Abstract
Triple-negative breast cancer (TNBC) represents a significant health concern for women worldwide, and the overproduction of MMP9 and CD151 is associated with various cancers, influencing tumour growth and progression. This study aimed to investigate how CD151 and MMP9 affect TNBC cell migration, apoptosis, proliferation, and invasion. Immunohistochemical experiments revealed that CD151 and MMP9 were positively expressed in triple-negative breast cancer, and lymph node metastasis, the histological grade, and CD151 and MMP9 expression were found to be independent prognostic factors for the survival of patients with triple-negative breast cancer. Cytological experiments indicated that the knockdown of CD151 or MMP9 slowed triple-negative breast cancer cell growth, migration, and invasion and increased the apoptosis rate. Compared with CD151 knockdown, double MMP9 and CD151 knockdown further promoted cell death and inhibited TNBC cell proliferation, migration, and invasion. Moreover, β-catenin and p-GSK-3β were significantly downregulated. In summary, simultaneously silencing CD151 and MMP9 further suppressed the proliferation, migration and invasion of TNBC cells and promoted their apoptosis. One possible strategy for inducing this effect is to block the GSK-3β/β-catenin pathway.
Similar content being viewed by others
Introduction
Breast tumours are a group of diseases that endanger women’s lives and health; they rank first among all female tumours in terms of incidence, with an incidence rate of 19.2%1, and are the leading cause of cancer-related deaths. Breast cancer can be categorized into several subtypes according to various factors, and triple-negative breast cancer (TNBC) is typically considered the basal-like breast cancer subtype. TNBC is a highly diverse and heterogeneous group of breast cancers lacking immunohistochemical expression of oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (Her2/neu). Based on guidelines from the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP)2, TNBC is most commonly detected in premenopausal women and obese people3 and typically presents as a high histological grade with a high risk of brain metastasis4. TNBC accounts for 15–20% of all breast cancer cases5.
The 24 members of the zinc-dependent endopeptidase family known as matrix metalloproteinases (MMPs) not only promote tumour metastasis but also regulate a variety of physiological processes6; furthermore, in patients with BC, MMP9 overexpression in tumour cells was linked to a decreased chance of survival, a larger tumour size, metastasis to lymph nodes, a greater clinical stage, a higher histological grade, and distant metastases7,8. Among them, MMP9 is a complex matrix metalloproteinase that is essential for remodelling of the extracellular matrix (ECM)9, metastasis, angiogenesis and apoptosis and has been implicated in tumour invasion, metastasis and tumour microenvironment regulation10,11. According to pertinent research, MMP9 is necessary for TNBC metastasis and functions in the degradation of the basement membrane to facilitate migration, invasion, and metastasis12. Increased MMP9 expression has been linked to tumour progression in both in vitro and in vivo tests conducted on humans and experimental cancer models13.
CD151 belongs to the tetratransmembrane protein (tetaspanin) family, which is extensively expressed in a variety of tissues, such as epidermal cells, endothelial cells, and platelets, and is localized on the surface of cell membranes; additionally, CD151 is thought to be a powerful driver of cell adhesion and behaviour, tumorigenesis and progression, angiogenesis, metastasis, cancer recurrence, and resistance to existing chemotherapeutic and targeted therapy14. CD151 is highly expressed at the protein and mRNA levels in breast cancer, colon cancer and hepatocellular carcinomas, and changes in its expression are significantly correlated with cancer growth, invasion and migration15. CD151 was shown to be highly expressed in TNBC cells and to promote the proliferation, invasion and migration of breast cancer cells16,17,18,19, and our previous study revealed that patients with TNBC exhibit high CD151 expression, which is associated with lymph node metastasis and the histological grade.
Previous research has indicated that β-catenin is differentially expressed in various forms of breast cancer, and the GSK-3β/β-catenin-related signalling pathway plays crucial roles in the regulation of breast cancer metastasis, cell proliferation and the EMT20. Notably, overexpression of CD151 in hepatocellular carcinoma promotes MMP9 expression through the PI3K/AKT/GSK-3β signalling pathway, which promotes hepatocellular carcinoma neoangiogenesis and increases the metastatic potential of tumours21. However, the manner in which CD151 and MMP9 work together in TNBC and whether these effects are related to the GSK-3β/β-catenin-related signalling pathway are unclear. Therefore, the purpose of this study was to investigate the effects of double knockdown of CD151 and MMP9 on the proliferation, migration, invasion, and apoptosis of TNBC cells and the related mechanisms.
Results
Validation of CD151 silencing in stably transduced cell lines
After MDA-MB-231 cells and MDA-MB-436 cells were infected with the CD151 shRNA lentivirus (sh#1, sh#2, and sh#3) or negative control (shNC) for 96 h, the infection efficiency was assessed via fluorescence microscopy. ShCD151 successfully infected TNBC cells (Fig. 1A, B). The infection efficiency was detected via qRT‒PCR and Western blot analyses. The findings revealed that the shCD151#3 group of both cell lines had considerably lower mRNA (Fig. 1C) and protein (Fig. 1D, E) expression levels than the control group (P < 0.001; shCD151#3 was used in all subsequent tests).
Validation of stably transfected cell lines in which CD151 or MMP9 was silenced. (A, B) Fluorescence and bright-field images of shCD151 lentivirus-infected MDA-MB-231 cells and MDA-MB-436 cells. (C) qRT‒PCR was used to detect the relative expression of the CD151 mRNA in MDA-MB-231 cells and MDA-MB-436 cells. (D) Western blotting was used to determine whether the cells expressed the CD151 protein. (E) Statistical analysis of the silencing of CD151 following the analysis of the relative cellular expression of the protein in MDA-MB-231 cells and MDA-MB-436 cells. (F) qRT‒PCR analysis of relative MMP9 mRNA expression in MDA-MB-231 cells and MDA-MB-436 cells. (G) Western blotting was used to determine whether the MDA-MB-231 cells and MDA-MB-436 cells expressed the MMP9 protein. (H) Statistical analysis of the relative protein expression in TNBC cells. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Validation of the efficacy of the small interfering RNA (siRNA) against MMP9
MMP9 siRNAs (si#1, si#2, and si#3) and a negative control (siNC) were transfected into MDA-MB-231 cells and MDA-MB-436 cells. QRT‒PCR and Western blot analyses revealed that siMMP9#3 produced considerably lower mRNA (Fig. 1F) and protein (Fig. 1G, H) expression levels in both cell lines than did the control (P < 0.001; siMMP9#3 was used in all subsequent experiments).
Simultaneous interference of MMP9 and CD151 in TNBC cells significantly inhibited their migration ability
The results of the cell scratch wound healing assay showed that the migratory ability of the shCD151 and siMMP9 groups of MDA-MB-231 cells and MDA-MB-436 cells was significantly lower than that of the negative control group (P < 0.0001); moreover, no significant difference was observed between the two negative control groups (P > 0.05). Compared to the effects on the shCD151 and siMMP9 groups, the inhibitory effect on cell migration was more obvious (P < 0.05) and statistically significant in the shCD151 + siMMP9 group (Fig. 2A, B).
We used flow cytometry, transwell, CCK-8, and scratch wound healing assays to determine how each treatment affected TNBC cells. (A, B) The scratch wound healing assay was used to evaluate the migratory capacity of each cell group. (C) The CCK-8 assay was used to measure the ability of each cell group to proliferate. (D, E, F) Transwell assays were used to measure the invasion and migration potential of each cell group. (G, H) A flow cytometry assay was used to determine which cell groups underwent apoptosis. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Note: The above experiments were performed for the shCD151 group with the shNC group as the control, the siMMP9 group with the siNC group as the control, and the shCD151 + siMMP9 group with the shCD151 group and siMMP9 group as controls.
Simultaneous inhibition of CD151 and MMP9 significantly inhibited TNBC cell proliferation
The effects of single-gene knockdown and double-gene knockdown on the proliferation of MDA-MB-231 cells were compared via cell proliferation assays and CCK-8 assays. The findings revealed that interference of one gene alone significantly decreased the proliferative ability of cells in the shCD151 and siMMP9 groups compared with that in the negative control group (P < 0.01); moreover, the negative control group did not exhibit any significant differences (P > 0.05). In contrast to that in the shCD151 and siMMP9 groups, the inhibitory effect on cell proliferation in the shCD151 + siMMP9 group was more obvious (P < 0.05) and statistically significant (Fig. 2C).
The capacity of TNBC cells to migrate and invade was further inhibited by interfering with the expression of CD151 and MMP9
Transwell assays demonstrated that the migration ability of MDA-MB-231 cells and MDA-MB-436 cells in the shCD151 and siMMP9 groups was significantly lower than that in the negative control group (P < 0.0001); moreover, the negative control group did not exhibit any significant differences (P > 0.05). Compared with that in the shCD151 and siMMP9 groups, the inhibitory effect on cell migration was significantly greater in the shCD151 + siMMP9 group (P < 0.01) (Fig. 2D, E). The results of the invasion assay were the same as those of the migration assay, and the inhibitory effect on cell invasion was obvious in the shCD151 + siMMP9 group. In addition, the invasion ability of the shCD151 + siMMP9 group was significantly greater than that of the shCD151 or siMMP9 alone groups (P < 0.001) (Fig. 2D, F).
Simultaneous knockdown of CD151 and MMP9 significantly promotes apoptosis in TNBC cells
Flow cytometry experiments showed a greater number of apoptotic cells in the shCD151-alone or siMMP9-alone groups than in the negative control group (P < 0.0001); moreover, the two negative control populations did not significantly differ (P > 0.05). Compared with those in the shCD151 and siMMP9 alone groups, the number of apoptotic cells was significantly greater in the shCD151 + siMMP9 group (P < 0.01) (Fig. 2G, H).
Knockdown of both CD151 and MMP9 may inhibit TNBC cell progression through the GSK-3β/β-catenin pathway
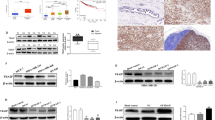
We further investigated the potential downstream pathways that may lead to the combined inhibition of tumour progression by CD151 and MMP9. Western blot results verified that CD151 protein expression in the shCD151 and shCD151 + siMMP9 groups was considerably lower than that in the control group (Fig. 3A, B, P< 0.0001), and MMP9 protein expression in the shCD151 + siMMP9 and siMMP9 groups was significantly lower than that in the control group (Fig. 3A, C, P< 0.0001). The results also showed that the shNC and siNC treatments had no effect on β-catenin protein expression (P > 0.05), whereas the shCD151, siMMP9, and shCD151 + siMMP9 treatments had inhibitory effects on β-catenin expression, but the shCD151 + siMMP9 treatment exhibited a stronger inhibitory effect (Fig. 3A, D, P< 0.001). In addition, shNC, shCD151, siNC, and siMMP9 showed discernible suppression of p-GSK-3β levels (P > 0.05), while shCD151 + siMMP9 exerted a very obvious inhibitory effect on p-GSK-3β expression (Fig. 3A, E, P< 0.001).
The impact of each treatment on the expression of the pertinent proteins in MDA-MB-231 cells was examined using Western blotting. (A, B) Expression of the CD151 protein. (A, C), Expression of the MMP9 protein. (A, D) Expression of the β-catenin protein. (A, E) Expression of the p-GSK-3β protein. **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns: not statistically significant.
Expression of CD151 and MMP9 in different breast tissues
CD151 was localized to the cell membrane and/or cytoplasm, and the expression of CD151 differed significantly among the three groups of TNBC tissues, NTNBC tissues, and normal breast tissues (P < 0.001). Similarly, MMP9 expression differed significantly among the three groups, and MMP9 was detected in the cell membrane and cytoplasm (P < 0.001) (Table 1, Fig. 4A). In this study, we analysed the correlation between clinicopathological characteristics and the expression of CD151 and MMP9 in TNBC tissues, and the results showed that lymph node metastasis and the histological grade affected the expression of CD151 and MMP9 (Tables 2 and 3). According to the findings of the one-way Kaplan‒Meier survival analysis, lymph node metastasis, histological grade, CD151 positivity, and MMP9 positivity were significantly associated with DFS in TNBC patients (P < 0.0001; Fig. 4B-E). Cox logistic analysis revealed that lymph node metastasis, histological grade, CD151 positivity, and MMP9 positivity were independent prognostic factors (Fig. 4F).
Factors affecting TNBC and prognostic analysis of survival. (A) Expression of CD151 and MMP9 in different breast tissues. EliVision two-step method (× 200). (B) Relationship between patient DFS and lymph node metastasis. (C) Relationship between patient DFS and histological grade. (D) Relationship between DFS and positive CD151 expression in patients. (E) Relationship between DFS and positive MMP9 expression in patients. (F) Multivariate analysis of factors affecting the DFS of TNBC patients. Note: All differences between survival curves in the graph are statistically significant (P < 0.0001).
Discussion
Triple-negative breast cancer cells exhibit greater immunogenicity, with greater immunoreactivity and an enrichment of immune pathway genes and higher levels of lymphocyte infiltration22. In terms of treatment, since proper hormone receptor therapy is unavailable, TNBC patients are not good candidates for conventional endocrine or targeted therapies, have a high risk of postoperative recurrence and metastasis, and still have the worst survival rate among patients with various types of breast cancer23,24,25. Thus, innovative and effective treatment options for TNBC patients are key clinical needs.
Clinical studies have shown that CD151 overexpression usually predicts poor outcomes for a variety of malignancies, such as gastric cancer26,27, prostate cancer28,29,30,31 and non-small cell lung cancer32,33,34,35. Over the last ten years, accumulating evidence has identified CD151 as a biologically diverse enzyme and a cause of cancer arising from human epithelial cells. It controls proliferation, the EMT, metabolism, cell‒cell junctions, tumour stem cells, and the tumour microenvironment15,36,37. These many roles demonstrate the pivotal role of CD151 in a variety of human malignancies. Breast cancer, colon cancer, and hepatocellular carcinomas exhibit high levels of CD151 mRNA and protein expression, and variations in CD151 expression are strongly correlated with cancer development, invasion, and migration31 and are significantly correlated with the stage, lymph node status or distant metastasis in breast cancer patients with a poor prognosis38. The present study employed in vitro experiments to confirm that downregulating CD151 prevents the invasion, migration, and proliferation of TNBC cells while simultaneously increasing apoptosis. Additionally, immunohistochemical staining revealed high expression of CD151 in TNBC tissues. In addition, the positive expression of CD151 and MMP9 affects the prognosis of patients with TNBC.
According to recent research, MMP9 plays a critical role in processes connected to the progression and malignancy of cancer39. A wide range of bioactive molecules are released or activated by MMP9; these molecules include growth factors, chemokines, cytokines, and stromal factors, which are involved in the immune response; angiogenesis; the development of the tumour microenvironment; and cell migration, differentiation, and survival. MMP9 is involved in apoptosis, proliferation, and angiogenesis and is frequently overexpressed in a range of cancer types. These processes are linked to the initiation and spread of cancer39, and their manifestation is strongly correlated with the tumour size, metastasis, clinical status and histological grade7, e.g., lung, breast or colorectal cancer. As a result, MMP9 is a prospective cancer therapeutic target. In the present study, the inhibition of MMP9 expression significantly inhibited the ability of TNBC cells to divide, move, and invade; it promoted apoptosis, and MMP9 was highly expressed in TNBC tissues.
A positive correlation between CD151 and MMP9 has been shown in hepatocellular carcinoma and osteosarcoma40, but a correlation has not been found between CD151 and MMP9 in breast cancer subtypes. This study showed for the first time that the downregulation of both CD151 and MMP9 had a more notable effect on the number of apoptotic cells and inhibited the capacity of TNBC cells to multiply, move, and invade than the downregulation of either factor alone. As a common critical signalling pathway in the genesis of cancer, the GSK-3β/β-catenin pathway participates in the metabolism and proliferation of tumour cells. Dysregulation of this system has been documented in a variety of cancer types, including gastric, liver and colon cancers41,42,43,44,45,46,47,48. Our study showed that dual downregulation of CD151 and MMP9 significantly inhibited the expression of the p-GSK-3β and β-catenin proteins, indicating that combined interference with CD151 and MMP9 may prevent TNBC cells from proliferating, migrating, and invading their environment and instead encourage apoptosis by blocking the signalling pathways related to GSK-3β/β-catenin. This analysis revealed, for the first time, the joint actions of CD151 and MMP9 in TNBC, which could provide a novel theoretical framework and therapeutic target for the management of this disease type and others.
This study has several limitations. First, the number of clinical samples used in the present study was limited, and additional paraffin tissue samples from TNBC patients are needed to verify our findings. In addition, we conducted only a preliminary study on the combined mechanism of action of CD151 and MMP9, and the upstream signalling pathways that cause the downregulation of the p-GSK-3β and β-catenin proteins have not yet been explored; these pathways will continue to be investigated in the future. Finally, this study investigated the combined mechanism of CD151 and MMP9 through in vitro experiments, and we will use in vivo animal models to further confirm our findings.
Materials and methods
Materials and reagents
The following cells and reagents were used: the human triple-negative breast cancer cell lines MDA-MB-231 and MDA-MB-436 (Wuhan Pricella Biotechnology Co., Ltd); MDA-MB-231 cell-specific medium (L15, DMEM) (Procell Life Science & Technology); MDA-MB-436 cell-specific medium (DMEM); Lipo8000™ transfection reagent (Beyotime, Shanghai, China); CD151 knockdown lentivirus and negative control (Obio Technology); MMP9 small interfering RNA and negative control (Obio Technology); reverse transcription kit; fluorescent quantitative PCR kit (Suzhou Novoprotein Scientific); CD151 antibody (Affinity Biosciences); CD151 antibody (Proteintech Group); MMP9 antibody (Proteintech Group); MMP9 antibody (Affinity Biosciences); p-GSK-3β antibody (Wanleibio); β-catenin antibody (Wanleibio); CCK-8 reagents (Beyotime Biosciences); and an Annexin V-FITC/PI apoptosis detection kit (Beyotime, Shanghai, China). Between 2018 and 2022, one hundred fifteen wax block specimens were collected from the First Affiliated Hospital of Bengbu Medical College’s Department of Pathology; these specimens included 38 triple-negative breast cancer samples, 38 non-triple-negative breast cancer samples, and 39 normal tissue samples. All patients who participated in the study signed informed consent forms. The study was authorized by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical University (approval no. 2021085), and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki (6th revision, 2008).
Cell culture, transfection and lentiviral infection
The cells were cultured in cell-specific complete medium (L15) and grown in an incubator at 37 °C without CO2 or in DMEM at 37 °C and 5% CO2. According to the lentiviral infection guidelines, cells from the logarithmic growth phase were obtained, infected and divided into the shNC group and shCD151 group 96 h after infection. A fluorescence microscope was used to monitor the infection efficiency, and 2.5 µg/mL puromycin was utilized for screening for viable cell lines. Cells were transfected according to the instructions of the Lipo8000™ transfection reagent and divided into the siNC group, siMMP9 group, and shCD151 + siMMP9 group. Sequences of CD151 and MMP9: ShNC: 5’-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3’, ShCD151#1: 5’-GCCTCAAGT. ACCTGCTGTTTACTCGAGTAAACAGCAGGTACTTGAGGC-3’, ShCD151#2: 5’-CCCTCAAGAGTGACTACATCACTCGAGTGAT. GTAGTCACTCTTGAGGG-3’, ShCD151#3: 5’-GCTGGAGATCATCGCTGGTATCTCGAGATACCAGCGATGATCTCCAGC-3’; siNC-S: 5’-UUCUCCGAACGUGUCACGUTT-3’, siNC-A: 5’-ACGUGACACGUUCGGAGAATT-3’; siMMP9#1-S: 5’-GCUGCU. UCUCCAGAAGCAACUTT-3’, siMMP9#1-A: 5’-AGUUGCUUCUGGAGAAGCAGCTT-3’; siMMP9#2-S: 5’-CGAGCUGACU. CGACGGUGAUGTT-3’, siMMP9#2-A: 5’-CAUCACCGUCGAGUCAGCUCGTT-3’; siMMP9#3-S: 5’-CGAACUUUGACAGC. GACAAGATT-3’, siMMP9#3-A: 5’-UCUUGUCGCUGUCAAAGUUCGTT-3’.
Real-time fluorescent quantitative PCR (qRT‒PCR)
Total RNA was extracted from cells using an RNA extraction sorbent column (Vazyme) and subsequently reverse-transcribed to cDNA using the NovoScript®Plus All-in-one 1st Strand cDNA Synthesis SuperMix (gDNA Purge) Kit (Novoprotein). With a Roche Applied Science LightCycler 480 system, real-time PCR was performed with the NovoStart® SYBR qPCR SuperMix Plus Kit (Novoprotein). The following PCR procedure was used: 95 °C for 1 min and 40 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 30 s. The comparative 2−ΔΔCt method was used to calculate the relative expression of the genes. β-Actin served as the internal reference. The primers used were as follows: MMP9, upstream 5′-AGTTCCCGGAGTGAGTTGAA-3′ and downstream 5′-CCCTTTCCTCCAGAACAGAA-3′; CD151, upstream 5′-CCTGCTCCTCATCTTCC-3′ and downstream 5′-CTGCCACAACAGTGTGGAACTC-3′; and β-actin, upstream 5′-GCCAACACACAGTGCTGTCTGG-3′ and downstream 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′.
Western blotting
With the aid of loading buffer and RIPA lysis buffer, all the cellular proteins were extracted. After separation on 12% SDS‒PAGE gels, the proteins were applied to PVDF membranes. The membranes were then incubated for 1 h and 30 min in 5% skim milk powder and overnight at 4 °C in primary antibody incubation solutions containing antibodies against CD151 (1:500), MMP9 (1:1000), and β-actin (1:1500). Afterwards, the cells were incubated for one hour with a goat anti-rabbit secondary antibody (1:5000). And photographed using Gel Doc 2000 (Bio-RAD) equipment, and then detected the protein expression of MMP9 and CD151 associated with β-actin using ImageJ software.
Cell scratch wound healing experiment
Cells from each group were seeded into six-well plates, and once the cell growth density exceeded 90%, the culture media was removed. The tip of a 200 μL pipette was used to draw two parallel lines on the bottom of the culture plate, the cells between the scratches were removed by washes with PBS, fresh basal medium was added, and images were captured under a microscope at 0 h and 48 h. And used Image J to measure the migratory capacity of the cells.
CCK-8 assay for cell proliferation
The cells in each group were seeded in each well of 96-well plates at a density of 2 × 103 cells/100 μL. After incubations for 24, 48, 72, or 96 h, 10 μL of CCK-8 solution was added to each well. After the cells were incubated in an incubator for two hours, an enzyme marker was used to measure the optical density (OD) at 450 nm.
Transwell assay
Before the invasion assay was started, Matrigel was diluted with serum-free DMEM at a ratio of 1:8, after which the inner chamber was filled with 50 μL of diluted Matrigel and the cells were placed in an incubator at 37 °C. After the Matrigel solidified, 200 μL of a cell suspension (105 cells) diluted in basal medium was added to the inner chamber, 500 μL of complete culture media was added to the outer chamber, and the cells were removed from the chamber after 24 h of incubation in the incubator. The Matrigel and cells in the inner chamber were gently removed with a cotton swab, fixed with 4% paraformaldehyde for 20 min, stained with crystal violet stain for 15 min, and observed and photographed under a microscope. Cells were then counted using Image J. The difference between the migration and invasion experiments was that the former contained Matrigel; the remaining conditions and steps were the same.
Flow cytometry experiments
Following the collection and digestion of the cells from each group, the cells were resuspended in PBS and centrifuged. Then, 195 μL of binding solution, 5 μL of Annexin V-FITC, and 10 μL of PI were added sequentially. Next, the cells were stained for 20 min at room temperature in the dark, after which flow cytometry was used to measure the percentage of apoptotic cells.
IHC staining
The collected paraffin-embedded samples were serially sectioned at a thickness of 4 μm, deparaffinized with xylene, baked for 2 h and dehydrated with gradient ethanol. Afterwards, the slides were placed in a 3% H2O2 solution for 10 min and rinsed three times for 3 min each with PBS buffer. The section box containing the citrate solution was boiled in a water bath at 100°C. Then, the sections were placed in the box and heated for 30 min, after which they were cooled to room temperature in cold water and rinsed slowly with PBS 3 times. When the sections were dry, they were blocked with goat serum. The CD151 and MMP9 antibodies were diluted to the recommended ratios (anti-CD151 = 1:200, anti-MMP9 = 1:200), added dropwise to the tissue and incubated at room temperature for 2 h or overnight at 4 °C. The slides were washed several times with PBS, and the liquid was shaken off from the slides, followed by the addition of the secondary antibody dropwise; the slides were left in a warm box for 30 min and then rinsed several times with PBS again. The DAB solution was added dropwise to the slides, and when a colour change occurred, the slides were observed under a microscope. The colour development was terminated by washing the slides with running water when a suitable tan colour appeared in the field of view. The next steps involved counterstaining the nuclei with a haematoxylin solution, washing the slides with differentiation buffer, washing them blue, dehydrating and then clearing them, and finally, the slices were sealed with cedar oil to observe the experimental results under a microscope.
Immunohistochemical staining was performed using the Envision two-step method. Paraffin sections from the study subjects immunohistochemically stained using antibodies against ER, PR, and Her-2 were reviewed, and the immunohistochemical staining results were evaluated by two or more pathologists who read the sections. CD151 showed positive cytosolic and/or cytoplasmic staining, and MMP9 predominantly showed positive cytoplasmic staining and appeared as a brownish-yellow or tan colour. Photographs were taken using an inverted microscope(IX71, Olympus, Tokyo, Japan). Based on the staining intensity, positive tumour cells were scored as colourless (0), yellowish (1), brownish (2), or tan (3), and a score of ≥ 2 was considered positive.
Statistical methods
The statistical analysis of the data was conducted with GraphPad Prism 8.0 and SPSS 23.0. Fisher’s exact test was used to compare categorical data. The clinicopathological characteristics that impact DFS were identified using Cox survival analysis, and the tumour-free survival rate was determined using the Kaplan‒Meier method. A difference was considered statistically significant at P < 0.05. (Supplementary informations).
Data availability
Data is provided within the manuscript or supplementary information files.
References
Feng, R. M. et al. Current cancer situation in China: Good or bad news from the 2018 Global Cancer Statistics. Cancer Commun. (Lond) 39(1), 22 (2019).
Wolff, A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline update. J. Clin. Oncol. 31(31), 3997–4013 (2013).
Metelková, A., Skálová, A. & Fínek, J. Breast cancer in young women—Correlation of clinical histomorphological, and molecular-genetic features of breast carcinoma in women younger than 35 years of age. Klin. Onkol. 30(3), 202–209 (2017).
Salimi, M. & Sedaghati, B. S. Integrity and quantity evaluation of plasma cell-free DNA in triple negative breast cancer. Avicenna J. Med. Biotechnol. 11(4), 334–338 (2019).
Hwang, S. Y., Park, S. & Kwon, Y. Recent therapeutic trends and promising targets in triple negative breast cancer. Pharmacol. Ther. 199, 30–57 (2019).
Javadian, M. et al. The role of microRNAs regulating the expression of matrix metalloproteinases (MMPs) in breast cancer development, progression, and metastasis. J. Cell Physiol. 234(5), 5399–5412 (2019).
Jiang, H. & Li, H. Prognostic values of tumoral MMP2 and MMP9 overexpression in breast cancer: A systematic review and meta-analysis. BMC Cancer 21(1), 149 (2021).
Yan, C. et al. Estimation of associations between MMP9 gene polymorphisms and breast cancer: Evidence from a meta-analysis. Int. J. Biol. Markers 37(1), 13–20 (2022).
Vu, H. T., Hoang, T. X. & Kim, J. Y. All-trans retinoic acid enhances matrix metalloproteinase 2 expression and secretion in human myeloid Leukemia THP-1 cells. Biomed. Res. Int. 2018, 5971080 (2018).
Raeeszadeh-Sarmazdeh, M., Do, L. D. & Hritz, B. G. Metalloproteinases and their inhibitors: Potential for the development of new therapeutics. Cells https://doi.org/10.3390/cells9051313 (2020).
Gonzalez-Avila, G. et al. Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer. Crit. Rev. Oncol. Hematol. 137, 57–83 (2019).
Mondal, S., Adhikari, N., Banerjee, S., Amin, S. A. & Jha, T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview. Eur J Med Chem. 194, 112260 (2020).
Gobin, E. et al. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 19(1), 581 (2019).
Matsuzaki, S. et al. Anti-glypican-1 antibody-drug conjugate exhibits potent preclinical antitumor activity against glypican-1 positive uterine cervical cancer. Int. J. Cancer 142(5), 1056–1066 (2018).
Huang, R., Li, J., Pan, F., Zhang, B. & Yao, Y. The activation of GPER inhibits cells proliferation, invasion and EMT of triple-negative breast cancer via CD151/miR-199a-3p bio-axis. Am. J. Transl. Res. 12(1), 32–44 (2020).
Marni, R., Malla, M., Chakraborty, A. & Malla, R. Proteomic profiling and ROC analysis identify CD151 and ELAVL1 as potential therapy response markers for the antiviral drug in resistant TNBC. Life Sci. 320, 121534 (2023).
Kgk, D. & Kumari, S. Marine natural compound cyclo(L-leucyl-L-prolyl) peptide inhibits migration of triple negative breast cancer cells by disrupting interaction of CD151 and EGFR signaling. Chem. Biol. Interact. 315, 108872 (2020).
Li, S. et al. Proteomic landscape of exosomes reveals the functional contributions of CD151 in triple-negative breast cancer. Mol. Cell Proteom. 20, 100121 (2021).
Marni, R., Kundrapu, D. B., Chakraborti, A. & Malla, R. Insight into drug sensitizing effect of diallyl disulfide and diallyl trisulfide from Allium sativum L. on paclitaxel-resistant triple-negative breast cancer cells. J. Ethnopharmacol. 296, 115452 (2022).
Vijay, G. V. et al. GSK3® regulates epithelial-mesenchymal transition and cancer stem cell properties in triple-negative breast cancer. Breast Cancer Res. 21(1), 37 (2019).
Shi, G. M. et al. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology 52(1), 183–196 (2010).
Cao, J. et al. Twist promotes tumor metastasis in basal-like breast cancer by transcriptionally upregulating ROR1. Theranostics 8(10), 2739–2751 (2018).
Luo, C. et al. Progress and prospect of immunotherapy for triple-negative breast cancer. Front Oncol. 12, 919072 (2022).
Zhao, L. et al. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Control Release 318, 1–15 (2020).
Sui, J. et al. Acid-labile polysaccharide prodrug via lapatinib-sensitizing effect substantially prevented metastasis and postoperative recurrence of triple-negative breast cancer. Nanoscale 12(25), 13567–13581 (2020).
Deng, Y., Cai, S., Shen, J. & Peng, H. Tetraspanins: Novel molecular regulators of gastric cancer. Front Oncol. 11, 702510 (2021).
Wang, Y. et al. Neutrophils promote tumor invasion via FAM3C-mediated epithelial-to-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 19(5), 1352–1368 (2023).
Kang, Z., Luo, Y., Xiao, E., Li, Q. & Wang, L. CD151 and prostate cancer progression: A review of current literature. Asia Pac. J. Clin. Oncol. 19(4), 434–438 (2023).
Han, R. et al. Integrin-associated CD151 is a suppressor of prostate cancer progression. Am. J. Transl. Res. 12(4), 1428–1442 (2020).
Palmer, T. D. et al. Integrin-free tetraspanin CD151 can inhibit tumor cell motility upon clustering and is a clinical indicator of prostate cancer progression. Cancer Res. 74(1), 173–187 (2014).
Erfani, S., Hua, H., Pan, Y., Zhou, B. P. & Yang, X. H. The context-dependent impact of integrin-associated CD151 and Other Tetraspanins on cancer development and progression: A class of versatile mediators of cellular function and signaling tumorigenesis and metastasis. Cancers (Basel) 13(9), 2005 (2021).
Tokuhara, T. et al. Clinical significance of CD151 gene expression in non-small cell lung cancer. Clin. Cancer Res. 7(12), 4109–4114 (2001).
Kwon, M. J. et al. Prognostic significance of CD151 overexpression in non-small cell lung cancer. Lung Cancer 81(1), 109–116 (2013).
Zhou, J. et al. Integrin 〈3/〈6 and 〈V are implicated in ADAM15-activated FAK and EGFR signalling pathway individually and promote non-small-cell lung cancer progression. Cell Death Dis. 13(5), 486 (2022).
Zhu, J. et al. CD151 drives cancer progression depending on integrin〈3®1 through EGFR signaling in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 40(1), 192 (2021).
Zhang, P. F. et al. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J. Cell Physiol. 234(3), 2788–2794 (2019).
Lin, W. H. et al. STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT-MET switch and cancer metastasis. Oncogene 40(4), 791–805 (2021).
Zhao, S. J. et al. CD151 promotes breast cancer metastasis by activating TGF-®1/Smad signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 22(21), 7314–7322 (2018).
Augoff, K., Hryniewicz-Jankowska, A., Tabola, R. & Stach, K. MMP9: A tough target for targeted therapy for cancer. Cancers (Basel) 14(7), 1847 (2022).
Zhang, Z. et al. CD151 knockdown inhibits osteosarcoma metastasis through the GSK-3®/®-catenin/MMP9 pathway. Oncol. Rep. 35(3), 1764–1770 (2016).
Ma, Y. et al. Protease activated receptor 2 signaling promotes self-renewal and metastasis in colorectal cancer through ®-catenin and periostin. Cancer Lett. 521, 130–141 (2021).
Wang, J. et al. Correction for: CD36 upregulates DEK transcription and promotes cell migration and invasion via GSK-3®/®-catenin-mediated epithelial-to-mesenchymal transition in gastric cancer. Aging (Albany NY) 14(8), 3720–3721 (2022).
Ma, B. et al. Effects of miR-330-3p on invasion, migration and EMT of gastric cancer cells by targeting PRRX1-mediated Wnt/®-catenin signaling pathway. OncoTargets Ther. 13, 3411–3423 (2020).
Zhang, B. et al. Proton pump inhibitor pantoprazole abrogates adriamycin-resistant gastric cancer cell invasiveness via suppression of Akt/GSK-®/®-catenin signaling and epithelial-mesenchymal transition. Cancer Lett. 356, 704–712 (2015).
Liu, H. et al. EPRS/GluRS promotes gastric cancer development via WNT/GSK-3®/®-catenin signaling pathway. Gastric Cancer 24(5), 1021–1036 (2021).
Pan, J. et al. CD36 mediates palmitate acid-induced metastasis of gastric cancer via AKT/GSK-3®/®-catenin pathway. J. Exp. Clin. Cancer Res. 38(1), 52 (2019).
Jin, C. et al. Acetyltransferase NAT10 regulates the Wnt/β-catenin signaling pathway to promote colorectal cancer progression via ac4C acetylation of KIF23 mRNA. J. Exp. Clin. Cancer Res. 41(1), 345 (2022).
Yang, L. et al. CircLIFR suppresses hepatocellular carcinoma progression by sponging miR-624-5p and inactivating the GSK-3®/®-catenin signaling pathway. Cell Death Dis. 13(5), 464 (2022).
Funding
This work was supported by an Anhui Medical University Foundation Grant (2023xkj159) and 512 Talents Development Project of Bengbu Medical University (by51202205).
Author information
Authors and Affiliations
Contributions
F.L. is responsible for cytology experiments and manuscript writingt; F.L. and L.C. are responsible for the collection of clinical data; F.L. and Q.X. designed the clinical trial and analysed the data; Z.F. provided clinical advice; N.L. made the pathologic diagnosis and supervised the report. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
The study was authorized by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical University (approval no. 2021085).
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, F., Chen, L., Xia, Q. et al. Combined knockdown of CD151 and MMP9 may inhibit the malignant biological behaviours of triple-negative breast cancer through the GSK-3β/β-catenin-related pathway. Sci Rep 14, 21786 (2024). https://doi.org/10.1038/s41598-024-71533-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71533-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.