Bypass-Activated subtype may not benefit from either MET-TKI or immunotherapy, and instead require additional investigation to identify alternative options of combinational strategy.
Regarding the unbalanced proportion of FGFR-Activated and Bypass-Activated subtypes between the discovery and validation cohort, this discrepancy may be explained by the clone evolution hypothesis during cancer progression. Activation of FGF-FGFR or WNT, ERBB2, MAPK pathways may promote oncogenesis from preinvasive to invasive stage47,48,49. Based on the results from this study, it can be hypothesized that there was co-occurrence of clones with METex14 and non-genetic subclone with FGFR2 overexpression (FGFR-Activated subtype) or by WNT, ERBB2, MAPK pathways (Bypass-Activated subtype) in part of METex14 NSCLC. During cancer progression, majority of cancer cells were putatively clones of minority by MET exon 14 clones, with diminished clones with FGF-FGFR or WNT, ERBB2, MAPK pathways, which may lead less FGFR-Activated or Bypass-Activated subtype in more advanced disease stages. This hypothesis deserves in-depth investigation and further validation in a larger sample size of patients.
This study has several limitations. Firstly, most of the patients were enrolled at early disease stages, with a larger majority of patients in stage I (111 of 126, 88.1%). The lower number of patients with later stage were collected. It may lead the unbalanced disease background between discovery and validation cohort. Due to the low incidence of METex14, it was timely unavailable to recruit more patients with METex14 advanced staged NSCLC in our center or in a multi-center approach. However, very little difference was observed in gene expression, tumor microenvironment, and signaling pathways between discovery and validation cohort. Meanwhile, constant subtype-specific biological features were observed between discovery and validation cohorts. This finding again strongly suggested the molecular subtypes were highly conservative across different stages of METex14 NSCLC. Additionally, although early-stage patients predominated in the discovery cohort, MET-Driven patients displayed advanced disease stages, aggressive clinicopathologic phenotypes and poor prognosis. Importantly, in the validation cohort, MET-driven subtype displayed a better response to MET inhibitors. Thus, transcriptomic signatures from early lung cancer patients illustrated the potential to predict outcomes of stage IV lung cancers. Secondly, only a limited number of patients received treatment of MET targeting TKIs in validation cohort. Further, frontline treatments and type of MET-TKIs used were also heterogenous. While mPFS (7.5 months) of these all 41 patients was highly comparable to published mPFS (6.9 month) of MET-TKI in Chinese NSCLC patients50. Highly comparable efficacy data suggested the potential bias would not largely influence the confidence of the result that MET-Driven subtype was independently associated with the clinical benefit of MET-TKIs. Nevertheless, a multi-center, prospective study is warranted to validate these preliminary results from this study. Especially, it would be more interesting to prospectively verify the potential correlation of molecular subtypes to the therapeutic efficacy of MET-TKI(s) in treatment-naïve advanced/metastatic METex14 NSCLC.
In summary, this study provided novel insight into inter-tumoral molecular heterogeneity of METex14 NSCLC. Four distinct transcriptional molecular subtypes were discovered of the first time with unique biological and clinical significance. A classifier with sufficient robustness was translated and validated in relapsed/metastatic patients for putative clinical practice. These findings together offer a novel prism for more precise therapeutic strategy for the patients with METex14 advanced/metastatic NSCLC.
Methods
Clinical samples
10,525 stage I-III and 3,528 stage IIIB-IV NSCLC patients receiving routine molecular diagnostics in the Department of Pathology of Shanghai Chest Hospital from 2018 to 2022 were screened. 132 METex14 skipping NSCLC surgical specimens at stage I-III (the discovery cohort) and 65 relapsed/metastatic METex14 NSCLC specimens (the independent validation cohort) were retrospectively collected. Genetic test results of METex14 were reviewed by RNA-based RT-PCR, in stage I-III patients, 3 cases did not meet the quality control requirements, 3 cases were negative, and the remaining 126 cases were positive. In relapsed/metastatic patients, 1 case did not meet the quality control requirements, and the remaining 64 cases were positive (Fig. 1, right). All tissue samples were collected in accordance with the informed consent policy. The research protocol was approved by the Ethics Committee of Shanghai Chest Hospital (No. IS2183), in accordance with the Declaration of Helsinki. All participants gave informed consent or requested waiver of informed consent prior to participation in this study. This study meets the requirements of the “Guidance of the Ministry of Science and Technology (MOST) for the Review and Approval of Human Genetic Resources”.
DNA and RNA extraction, library construction, targeted sequencing, and data analysis
Targeted RNA sequencing covering transcripts of 2,660 onco-immunological genes and comprehensive genomic profiling in 571 cancer-related genes were performed on 126 specimens of discovery cohort (Fig. 1, left panel). Only targeted RNA sequencing was performed on 64 biopsy specimens of validation cohort (Fig. 1, right panel). Genomic DNA and RNA extraction refer to previously published literature, using AmoyDx FFPE DNA and RNA Kit (Amoy Diagnostics, Xiamen, China)51,52. DNA and RNA concentration was measured by Qubit (Thermo Fisher Scientific, Waltham, MA, USA). Fragment length was assessed using an Agilent 2100 Bioanalyzer and DNA HS Kit (Cat. # 5067-1504/5067-1511, Agilent). DNA was sheared into 200–250 bp fragments using Covaris LE220 (Woburn, MA, USA) and indexed NGS libraries were prepared by end repairing, A-tailing, adaptor ligation, and amplification procedures using NEBNext® Ultra™ II DNA Prep Kit (Cat. #E7645, NEB). DNA libraries were captured by AmoyDx® Master Panel, which contains 571 genes for DNA mutation (supporting single-nucleotide variation [SNV], insertion/deletion [Indel], Fusion, copy number variation, microsatellite instability and tumor mutation burden [TMB]) detection. Captured products were amplified and quantified by a Quantus fluorometer. Library size was assessed using Agilent 2100 Bioanalyzer. After pooling, libraries were then sequenced on Illumina NovaSeq 6000 instrument (Illumina) with 2 × 150 bp pair-end reads. RNA was fragmented at 95 °C for 0–15 min according to the DV200 value estimated by the Agilent 2100 Bioanalyzer System. RNA fragments then undergo reverse transcription, complementary RNA synthesis, and strand-specific library preparation using NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (Cat. #E7760L, NEB). For RNA-seq, cDNA libraries were generated using a TruSeq RNA Sample Preparation kit (Illumina) according to the manufacturer’s protocol, and sequenced on Illumina NovaSeq 6000 (San Diego, CA, USA) using a panel consisting of 2660 onco-immunology genes (AmoyDx, Xiamen, China). Sequencing data were analyzed and annotated with an in-house developed pipeline. A set of experimental and data quality control parameters were set up.
Variants calling, annotation and filtration
Single nucleotide variants (SNVs) were identified and recorded by SSBC-VarScan (version 1.3.0; AmoyDx, Xiamen, China, inhouse). Insertions/deletions (Indels) were called by Indel Caller (version 0.2.1; AmoyDx, Xiamen, China, inhouse). All variants identified were annotated with Annotator (version 0.3.6; AmoyDx, Xiamen, China, inhouse). Variants classification and criteria for filtering SNVs, Indels and fusion were provided in reference53.
The average depth of sequencing in the coverage area of tumor hotspots and non-hotspots was 1957.4 × and 830.5 ×, respectively. Sequencing data were first cleaned to remove sequencing adaptors and low-quality reads (quality < 15) or poly-N with Trimmomatic (Trimmomatic, RRID:SCR_011848) and mapped to the human reference genome, version 19 (hg19) using the Burrows-Wheeler Aligner. PCR duplicates were marked and removed using Mark Duplicates from the Genome Analysis Toolkit (GATK, RRID:SCR_001876). Base Quality Score Recalibration was performed using GATK’s BaseRecalibrator and ApplyBQSR. After correction, a bam file was written. The Indels and single-nucleotide polymorphisms were compared by Mutect2 and FilterMutectCalls of GATK to obtain the final vcf file. Analysis of variance was used to annotate the vcf files. SNVs and Indels called were further filtered using the following criteria53: (i) minimum ≥ 5 variant supporting reads and ≥ 5% variant allele frequency supporting the variant, (ii) filtered if present in > 2% population frequency in the 1000 g or ExAC or GnomAD (GnomAD, RRID:SCR_014964) database, (iii) filtered if variants not located in CDS region, (iv) filtered if variants were not annotated as (likely/predicted) oncogenic in the OncoKB database. These filtered variants were functional and were used for subsequent data analysis.
Transcriptome annotation and gene quantification
We mapped RNA-seq paired-end reads to the Homo sapiens genome assembly GRCh37 (hg19) using STAR32 (version 020,201) with transcriptome annotation (Genecode version 20) and gene quantification was performed using RSEM 33 (version v1.2.28). Coding region reads were converted to the form of Fragments Per Kilobase Million (FPKM) to measure expressional level of each gene.
Gene set and pathway enrichment analysis
Gene Set Enrichment Analysis (GSEA) was performed by R package clusterProfiler (v 4.2.2), with an adjusted P value < 0.05 as statistically significant. Gene Set Variation Analysis (GSVA) from GSVA (v 1.42.0) R package was used to evaluate enrichment scores of functional terms in GO, KEGG, Hallmark, Reactome, and several immune related signatures in NSCLC Samples.
Consensus clustering
Log2(FPKM + 1) data matrix was first quantile normalized, followed by GSVA enrichment of terms in GO, KEGG, Hallmark, and Reactome from MsigDB database54,55. Consensus clustering was then utilized to identify the different subtypes by R package ConsensusClusterPlus (v 1.58.0) based on the GSVA matrix. We start with 2 consensus clusters until 6 consensus clusters build the cumulative distribution function (CDF). The optimal number of clusters is determined by both CDF and the consensus matrix.
Mutational signatures
Mutational signatures of gene mutation from the gene panel were calculated by SigMA (v 1.6) R package, in which signatures 1 to 30 were obtained from the COSMIC database (https://cancer.sanger.ac.uk/signatures/signatures_v2/), and signatures APOBEC and smoking were clustered based on the COSMIC signatures56. Somatic SNV and small deletion and insertion, were included for calculation.
T-Distributed Stochastic Neighbor Embedding (t-SNE)
Applying the unsupervised nonlinear dimensionality reduction method t-SNE57, how tumors in the discovery cohort were localized to each other was investigated in the dimensionality reduction space based on CNVs that differed significantly among the four subtypes.
Differential gene expression and pathway enrichment
Analysis of differential gene expression (DEG) in each subtype was performed using Limma (v 3.50.0) package, and different pathway enrichment score was analyzed by Wilcoxon test, with false discovery rate (FDR) < 0.05 as threshold58.
In silico Tumor microenvironment characterization & Deconvolution of infiltrated immune cells
Several TME signatures, including 29 TME gene expression signatures59, 28 immune gene sets60, Danaher61, and AO62 were scored by GSVA. MCPcounter63 and xCell score64 was analyzed by R packages MCP counter (v 1.2.0) and xCell (v 1.1.0), respectively.
Classifier construction
Univariate logistic regression was performed for each cluster in the sample set of 126 stage I-III NSCLC patients. The top 10 genes with the highest odds ratio (OR) values and the bottom 10 genes with the lowest OR values were selected for classifier construction. 39 genes were screened out via Lasso-logistic regression and a multivariable logistic regression classifier was constructed. The classifier was further validated internally by 200 iterations of 5-fold cross-validation and leave-one-out cross-validation. The molecular subtype was predicted by maximal likelihood given by this trained classifier model in 64 relapsed/metastatic NSCLC samples.
RT-PCR analysis for METex14 transcripts
RNA was converted to cDNA using M-MLV retro-transcriptase (Thermo Fisher Scientific) and oligo-dT primers, and METex14 was amplified using Hot-Start Taq polymerase (Qiagen) in a 20 μL reaction and visualized in agarose gels. Primers sequences were as follows: forward (exon 13) 5’-TTTTCCTGTGGCTGAAAAAGA-3’ and reverse (exon 15) 5’-GGGGACATGTCTGTCAGAGG-3’. Amplification generated a 246-bp band for wild-type (WT) MET RNA and a 106-bp band for METex14. Positivity for METex14 was determined using a real-time reverse transcription-polymerase chain reaction (RT-PCR) assay (AmoyDx, Xiamen, China) performed at laboratories.
Multiplex immunofluorescence
Multiplex immunofluorescence (mIF) staining was performed to visualize the expression of CD8, GZMK, PD-L1, CD4, CK, CD20 and FOXP3 in tumor tissues. Three consecutive sections (3 μm-thick) were obtained from paraffin blocks. One section was used for H&E staining and the other two sections were used for mIF staining65. The primary antibodies used for each staining were Rabbit monoclonal CD4 (Abcam Cat# ab133616, RRID: AB_2750883), Rabbit monoclonal CD8 alpha (Abcam Cat# ab245118, RRID: AB_3068617), Rabbit monoclonal Granzyme K (Abcam, ab282703), Rabbit monoclonal PD-L1 (Abcam Cat# ab213524, RRID: AB_2857903), Mouse monoclonal Cytokeratin (Long Island Antibody, #349), Rabbit monoclonal CD20 (Abcam Cat# ab78237, RRID: AB_1640323), Mouse monoclonal FOXP3 (Abcam Cat# ab20034, RRID: AB_445284). The concentration and staining order of the antibodies used in this study were optimized beforehand. After all sequential staining steps, nuclei acids were stained with DAPI.
Statistical analysis
Demographic characteristics and mutational landscape of patients were analyzed using descriptive statistics. Chi-square or Fisher’s exact probability tests were performed when the rate or percentage was compared for significance. Nonparametric Wilcoxon rank sum tests were used for the comparison of medians between the two datasets. ANOVA tests were utilized for the comparison among three or more groups. Kaplan–Meier survival analysis was used to evaluate the association between subtypes and progression-free survival. Survival curves were compared by using the log-rank test. A multivariable logistic regression model based on differentiated expressed genes in each subtype was used to build a subtype classifier. k-fold cross-validation and leave-one-out cross-validation were used to determine subtype classifier performance. Univariate and multivariate Cox regression analysis was performed to figure out the potential risk factors. All statistical analyses were performed with the R software (R Project for Statistical Computing, RRID:SCR_001905). P value < 0.05 was considered statistically significant.
Data availability
The raw sequencing data generated in this study have been deposited in the GSA for Human (Genome Sequence Archive for Human in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, https://ngdc.cncb.ac.cn/gsa-human/) under the accession code HRA007761. Any additional information required to reanalyze the data reported in this paper is available from the corresponding authors upon request.
Code availability
This paper does not report the original code.
References
Li, S. et al. Comprehensive characterization of oncogenic drivers in Asian Lung Adenocarcinoma. J. Thorac. Oncol. 11, 2129–2140 (2016).
Lee, G. D. et al. MET Exon 14 skipping mutations in Lung Adenocarcinoma: Clinicopathologic implications and prognostic values. J. Thorac. Oncol. 12, 1233–1246 (2017).
Frampton, G. M. et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 5, 850–859 (2015).
Schrock, A. B. et al. Characterization of 298 patients with lung cancer harboring MET Exon 14 Skipping Alterations. J. Thorac. Oncol. 11, 1493–1502 (2016).
Wolf, J. et al. Capmatinib in MET Exon 14-mutated or MET-amplified non-small-cell lung cancer. N. Engl. J. Med. 383, 944–957 (2020).
Paik, P. K. et al. Tepotinib in non-small-cell lung cancer with MET Exon 14 skipping mutations. N. Engl. J. Med. 383, 931–943 (2020).
Lu, S. et al. Once-daily savolitinib in Chinese patients with pulmonary sarcomatoid carcinomas and other non-small-cell lung cancers harbouring MET exon 14 skipping alterations: a multicentre, single-arm, open-label, phase 2 study. Lancet Respir. Med. 9, 1154–1164 (2021).
Drilon, A. et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat. Med. 26, 47–51 (2020).
Onozato, R. et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J. Thorac. Oncol. 4, 5–11 (2009).
Marks, J. A. et al. Updated molecular analysis of MET exon 14 skipping mutations (METex14) in non-small cell lung cancer (NSCLC). J. Clin. Oncol. 41, 9095–9095 (2023).
Schuler, M. et al. Molecular correlates of response to capmatinib in advanced non-small-cell lung cancer: clinical and biomarker results from a phase I trial. Ann. Oncol. 31, 789–797 (2020).
Fischer, A. et al. Mutational landscape and Expression of PD-L1 in patients with non-small cell lung cancer harboring genomic alterations of the MET gene. Target. Oncol. 17, 683–694 (2022).
Kim, S. Y. et al. Characterization of MET Exon 14 Skipping Alterations (in NSCLC) and identification of potential therapeutic targets using whole transcriptome sequencing. JTO Clin. Res. Rep. 3, 100381 (2022).
Garassino, M. C. et al. Real-world treatment patterns and outcomes of first-line immunotherapy among patients with advanced nonsquamous NSCLC harboring BRAF, MET, or HER2 Alterations. JTO Clin. Res. Rep. 4, 100568 (2023).
Cai, R. et al. To be, or not to be: the dilemma of immunotherapy for non-small cell lung cancer harboring various driver mutations. J. Cancer Res. Clin. Oncol. 149, 10027–10040 (2023).
Akers, K. G., Oskar, S., Zhao, B., Frederickson, A. M. & Arunachalam, A. Clinical outcomes of PD-1/PD-L1 inhibitors among patients with advanced or metastatic non-small cell lung cancer with BRAF, ERBB2/HER2, MET, or RET Alterations: A systematic literature review. J. Immunother., https://doi.org/10.1097/CJI.0000000000000500 (2023).
Gay, C. M. et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 39, 346–360.e347 (2021).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Jiang, Y. Z. et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell 35, 428–440.e425 (2019).
Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540–556.e525 (2017).
Sabari, J. K. et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann. Oncol. 29, 2085–2091 (2018).
Baba, K. et al. Efficacy of pembrolizumab for patients with both high PD-L1 expression and an MET exon 14 skipping mutation: A case report. Thorac. Cancer 10, 369–372 (2019).
Reis, H. et al. MET expression in advanced non-small-cell lung cancer: effect on clinical outcomes of chemotherapy, targeted therapy, and immunotherapy. Clin. Lung cancer 19, e441–e463 (2018).
Mooi, J. K. et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: molecular analysis of the AGITG MAX clinical trial. Ann. Oncol. 29, 2240–2246 (2018).
Liu, Y. et al. Subtyping-based platform guides precision medicine for heavily pretreated metastatic triple-negative breast cancer: The FUTURE phase II umbrella clinical trial. Cell Res. 33, 389–402 (2023).
Fan, L. et al. Optimising first-line subtyping-based therapy in triple-negative breast cancer (FUTURE-SUPER): a multi-cohort, randomised, phase 2 trial. Lancet Oncol. 25, 184–197 (2024).
Yu, Y. et al. Gumarontinib in patients with non-small-cell lung cancer harbouring MET exon 14 skipping mutations: a multicentre, single-arm, open-label, phase 1b/2 trial. EClinicalMedicine 59, 101952 (2023).
Dominguez, C. X. et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15(+) Myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 10, 232–253 (2020).
Tang, H. et al. Integrated microenvironment-associated genomic profiles identify LRRC15 mediating recurrent glioblastoma-associated macrophages infiltration. J. Cell. Mol. Med. 25, 5534–5546 (2021).
Xu, Z. et al. Incidence and PD-L1 Expression of MET 14 Skipping in Chinese population: a non-selective NSCLC cohort study using RNA-based sequencing. Onco Targets Ther. 13, 6245–6253 (2020).
Zhang, M., Liu, K. & Wang, M. Development of cancer immunotherapy based on PD-1/PD-L1 pathway blockade. RSC Adv. 9, 33903–33911 (2019).
Mayenga, M. et al. Durable responses to immunotherapy of non-small cell lung cancers harboring MET exon-14-skipping mutation: A series of 6 cases. Lung cancer 150, 21–25 (2020).
Marek, L. et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol. Pharm. 75, 196–207 (2009).
Semrad, T. J. & Mack, P. C. Fibroblast growth factor signaling in non-small-cell lung cancer. Clin. Lung Cancer 13, 90–95 (2012).
Hu, H. et al. Three subtypes of lung cancer fibroblasts define distinct therapeutic paradigms. Cancer Cell 39, 1531–1547 e1510 (2021).
Svaton, M. et al. Aggressive pulmonary adenocarcinoma with new FGFR translocation and cMET mutation not responsive to crizotinib and nintedanib treatment: a case report. Transl. Cancer Res. 9, 2082–2088 (2020).
Kono, S. A., Marshall, M. E., Ware, K. E. & Heasley, L. E. The fibroblast growth factor receptor signaling pathway as a mediator of intrinsic resistance to EGFR-specific tyrosine kinase inhibitors in non-small cell lung cancer. Drug Resist. Update 12, 95–102 (2009).
Ellis, L. M. & Hicklin, D. J. Pathways mediating resistance to vascular endothelial growth factor-targeted therapy. Clin. Cancer Res. 14, 6371–6375 (2008).
Sanchez-Guixe, M. et al. High FGFR1-4 mRNA expression levels correlate with response to selective FGFR inhibitors in breast cancer. Clin. Cancer Res. 28, 137–149 (2022).
Santarpia, M. et al. A narrative review of MET inhibitors in non-small cell lung cancer with MET exon 14 skipping mutations. Transl. Lung Cancer Res. 10, 1536–1556 (2021).
Jamme, P. et al. Alterations in the PI3K pathway drive resistance to MET inhibitors in NSCLC harboring MET Exon 14 skipping mutations. J. Thorac. Oncol. 15, 741–751 (2020).
Rotow, J. K. et al. Co-occurring alterations in the RAS-MAPK pathway limit response to MET inhibitor treatment in MET Exon 14 skipping mutation-positive lung cancer. Clin. Cancer Res 26, 439–449 (2020).
Lu, D. et al. MET Exon 14 splice-site mutations preferentially activate KRAS signaling to drive Tumourigenesis. Cancers 14, https://doi.org/10.3390/cancers14061378 (2022).
Liang, Q. et al. MET exon 14 skipping mutation drives cancer progression and recurrence via activation of SMAD2 signalling. Br. J. Cancer, https://doi.org/10.1038/s41416-023-02495-5 (2023).
Black, J. R. M. & McGranahan, N. Genetic and non-genetic clonal diversity in cancer evolution. Nat. Rev. Cancer 21, 379–392 (2021).
Sanchez-Vega, F. et al. Oncogenic signaling pathways in the cancer genome Atlas. Cell 173, 321–337.e310 (2018).
Chen, H. et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat. Commun. 10, 5472 (2019).
Paterson, A. L. et al. Characterization of the timing and prevalence of receptor tyrosine kinase expression changes in oesophageal carcinogenesis. J. Pathol. 230, 118–128 (2013).
Silva, A. L. et al. Boosting Wnt activity during colorectal cancer progression through selective hypermethylation of Wnt signaling antagonists. BMC Cancer 14, 891 (2014).
Lu, S. et al. Long-term efficacy, safety, and subgroup analysis of Savolitinib in Chinese patients With NSCLCs harboring MET Exon 14 skipping alterations. JTO Clin. Res. Rep. 3, 100407 (2022).
Liu, L. et al. Construction of TME and Identification of crosstalk between malignant cells and macrophages by SPP1 in hepatocellular carcinoma. Cancer Immunol. Immunother. : CII 71, 121–136 (2022).
Bao, X. et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J. Immunother. Cancer 8, https://doi.org/10.1136/jitc-2020-001437 (2020).
Han, X. et al. Short-term dynamics of circulating tumor DNA predicting efficacy of sintilimab plus docetaxel in second-line treatment of advanced NSCLC: biomarker analysis from a single-arm, phase 2 trial. J. Immunother. Cancer 10, https://doi.org/10.1136/jitc-2022-004952 (2022).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Gulhan, D. C., Lee, J. J., Melloni, G. E. M., Cortes-Ciriano, I. & Park, P. J. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat. Genet 51, 912–919 (2019).
Li, W., Cerise, J. E., Yang, Y. & Han, H. Application of t-SNE to human genetic data. J. Bioinform. Comput. Biol. 15, 1750017 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Bagaev, A. et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 39, 845–865 e847 (2021).
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013).
Danaher, P. et al. Gene expression markers of Tumor Infiltrating Leukocytes. J. Immunother. Cancer 5, 18 (2017).
Bareche, Y. et al. Leveraging big data of immune checkpoint blockade response identifies novel potential targets. Ann. Oncol. 33, 1304–1317 (2022).
Becht, E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218 (2016).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017).
Yagi, Y. et al. Three-dimensional histologic, immunohistochemical, and multiplex immunofluorescence analyses of dynamic vessel co-option of spread through air spaces in lung Adenocarcinoma. J. Thorac. Oncol. 15, 589–600 (2020).
Acknowledgements
This work was supported by National Natural Science Foundation of China No. 82003154 (to C.X.) and No. 82372669 (to Y.C.H.), and Shanghai Municipal Health Commission Intelligent Medical Special Project 2018ZHYL0213 (to Y.C.H.). We thank the members of Qian Li, Yanhua Chen, and Tianlin Sun for the discussion and helpful comments on the article.
Author information
Authors and Affiliations
Contributions
S.N.C. and T.H. were involved in data curation, visualization, writing, review, and editing. J.K.Z. and Q.Z. were involved in data generation curation and analysis. J.W. contributed to data curation, analysis, and interpretation. Z.H. contributed to manuscript writing and editing. C.X. contributed to Funding and supervision. R.Y.Z. was involved in data analysis and manuscript editing. C.B.Z. and S.L. were involved in conceptualization, data analysis, manuscript writing, and editing. Y.C.H. contributed to conceptualization, data analysis, funding and supervision, and manuscript editing. All authors approved the article for submission and publication.
Corresponding authors
Ethics declarations
Competing interests
T.H., J.W., Z.H., and C.B.Z. were employed by Amoy Diagnostics Co., Ltd. None of the other authors have any conflicts of interest to declare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, S., Hu, T., Zhao, J. et al. Novel molecular subtypes of METex14 non-small cell lung cancer with distinct biological and clinical significance. npj Precis. Onc. 8, 159 (2024). https://doi.org/10.1038/s41698-024-00642-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-024-00642-6