Abstract
Steroid-induced osteonecrosis of the femoral head (SONFH) is a debilitating condition caused by long-term corticosteroid use, leading to impaired blood flow and bone cell death. The disruption of cellular processes and promotion of apoptosis by endoplasmic reticulum stress (ERS) is implicated in the pathogenesis of SONFH. We identified ERS-associated genes in SONFH and investigated their potential as therapeutic targets. We analysed the GSE123568 GEO dataset to identify differentially expressed genes (DEGs) related to ERS in SONFH. We conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, identified hub genes by protein-protein interaction (PPI) analyses, and evaluated their functions by gene set enrichment analysis (GSEA). We constructed mRNA–miRNA networks, identified potential therapeutics, and assessed immune cell infiltration. We performed cross-validation using the GEO dataset GSE74089, qRT-PCR on clinical samples from patients with SONFH and controls, and a receiver operating characteristic (ROC) curve analysis to assess the diagnostic performance of the hub genes. We identified 195 ERS-related genes in SONFH, which were primarily involved in oxidative stress, immune responses, and metabolic pathways. The PPI network suggested CXCL8, STAT3, IL1B, TLR4, PTGS2, TLR2, CASP1, CYBB, CAT, and HOMX1 to be key hub genes, which were shown by GSEA to be involved in biological pathways related to metabolism, immune modulation, and cellular integrity. We also identified 261 microRNAs (miRNAs) as well as drugs such as dibenziodolium and N-acetyl-L-cysteine that modulated inflammatory responses in SONFH. Twenty-two immune cell subtypes showed significant correlations, such as a positive correlation between activated mast cells and Tregs, and patients with SONFH had fewer dendritic cells than controls. The hub genes CYBB and TLR4 showed significant correlations with M1 macrophages and CD8 T cells, respectively. Cross-validation and qRT-PCR confirmed the upregulation of STAT3, IL1B, TLR2, and CASP1 in patients with SONFH, validating the bioinformatics findings. An ROC curve analysis confirmed the diagnostic potential of the hub genes. The top 10 hub genes show promise as ERS-related diagnostic biomarkers for SONFH. We discovered that 261 miRNAs, including hsa-miR-23, influence these genes and identified potential therapeutics such as dibenziodolium and simvastatin. Immune profiling indicated altered immune functions in SONFH, with significant correlations among immune cell types. Validation confirmed the upregulation of STAT3, IL1B, TLR2 and CASP1, which had diagnostic potential. The findings suggest potential diagnostic markers and therapeutic targets for SONFH.
Similar content being viewed by others
Introduction
Osteonecrosis of the femoral head (ONFH) is a debilitating condition characterised by ischaemia and cellular death in the femoral head, leading to structural and functional changes in the bone1,2,3. The aetiology of OFNH can be traumatic or non-traumatic4. Hormone use, chronic alcohol consumption, and a history of trauma are implicated as triggers of ONFH5. Steroid use is the most prevalent non-traumatic cause of ONFH6.
Steroid-induced ONFH (SONFH) occurs predominantly as a result of excessive use of corticosteroids in patients with immune-related diseases7,8. The effects of glucocorticoids (GCs) are important in SONFH. GCs can induce lipid metabolism disorders, leading to intraosseous hypertension, impairing the blood supply to the femoral head and initiating necrosis9. In addition, GCs can directly induce apoptosis of osteoblasts and osteocytes, which are responsible for bone formation and maintenance, thus accelerating bone loss and necrosis10. Moreover, GCs influence autophagy, which is essential for maintaining cellular homeostasis, by altering the balance between autophagy activation and inhibition11. This may contribute to the impairment of osteocyte function and increase susceptibility to ONFH. Furthermore, GCs reportedly regulate ferroptosis, a type of cell death driven by iron-dependent lipid peroxidation. Excess GC exposure promotes ferroptosis, which may aggravate bone loss and contribute to the progression of SONFH12. Critically, GCs activate endoplasmic reticulum stress (ERS), exacerbating the disruption of osteogenesis and angiogenesis, which are key factors in disease development13.
ERS occurs when the ER (the site of protein folding, lipid synthesis, and calcium storage) is overwhelmed by unfolded or misfolded proteins, triggering the unfolded protein response (UPR)13. The UPR is mediated by three key pathways: IRE1, which enhances the ability of the ER to manage misfolded proteins by splicing XBP1 mRNA; ATF6, which upon ER stress travels to the Golgi apparatus to be cleaved and induce the expression of UPR target genes; and PERK, which phosphorylates eIF2α to reduce protein synthesis and lessen the ER load, and activates stress response genes14,15. These pathways restore ER function by reducing protein misfolding, enhancing protein folding, and degrading misfolded proteins. If this balance is not restored, prolonged ERS can lead to apoptosis, exacerbating bone cell dysfunction and accelerating the progression of SONFH16.
In this study, we used bioinformatics methods to identify differentially expressed genes (DEGs) associated with ERS in SONFH. We analysed the roles of key hub genes, investigated their interactions with regulatory microRNAs (miRNAs), and identified therapeutics that could target these genes. In addition, we examined immune cell infiltration to understand the immunological landscape of SONFH. We validated the findings using external datasets and clinical samples. Our results will facilitate the development of targeted therapeutics for SONFH.
Materials and methods
Bioinformatics
Data collection
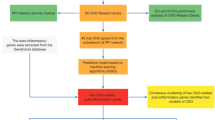
We downloaded the GSE123568 dataset from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), and compared gene expression levels in 30 patients with SONFH and 10 non-affected controls post-steroid treatment17. Next, we extracted genes associated with ERS from the GeneCards database (https://www.genecards.org)18. Using a relevance threshold score of 10, we identified 2176 ERS-related genes (ERGs).
Analysis of differentially expressed genes
We used DESeq2 in R to identify genes in the GSE123568 dataset with significant changes in expression, defined as an absolute fold-change > 0.75 and an adjusted p < 0.0519; these were termed DEGs. The DEGs intersecting with 2176 ERGs were classified as differentially expressed ERS-related genes (DE-ERGs).
Functional enrichment analysis
We conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to evaluate the functions of the DE-ERGs20,21,22. GO analysis categorises DE-ERGs into Biological Processes (BP), Cellular Components (CC), and Molecular Functions (MF). KEGG analysis maps DE-ERGs to broader biological pathways. We used the clusterProfiler package to identify significant enrichments (adjusted p < 0.05 and Q < 0.2) and visualised the results with ggplot223.
Screening of hub genes
We used the STRING database (https://string-db.org/) to analyse protein–protein interactions (PPIs) related to DE-ERGs24. The dataset was imported into Cytoscape, and we used the cytoHubba plugin to identify hub genes with crucial connections in the ERS PPI network25. We used the MCODE plugin in Cytoscape to identify significant clusters and refined the focus to the top 10 hub genes.
Single-gene gene set enrichment analysis
We conducted a gene set enrichment analysis (GSEA) to evaluate the associations between the hub genes with biological pathways or processes. Using the Molecular Signatures Database (MSigDB) (https://www.gseamsigdb.org/gsea/msigdb/index.jsp), we linked the hub genes to biological functions26. This approach is underpinned by stringent criteria (adjusted p < 0.05 and false discovery rate [FDR] < 0.25). We used the ggplot2 package in R to visualise the contributions of the hub genes to ERS and other stress-response mechanisms.
Construction of the mRNA–miRNA network
We identified miRNAs that may regulate the top 10 genes implicated in ERS using miRWalk (http://mirwalk.umm.uni-heidelberg.de/)27. By visualising these interactions in Cytoscape, we constructed an mRNA–miRNA regulatory network.
Targeted drug prediction
We used Enrichr (https://maayanlab.cloud/Enrichr) to identify drugs linked to key genes in ERS28,29. We input the top 10 hub genes into Enrichr, applied the Drug Signatures Database (DSigDB), and prioritised the results by p-values to identify the top 10 drugs associated with the hub genes.
Analysis of immune cell infiltration
We used the CIBERSORT algorithm (https://cibersortx.stanford.edu/) to analyse immune cell infiltration30. Using a permutation count of 100 and a p < 0.05, we evaluated the populations of immune cell types. We used the vioplot package to visualise the abundances of the immune cell types. The corrplot package was employed to generate correlation matrices to provide insight into the relationships between hub genes and immune cell types.
Cross-validation using an external dataset
To validate the findings from the GSE123568 dataset, we performed cross-validation31 of DE-ERG expression levels in the GSE74089 dataset. This dataset includes gene expression profiles of hip cartilage from patients with necrosis of the femoral head. By comparing the expression levels of the DE-ERGs in GSE74089 and GSE123568, we validated the identified hub genes. A value of p < 0.05 was considered indicative of statistical significance.
Validation of clinical samples
Clinical samples
This study complied with the Declaration of Helsinki and was approved by the Medical Ethics Committee of Qingdao University Affiliated Hospital. All of the participants provided informed consent. The inclusion criteria were adults 18–60 years of age diagnosed via radiographic evidence with femoral neck fractures or SONFH. The exclusion criteria were previous conditions that could influence immune response or gene expression, such as systemic infections or autoimmune diseases; immunosuppressive therapy within 6 months; or a history of metabolic bone diseases unrelated to our condition of interest, e.g., osteoporosis. We collected peripheral blood samples from three patients with SONFH and three healthy adults (following steroid administration).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Peripheral blood was collected into EDTA-treated tubes and centrifuged to segregate the cellular fraction from plasma. White blood cells were lysed using TRIzol reagent, releasing RNA and denaturing proteins and DNA. RNA was isolated from the aqueous phase, precipitated using alcohol, and the pellet was washed with ethanol to remove impurities. After resuspension in RNase-free water, RNA purity and concentration were determined. We used the qPCR HiScript III RT SuperMix Kit to convert RNA into cDNA according to the manufacturer’s protocol. cDNA was used for qRT-PCR with the ChamQ Universal SYBR qPCR Master Mix Kit, with ACTB mRNA as the normalisation control. Each sample underwent qRT-PCR in triplicate, together with three biological replicates per group. The primer sequences are listed in Table 1.
Validation of hub genes by receiver operating characteristic analysis
Using the R package pROC, we conducted ROC curve analyses on the GSE123568 dataset to evaluate the diagnostic accuracy of hub genes for SONFH32. We assessed model performance by calculating areas under the curve (AUCs); AUCs > 0.6 indicated statistical significance.
Results
DE-ERGs in SONFH
We identified 1220 DEGs, including 754 upregulated and 466 downregulated genes, in the GSE123568 dataset (Fig. 1A). By intersecting these DEGs with ERS-related genes scoring > 10 in the GeneCards database, we identified 195 DE-ERGs (Fig. 1B). The expression levels of the top 100 intersecting genes of the SONFH and control groups are shown in Fig. 1C.
Analysis and identification of differentially expressed endoplasmic reticulum stress-related genes (DE-ERGs) in SONFH and control samples. (A) Volcano map of the distribution of the 1220 DEGs in the GSE123568 dataset. Red, upregulated DEGs; blue, downregulated DEGs. (B) Venn diagram of 195 DE-ERGs. (C) Heatmap of changes in expression of the top 100 DE-ERGs. Blue, downregulation; red, upregulation.
GO and KEGG enrichment analyses of DE-ERGs
The GO and KEGG analyses identified 1987 GO terms and 117 KEGG pathways related to SONFH. The GO analysis yielded 1682 BPs, 137 CCs, and 158 MFs (Fig. 2), including BPs such as response to oxidative stress (OS), peptide-hormone signalling, and regulation of defence responses. CCs included structural features such as membrane microdomains and mitochondrial outer membranes. MFs were dominated by kinase activity and binding mechanisms. KEGG pathway analysis linked these DE-ERGs to the lipid and atherosclerosis, NOD-like receptor signalling, and FoxO signalling pathways.
Construction of the PPI network and identification of hub genes
We constructed a PPI network for the 195 DE-ERGs using the STRING database with visualisation in Cytoscape (Fig. 3). The MCODE plugin revealed three critical gene clusters. Cluster 1 included genes involved in inflammation and immune response, Cluster 2 contained genes linked to OS and innate immunity, and Cluster 3 included lipid metabolism enzymes. The cytoHubba plugin identified CXCL8, STAT3, IL1B, TLR4, PTGS2, TLR2, CASP1, CYBB, CAT, and HMOX1 as hub genes. Nine genes were upregulated, and one gene (CAT) was downregulated.
In the upregulated gene network, coexpression interactions predominated at 66.92%, followed by colocalisation at 18.35%, and physical interactions at 15.43%. Predicted interactions were noted at 4.46%, with pathway involvement at 4.14%, genetic interactions at 0.46%, and shared protein domains at 0.23%. The network of the downregulated gene, CAT, was characterised by physical interactions, which constituted 77.64% of the connections. Coexpression accounted for 8.1%, with predicted interactions at 5.37%, colocalisation at 3.63%, genetic interactions at 2.87%, pathway involvement at 1.88%, and shared protein domains at 0.60%.
Potential mechanisms of hub genes explored by GSEA
We performed GSEA for each of the hub genes (Fig. 4). CXCL8 showed enrichment in Thyroid Cancer and Galactose Metabolism, and negative enrichment in SNARE Interactions in Vesicular Transport and Proteasome. STAT3 showed negative enrichment in Epithelial Cell Signalling in Helicobacter pylori Infection and Prostate Cancer. IL1B showed positive enrichment in Insulin Signalling Pathway and NOD-like Receptor Signalling Pathway, and TLR4 exhibited negative enrichment in T Cell Receptor Signalling Pathway and ERBB Signalling Pathway. GSEA of PTGS2 indicated negative enrichment in, for instance, NOD-like Receptor Signalling Pathway and Apoptosis. TLR2 was negatively enriched in Vibrio cholerae Infection and Toll-like Receptor Signalling Pathway. CASP1 showed a mix of positive enrichment in pathways such as Pentose and Glucuronate Interconversions and negative enrichment in Lysosome and SNARE Interactions in Vesicular Transport. GSEA of CYBB revealed negative enrichment in Autoimmune Thyroid Disease and Lysosome. CAT was positively enriched in Valine, Leucine, and Isoleucine Degradation and Glyoxylate and Dicarboxylate Metabolism. HMOX1 exhibited negative enrichment in Selenoamino Acid Metabolism and Glycerolipid Metabolism.
Hub gene-miRNA regulatory network and drugs
The mRNA–miRNA network showed that 261 miRNAs modulated the expression of the top 10 hub genes associated with ERS (Fig. 5). These included hsa-miR-23, which is reportedly correlated with osteogenic and adipogenic functions33,34,35,36. In addition, dibenziodolium and N-acetyl-L-cysteine, together with simvastatin and curcumin (which modulated inflammatory responses), potentially interacted with the hub genes.
Immune infiltration and hub gene-immune cell interactions
Immune infiltration analysis using the GSE123568 dataset suggested the presence of diverse immune-cell subpopulations. The relative percentages of 22 immune-cell subtypes are shown in Fig. 6. The correlation analysis indicated significant interactions between immune-cell types. For instance, activated mast cells were strongly positively correlated with Tregs and resting dendritic cells and resting NK cells were negatively correlated with gamma-delta T cells.
Immune infiltration and hub gene-immune cell interactions. (A) Relative percentages of 22 subpopulations of immune cells in 40 samples from the GSE123568 dataset. (B) Spearman correlation analysis among immune cells. Red, positive correlation; blue, negative correlation. (C) Boxplots of the expression of immune cells between the two groups. (D) Correlation map of hub genes and immune infiltration. Yellow, positive correlation; blue, negative correlation. The darker the colour the stronger the correlation (*p < 0.05).
Immune-landscape analysis showed that the proportion of dendritic cells was significantly smaller in the SONFH group compared to the control group, suggesting suppression or deficit in antigen presentation or immune priming in SONFH.
We analysed the correlation between the hub genes identified in the PPI network and immune cell infiltration. The gene CYBB showed a significant positive correlation with M1 macrophages and a negative correlation with activated NK cells, indicating an inverse relationship with adaptive immune responses. TLR4 showed a significant negative correlation with CD8 T cells.
Validation of hub genes in the GSE123568 dataset
The cross-validation results confirm that the expression levels of the 10 hub genes in the GSE74089 dataset align with those in the GSE123568 dataset. Relative to the control group, the SONFH group exhibited upregulation of CXCL8, STAT3, IL1B, TLR4, PTGS2, TLR2, CASP1, CYBB, and HMOX1, and downregulation of CAT (Fig. 7). qRT-PCR analysis of samples from three patients with SONFH and three healthy adults confirmed these findings at the mRNA level, reinforcing the reproducibility of the GSE123568 data. Compared to the control group, there was significant upregulation of STAT3 (p = 0.0002), IL1B (p = 0.0396), TLR2 (p = 0.0054), and CASP1 (p = 0.0264) in the SONFH group, which agrees with the bioinformatics analysis (Fig. 8). To validate the predictive significance of the 10 hub genes, we performed an ROC curve analysis on the GSE123568 dataset. All of the hub genes had statistically significant AUC values > 0.6 (Fig. 9). PTGS2 (AUC = 0.805) showed the greatest diagnostic efficacy, followed by STAT3 (AUC = 0.815).
Discussion
Our findings provide insight into the molecular framework of SONFH by identifying and characterising DE-ERGs. Analysis of the GSE123568 dataset revealed a large number of DE-ERGs, thereby enhancing our understanding of the cellular mechanisms triggered by steroid exposure. The study that originally generated the GSE123568 dataset focused on identifying dynamic biomarkers for different stages of non-traumatic ONFH (NONFH) using a transcriptional regulatory network17. Specifically, it highlighted the alterations in gene expression across various stages of the disease and identified key regulatory genes and pathways involved in NONFH progression. However, our study specifically investigates the role of ERS in SONFH, which was not addressed in the original study. Our research fills this gap by specifically investigating the intersection of ERS-related genes with the DEGs identified from the GSE123568 dataset. The identification of 1220 DEGs (754 upregulated and 466 downregulated) highlights the effects of steroids on gene expression in SONFH. Among these, the intersection with ERS-related genes resulting in 195 DE-ERGs suggests a significant subset of genes that may play crucial roles in the pathophysiology of SONFH through ERS mechanisms. This focused analysis provides a more detailed understanding of how ERS contributes to the cellular and molecular disruptions observed in SONFH, thereby offering new insights that were not explored in the original study.
GO and KEGG enrichment analyses provided insight into the BP and pathways enriched for these DE-ERGs. The significant enrichment in BP (e.g., response to OS, peptide hormone signalling, and regulation of defence responses) aligns with pathological features of SONFH, including OS and inflammatory responses. GC-induced OS promotes apoptosis of osteocytes, primarily by increasing NOX-mediated production of ROS37. Similarly, the exacerbation of inflammatory processes by BMP-2–mediated downregulation of IL-34, contributes to osteoclast differentiation and the progression of ONFH38. Moreover, the CC enrichment in membrane microdomains and mitochondrial outer membranes suggests changes in cellular architecture that could influence signal transduction and metabolic processes crucial for cell survival and function. KEGG pathway analysis mapped DE-ERGs to pathways implicated in SONFH, such as lipid metabolism and atherosclerosis. ERS is important in this context because it can be exacerbated by intracellular lipid imbalances39. Dysfunctions in lipid metabolism can lead to the accumulation of misfolded proteins in the ER, triggering stress responses and ultimately apoptosis and bone-tissue degradation40. Moreover, the atherosclerotic processes identified by the pathway analysis indicate that lipid accumulation affects cellular health not only directly but also indirectly by impairing blood flow, thereby exacerbating the hypoxia that leads to osteonecrosis. In addition, identification of the NOD-like receptor signalling and FoxO signalling pathways underscores the roles of DE-ERGs in modulating immune responses and apoptosis41. This emphasises their potential as therapeutic targets and highlights the complex interplay between metabolic dysfunction and immune-regulatory mechanisms in SONFH.
The PPI network analysis provided insight into the molecular interactions and pathways linked to the pathophysiology of SONFH. The critical clusters in the PPI network, which were related to inflammation, immune response, OS, and lipid metabolism, indicates steroids affect the activities and interactions of these pathways in SONFH. The cytoHubba plugin identified CXCL8, STAT3, IL1B, TLR4, PTGS2, TLR2, CASP1, CYBB, CAT, and HMOX1 as hub genes. These genes regulate cytokine signalling, immune responses, and stress mechanisms, which are important for maintaining cellular homeostasis and responding to external stressors including steroids. Their central positions in the network implicate them in both normal cellular functions and in pathological conditions, potentially driving the processes that lead to bone degradation in SONFH. GSEA of hub genes highlighted their effects on metabolic regulation, immune modulation, and cellular integrity in SONFH. These genes are multifunctional: CXCL8 affects metabolic and stress management processes, STAT3 influences cellular growth and response to infection, and IL1B integrates metabolic and inflammatory pathways, which are key in the pathogenesis of SONFH. TLR4 and PTGS2 modulate immune responses and apoptosis, suggesting that they have potential as therapeutic targets to reduce osteocyte damage. The involvement of CASP1 in metabolic processing and cellular clean-up, together with the functions of CYBB and TLR2 in immune homeostasis, highlight the complex interplay of responses to steroid stress. The functions of CAT and HMOX1 in metabolic pathways suggest strategies to enhance cellular resilience against SONFH.
The regulation by miRNAs of the ERS-induced unfolded protein response suggests that their effects on pathways that govern cell survival or death influence cellular-stress outcomes42. Indeed, the fact that the mRNA-miRNA network centred on hub genes intersecting with ERS revealed the importance of miRNAs in SONFH. Among them, miR-23a modulates pathways critical for bone health. miR-23a inhibits osteogenesis by targeting LRP5, which is crucial for bone homeostasis, and icariin stimulates bone formation by modulating this pathway, suggesting therapeutic potential34,35. In addition, miR-23a modulates apoptosis and differentiation via LINC00473 in a PLGA hydrogel model, suggesting its importance for bone health36. Ganesan et al. found that downregulation of miR-23a led to upregulation of TLR2, enhancing the activity of autophagy-related pathways43. Lingam et al. reported that CXCL8 and miR-23a respond to inflammation and hypoxia44. Our predictions suggest that targeting miR-23a could improve SONFH outcomes by modulating TLR2 and CXCL8. These hub genes, which are significantly modulated by miRNAs, provide insight into the complex molecular mechanisms of SONFH and are potential targets for miRNA-based therapies.
Next, we identified potential therapeutic agents that could modulate these hub genes. Several drugs, including dibenziodolium and N-acetyl-L-cysteine, which have anti-inflammatory and antioxidant activity, respectively, potentially interacted with key hub genes. Dibenziodolium reduces inflammation-driven exacerbation, and N-acetyl-L-cysteine alleviates OS and inflammation, which are important in the pathogenesis of SONFH37. Similarly, simvastatin shows therapeutic promise for SONFH because of its anti-inflammatory activity and inhibition of the GC receptor45. Curcumin inhibits M1-type macrophage infiltration and osteocyte apoptosis in the femoral head, which are linked to the development of SONFH46. In combination, these drugs show greater therapeutic efficacy for SONFH, and may improve patient outcomes by modulating the expression of hub genes.
The significant correlations between immune cells and hub genes suggest immune modulation to be a pivotal component of therapeutic strategies. The positive correlation between activated mast cells, Tregs, and resting dendritic cells suggests that a protective regulatory mechanism is disrupted in SONFH. Therapeutic strategies that enhance Treg function or stabilise mast-cell activity could mitigate inflammatory processes in SONFH. Enhancing the function of Tregs to suppress inflammatory responses and pro-inflammatory cells (such as Th17) can restore the immune balance and reduce tissue damage, thereby promoting the repair and regeneration of bone tissue47. Conversely, the reduction in dendritic cells suggests impairment of immune priming, indicating that therapies boosting dendritic cell function could restore effective immune responses. He et al. showed that resting dendritic cells reduce osteonecrosis inflammation and promote bone repair by modulating antigen presentation and cytokine signalling, particularly by inhibiting the IL-17 pathway48.
Macrophage polarisation into the pro-inflammatory M1 or anti-inflammatory M2 phenotype in response to environmental cues is pivotal for bone homeostasis, and its dysregulation can lead to inflammatory responses that disrupt bone metabolism49. Furthermore, activated NK cells and CD8+ T cells are components of the cell-mediated cytotoxic immune response, and are important for clearing damaged cells. However, their overactivation may lead to detrimental effects on bone tissue, indicating the need for a balanced immune response to maintain skeletal integrity and prevent exacerbation of osteonecrosis50,51. The positive correlations of TLR4, CYBB, and CASP1 with M1 macrophages suggest a pro-inflammatory phenotype that could exacerbate bone loss and turnover. Targeting these genes to modulate M1 macrophage activity may attenuate this pro-inflammatory state and induce a shift towards bone preservation and repair. Conversely, the negative correlations of TLR4, TLR2, IL1B, HMOX1, CYBB, and CASP1 with activated NK cells suggest a protective mechanism against the cytotoxicity typically induced by these immune cells. Similarly, the negative correlations of these hub genes, together with STAT3 and PTGS2, with CD8 T cells suggest a dampening of cytotoxic T-cell activity, which could reduce bone tissue damage in SONFH. Therefore, adjusting hub gene activity could correct the immune imbalance in SONFH, reducing pro-inflammatory effects and fortifying defences against overactive immune responses, a promising direction for targeted therapy development.
Finally, we validated the expression levels of 10 hub genes using the GSE74089 dataset, confirming the accuracy of our findings from GSE123568. To further substantiate these findings and directly link them to our study’s focus on ERS-related genes in SONFH, we conducted qRT-PCR analysis on peripheral blood samples collected from three patients with SONFH and three healthy adults following steroid administration. This experimental design aimed to measure the expression levels of these hub genes in a clinical context, providing a more accurate reflection of their roles in SONFH and their association with ERS.
The qRT-PCR process involved several critical steps: isolating total RNA from the blood samples, converting it into cDNA using reverse transcription, and then amplifying specific gene targets using quantitative PCR to measure their expression levels. This rigorous approach ensured that our results were both reliable and relevant to the pathophysiological conditions of SONFH, particularly concerning ERS. The qRT-PCR analysis yielded consistent results for four hub genes: STAT3, IL1B, TLR2, and CASP1. These genes were identified as significantly altered in SONFH patients compared to healthy controls, underscoring their potential as biomarkers and therapeutic targets. Researches have reported that, STAT3 exacerbates SONFH by promoting osteoclast differentiation52, IL1B polymorphisms affect genetic susceptibility53, TLR2 enhances BMSC-mediated angiogenesis and osteogenesis54, and CASP1 affects osteogenic functions via the ROS/NLRP3 pathway55. In an ROC curve analysis, the 10 genes had AUC values > 0.6, underscoring their diagnostic potential. This indicates that the other hub genes might still be involved in the pathophysiology of SONFH, even though their qRT-PCR results were not consistent.
This validation step, by bridging bioinformatics predictions with clinical sample analysis, strengthens our findings and confirms the relevance of these hub genes in the pathophysiology of SONFH, specifically in the context of ERS. Thus, our qRT-PCR results not only support the bioinformatic data but also provide a tangible link to clinical outcomes and the role of ERS in SONFH, making them crucial for developing targeted therapies for this condition.
Strengths and limitations
This study provides valuable insights into the molecular framework of SONFH by identifying and characterizing DE-ERGs. Our comprehensive analysis of the GSE123568 dataset revealed significant gene expression changes associated with steroid exposure, offering a robust foundation for understanding the cellular mechanisms involved in SONFH. The use of GO and KEGG enrichment analyses, PPI network analysis, and validation with the GSE74089 dataset further strengthens the reliability of our findings.
However, the study has limitations. Firstly, the lack of randomization may introduce selection bias and affect the internal validity of our findings. Future studies should include randomized controlled trials to minimize bias and enhance the robustness of the results. Additionally, the generalizability of our findings is constrained by the specific characteristics of our study population, which was drawn from a single institution. This may not fully represent the broader population of patients with SONFH. Future studies should include more diverse and larger sample sizes from multiple centers to improve the applicability of the findings. The small sample size for qRT-PCR analysis may also limit the generalizability of the results. Future research should include in vitro validation of the predicted miRNA regulatory mechanisms and the therapeutic potential of the identified drugs to gain deeper insights into their clinical implications.
Conclusion
Using the GSE123568 dataset, we identified 195 DE-ERGs in SONFH, which were related to oxidative stress, immune responses, and metabolic pathways. GO and KEGG analyses, and the PPI network, identified CXCL8, STAT3, IL1B, TLR4, PTGS2, TLR2, CASP1, CYBB, CAT, and HMOX1 as key hub genes, which regulate metabolic, immune, and cellular integrity processes. The identification of 261 miRNAs, particularly hsa-miR-23, emphasised the complexity of the regulation of genes that modulate bone metabolism. Moreover, we found that dibenziodolium, N-acetyl-L-cysteine, simvastatin, and curcumin target inflammatory pathways important in the pathophysiology of SONFH. Twenty-two immune cell subtypes showed significant interactions, notably activated mast cells and Tregs, and there was a reduction in the population of dendritic cells, indicating altered immune function in SONFH. In addition, the positive and negative correlations of hub genes with immune cell types underscore their potential as therapeutic targets. Validation by cross-dataset comparison, qRT-PCR and ROC analysis confirmed the upregulation of STAT3, IL1B, TLR2, and CASP1. Further validation by ROC curve analysis confirmed that these hub genes are reliable diagnostic markers. The findings provide molecular insight into SONFH and will facilitate the development of targeted interventions.
Data availability
Publicly available datasets were analyzed in this study. The data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE123568;https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE74089. All data are available from the corresponding author on reasonable request.
References
Mont, M. A. & Hungerford, D. S. Non-traumatic avascular necrosis of the femoral head. J. Bone Jt. Surg. Am. Vol.77 (3), 459–474. https://doi.org/10.2106/00004623-199503000-00018 (1995).
Moya-Angeler, J., Gianakos, A. L., Villa, J. C., Ni, A. & Lane, J. M. Current concepts on osteonecrosis of the femoral head. World J. Orthop.6 (8), 590–601. https://doi.org/10.5312/wjo.v6.i8.590 (2015).
Assouline-Dayan, Y., Chang, C., Greenspan, A., Shoenfeld, Y. & Gershwin, M. E. Pathogenesis and natural history of osteonecrosis. Semin. Arthritis Rheum.32 (2), 94–124 (2002).
Zhao, D. et al. Guidelines for clinical diagnosis and treatment of osteonecrosis of the femoral head in adults (2019 version). J. Orthop. Transl.21, 100–110. https://doi.org/10.1016/j.jot.2019.12.004 (2020).
Hines, J. T. et al. Osteonecrosis of the femoral head: an updated review of ARCO on pathogenesis, staging and treatment. J. Korean Med. Sci.36 (24), e177. https://doi.org/10.3346/jkms.2021.36.e177 (2021).
Kerachian, M. A., Séguin, C. & Harvey, E. J. Glucocorticoids in osteonecrosis of the femoral head: a new understanding of the mechanisms of action. J. Steroid Biochem. Mol. Biol.114 (3–5), 121–128. https://doi.org/10.1016/j.jsbmb.2009.02.007 (2009).
Wang, A., Ren, M. & Wang, J. The pathogenesis of steroid-induced osteonecrosis of the femoral head: a systematic review of the literature. Gene671, 103–109. https://doi.org/10.1016/j.gene.2018.05.091 (2018).
Li, Y., Zhang, J., Zhao, Y., Tian, R. & Yang, P. A novel animal model of osteonecrosis of the femoral head based on 3D printing technology. J. Orthop. Surg. Res.18 (1), 564. https://doi.org/10.1186/s13018-023-04050-7 (2023).
Martel, D. et al. 3T chemical shift-encoded MRI: detection of altered proximal femur marrow adipose tissue composition in glucocorticoid users and validation with magnetic resonance spectroscopy. J. Magn. Reson. Imaging50 (2), 490–496. https://doi.org/10.1002/jmri.26586 (2019).
Chotiyarnwong, P. & McCloskey, E. V. Pathogenesis of glucocorticoid-induced osteoporosis and options for treatment. Nat. Rev. Endocrinol.16 (8), 437–447. https://doi.org/10.1038/s41574-020-0341-0 (2020).
Zhu, L. et al. Parathyroid hormone (PTH) induces autophagy to protect osteocyte cell survival from dexamethasone damage. Med. Sci. Monit.23, 4034–4040. https://doi.org/10.12659/msm.903432 (2017).
Sun, F., Zhou, J. L., Liu, Z. L., Jiang, Z. W. & Peng, H. Dexamethasone induces ferroptosis via P53/SLC7A11/GPX4 pathway in glucocorticoid-induced osteonecrosis of the femoral head. Biochem. Biophys. Res. Commun.602, 149–155. https://doi.org/10.1016/j.bbrc.2022.02.112 (2022).
Wu, T., Jiang, Y., Shi, W., Wang, Y. & Li, T. Endoplasmic reticulum stress: a novel targeted approach to repair bone defects by regulating osteogenesis and angiogenesis. J. Transl. Med.21 (1), 480. https://doi.org/10.1186/s12967-023-04328-8 (2023).
Wiseman, R. L., Mesgarzadeh, J. S. & Hendershot, L. M. Reshaping endoplasmic reticulum quality control through the unfolded protein response. Mol. Cell82 (8), 1477–1491. https://doi.org/10.1016/j.molcel.2022.03.025 (2022).
Yoshida, H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell107 (7), 881–891. https://doi.org/10.1016/s0092-8674(01)00611-0 (2001).
Tao, S. C. et al. Exosomes derived from human platelet-rich plasma prevent apoptosis induced by glucocorticoid-associated endoplasmic reticulum stress in rat osteonecrosis of the femoral head via the Akt/Bad/Bcl-2 signal pathway. Theranostics7 (3), 733–750. https://doi.org/10.7150/thno.17450 (2017).
Jia, Y. et al. Identification and assessment of novel dynamic biomarkers for monitoring non-traumatic osteonecrosis of the femoral head staging. Clin. Transl. Med.13 (6), e1295. https://doi.org/10.1002/ctm2.1295 (2023).
Rebhan, M., Chalifa-Caspi, V., Prilusky, J. & Lancet, D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet.13 (4), 163. https://doi.org/10.1016/s0168-9525(97)01103-7 (1997).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15 (12), 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28 (11), 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51 (D1), D587–D592. https://doi.org/10.1093/nar/gkac963 (2023).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28 (1), 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics16 (5), 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Franceschini, A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res.41, D808–D815. https://doi.org/10.1093/nar/gks1094 (2013).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13 (11), 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics (Oxford, England)27 (12), 1739–1740. https://doi.org/10.1093/bioinformatics/btr260 (2011).
Dweep, H., Sticht, C., Pandey, P. & Gretz, N. miRWalk–database: prediction of possible miRNA binding sites by walking the genes of three genomes. J. Biomed. Inform.44 (5), 839–847. https://doi.org/10.1016/j.jbi.2011.05.002 (2011).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform.14, 128. https://doi.org/10.1186/1471-2105-14-128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res.44 (W1), W90–W97. https://doi.org/10.1093/nar/gkw377 (2016).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods12 (5), 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Liang, X. Z. et al. Identification of potential autophagy-related genes in steroid-induced osteonecrosis of the femoral head via bioinformatics analysis and experimental verification. J. Orthop. Surg, Res.17 (1), 86. https://doi.org/10.1186/s13018-022-02977-x (2022).
Robin, X. et al. pROC: an open-source package for R and S + to analyze and compare ROC curves. BMC Bioinform.12, 77. https://doi.org/10.1186/1471-2105-12-77 (2011).
Li, T. et al. MicroRNA expression profile of dexamethasone-induced human bone marrow-derived mesenchymal stem cells during osteogenic differentiation. J. Cell. Biochem.115 (10), 1683–1691. https://doi.org/10.1002/jcb.24831 (2014).
Li, T. et al. microRNA-23a inhibits osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting LRP5. Int. J. Biochem. Cell Biol.72, 55–62. https://doi.org/10.1016/j.biocel.2016.01.004 (2016).
Xu, Y., Jiang, Y., Jia, B., Wang, Y. & Li, T. Icariin stimulates osteogenesis and suppresses adipogenesis of human bone mesenchymal stem cells via miR-23a-mediated activation of the Wnt/β-catenin signaling pathway. Phytomed. Int. J. Phytother. Phytopharmacol.85, 153485. https://doi.org/10.1016/j.phymed.2021.153485 (2021).
Xu, Y. et al. LINC00473-modified bone marrow mesenchymal stem cells incorporated thermosensitive PLGA hydrogel transplantation for steroid-induced osteonecrosis of femoral head: a detailed mechanistic study and validity evaluation. Bioeng. Transl. Med.7 (2), e10275. https://doi.org/10.1002/btm2.10275 (2022).
Zhang, X. et al. Dexamethasone induced osteocyte apoptosis in steroid-induced femoral head osteonecrosis through ROS-mediated oxidative stress. Orthop. Surg.16 (3), 733–744. https://doi.org/10.1111/os.14010 (2024).
Wang, M. et al. Bone morphogenetic protein 2 controls steroid-induced osteonecrosis of the femoral head via directly inhibiting interleukin-34 expression. J. Mol. Endocrinol.68 (1), 1–9. https://doi.org/10.1530/jme-21-0163 (2021).
Morishita, Y. et al. Cell death-associated lipid droplet protein CIDE-A is a noncanonical marker of endoplasmic reticulum stress. JCI Insight6 (7), 980. https://doi.org/10.1172/jci.insight.143980 (2021).
Yi, J. et al. Lipid metabolism disorder promotes the development of intervertebral disc degeneration. Biomed. Pharmacother.166, 115401. https://doi.org/10.1016/j.biopha.2023.115401 (2023).
Sun, F., Zhou, J. L., Wei, S. X., Jiang, Z. W. & Peng, H. Glucocorticoids induce osteonecrosis of the femoral head in rats via PI3K/AKT/FOXO1 signaling pathway. PeerJ10, e13319. https://doi.org/10.7717/peerj.13319 (2022).
Maurel, M. & Chevet, E. Endoplasmic reticulum stress signaling: the microRNA connection. Am. J. Physiol. Cell. Physiol.304 (12), C1117–C1126. https://doi.org/10.1152/ajpcell.00061.2013 (2013).
Ganesan, S. et al. Stromal cells downregulate miR-23a-5p to activate protective autophagy in acute myeloid leukemia. Cell Death Dis.10 (10), 736. https://doi.org/10.1038/s41419-019-1964-8 (2019).
Lingam, I. et al. Serial blood cytokine and chemokine mRNA and microRNA over 48 h are insult specific in a piglet model of inflammation-sensitized hypoxia-ischaemia. Pediatr. Res.89 (3), 464–475. https://doi.org/10.1038/s41390-020-0986-3 (2021).
Yu, Y., Lin, L., Liu, K., Jiang, Y. & Zhou, Z. Effects of simvastatin on cartilage homeostasis in steroid-induced osteonecrosis of femoral head by inhibiting glucocorticoid receptor. Cells11 (24), 945. https://doi.org/10.3390/cells11243945 (2022).
Jin, S. et al. Curcumin prevents osteocyte apoptosis by inhibiting M1-type macrophage polarization in mice model of glucocorticoid-associated osteonecrosis of the femoral head. J. Orthop. Res.38 (9), 2020–2030. https://doi.org/10.1002/jor.24619 (2020).
Kikuiri, T. et al. Cell-based immunotherapy with mesenchymal stem cells cures bisphosphonate-related osteonecrosis of the jaw-like disease in mice. J. Bone Miner. Res.25 (7), 1668–1679. https://doi.org/10.1002/jbmr.37 (2010).
He, J., Zhou, Q., Jia, X., Zhou, P. & Chen, L. Immune-related expression profiles of bisphosphonates-related osteonecrosis of the jaw in multiple myeloma. Die Pharmazie76 (4), 159–164. https://doi.org/10.1691/ph.2021.01013 (2021).
Hu, K., Shang, Z., Yang, X., Zhang, Y. & Cao, L. Macrophage polarization and the regulation of bone immunity in bone homeostasis. J. Inflamm. Res.16, 3563–3580. https://doi.org/10.2147/jir.S423819 (2023).
Weitzmann, M. N., Vikulina, T., Roser-Page, S., Yamaguchi, M. & Ofotokun, I. Homeostatic expansion of CD4 + T cells promotes cortical and trabecular bone loss, whereas CD8 + T cells induce trabecular bone loss only. J. Infect. Dis.216 (9), 1070–1079. https://doi.org/10.1093/infdis/jix444 (2017).
Wu, L. et al. Natural killer cells infiltration in the joints exacerbates collagen-induced arthritis. Front. Immunol.13, 860761. https://doi.org/10.3389/fimmu.2022.860761 (2022).
Wang, F. et al. IL-34 aggravates steroid-induced osteonecrosis of the femoral head via promoting osteoclast differentiation. Immune Netw.22 (3), e25. https://doi.org/10.4110/in.2022.22.e25 (2022).
Yu, Y. et al. Genetic polymorphisms in IL1B predict susceptibility to steroid-induced osteonecrosis of the femoral head in Chinese Han population. Osteoporos. Int.30 (4), 871–877. https://doi.org/10.1007/s00198-019-04835-9 (2019).
Zhou, Q. et al. The use of TLR2 modified BMSCs for enhanced bone regeneration in the inflammatory micro-environment. Artif. Cells Nanomed. Biotechnol.47 (1), 3329–3337. https://doi.org/10.1080/21691401.2019.1626867 (2019).
Luo, J. et al. CGRP-loaded porous microspheres protect BMSCs for alveolar bone regeneration in the periodontitis microenvironment. Adv. Healthc. Mater.12 (28), e2301366. https://doi.org/10.1002/adhm.202301366 (2023).
Acknowledgements
We would like to acknowledge the GEO (GSE123568 and GSE74089) network for providing data. We gratefully acknowledge Textcheck for their English language editing services. The English in this document has been checked by two native-speaking professional editors. For a certificate, please see: http://www.textcheck.com/certificate/2K7SOE.
Funding
This work was supported by grants from National Natural Science Foundation of China; Grant number: 82272489, 82203588; TaiShan Scholars Project Special Fund; Grant number: NO.tsqn202306396; Qingdao Science and Technology Benefiting the People Demonstration Special Project; Grant number: 24-1-8-smjk-3-nsh. The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
TW, WS and YZ contributed to the bioinformatics analysis and wrote the original manuscript. SG and HT collected the literature and participated in the qRT-PCR test. YJ and WL performed the statistical analysis. YW and TL designed the study and revised the manuscript. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This study was supported and approved by the Institutional Review Board (IRB) of The Affiliated Hospital of Qingdao University (QYFY WZLL 28701). All clinical samples patients were informed of the purpose of the study and signed the consent form.
Consent for publication
The manuscript was read and approved for publication by all authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, T., Shi, W., Zhou, Y. et al. Identification and validation of endoplasmic reticulum stress-related genes in patients with steroid-induced osteonecrosis of the femoral head. Sci Rep 14, 21634 (2024). https://doi.org/10.1038/s41598-024-72941-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-72941-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.