Abstract
Osteoporosis is usually caused by excessive bone resorption and energy metabolism plays a critical role in the development of osteoporosis. However, little is known about the role of energy metabolism-related genes in osteoporosis. This study aimed to explore the important energy metabolism-related genes involved in the development of osteoporosis and develop a diagnosis signature for osteoporosis. The GSE56814, GSE62402, and GSE7158 datasets were downloaded from the NCBI Gene Expression Omnibus. The intersection of differentially expressed genes between high and low levels of body mineral density (BMD) and genes related to energy metabolism were screened as differentially expressed energy metabolism genes (DE-EMGs). Subsequently, a DE-EMG-based diagnostic model was constructed and differential expression of genes in the model was validated by RT-qPCR. Furthermore, a receiver operating characteristic curve and nomogram model were constructed to evaluate the predictive ability of the diagnostic model. Finally, the immune cell types in the merged samples and networks associated with the selected optimal DE-EMGs were constructed. A total of 72 overlapped genes were selected as DE-EMGs, and a five DE-EMG based diagnostic model consisting B4GALT4, ADH4, ACAD11, B4GALT2, and PPP1R3C was established. The areas under the curve of the five genes in the merged training dataset and B4GALT2 in the validation dataset were 0.784 and 0.790, respectively. Moreover, good prognostic prediction ability was observed using the nomogram model (C index = 0.9201; P = 5.507e−14). Significant differences were observed in five immune cell types between the high- and low-BMD groups. These included central memory, effector memory, and activated CD8 T cells, as well as regulatory T cells and activated B cells. A network related to DE-EMGs was constructed, including hsa-miR-23b-3p, DANCR, 17 small-molecule drugs, and two Kyoto Encyclopedia of Genes and Genomes pathways, including metabolic pathways and pyruvate metabolism. Our findings highlighted the important roles of DE-EMGs in the development of osteoporosis. Furthermore, the DANCR/hsa-miR-23b-3p/B4GALT4 axis might provide novel molecular insights into the process of osteoporosis development.
Similar content being viewed by others
Introduction
Osteoporosis is an age-related chronic bone disease characterized by low bone mineral density (BMD). This disease occurs widely in all racial groups and causes more than 8.9 million fractures per year. Moreover, the fracture risk continues to increase, especially in Asia1,2. Among elderly and middle-aged Chinese residents, 33.49% develop osteoporosis3. The disease commonly manifests in individuals aged 50 years and often goes unnoticed until a fracture occurs. Excessive bone resorption, leading to an imbalance in bone remodeling, has been a common reason for disease development. In addition, late diagnosis greatly increases the risk of poor overall bone health and growth, including osteoporotic fractures in primary areas such as the hip, spine, and wrist4. Therefore, there is an urgent need to identify improved treatment strategies and biomarkers for an early diagnosis.
Bone resorption is mainly controlled by the number and activity of osteoclasts. Similarly, osteoblasts are bone-forming cells that are important for skeleton maintenance and growth. The dynamic processes of energy generation have attracted considerable attention in recent years5. Anabolic treatments have been accepted for osteoporosis, leading to the further exploration of substrate utilization by osteoblasts6. Several pathways related to energy metabolism have been identified in osteoporosis. Wnt signaling is a critical mechanism for increasing aerobic glycolysis and bone accrual. Wnt10b, Wnt7b, and Wnt3a could promote osteoblast differentiation and stimulate lactate production and glucose consumption7,8. Moreover, the functions of mTORC and mTOR in bone formation, which contribute to nutritional coordination, have also been demonstrated9. Organ transplant patients receiving mTOR inhibitors as immunosuppressive agents have a higher incidence of osteoporosis10. However, there remains a limited understanding of the role of energy metabolism-related genes in bone.
This study combined expression profile data from multiple blood tissues with different levels of body mineral density (BMD). First, genes related to energy metabolism (EM) were obtained from the literature11,12,13, and EM genes related to osteoporosis were screened in the expression profile. Diagnostic EM genes and related immune microenvironment signatures were subsequently analyzed using optimization algorithms, and a disease diagnosis model was constructed. A flowchart of this study is shown in Fig. 1.
Materials and methods
Data source
Osteoporosis-related datasets, including GSE56814, GSE62402, and GSE715814,15,16 were downloaded from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/)17. The GSE56814 dataset includes peripheral blood monocytes from 42 and 31 patients with high and low BMD, respectively. GSE62402 included peripheral blood monocytes from five patients with high BMD and five patients with low BMD. These two datasets were sequenced using the [HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array. GSE7158 included peripheral blood monocytes from 14 and 12 patients with high and low BMD, respectively. The data were based on the [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array.
Selection of differentially expressed EM genes
The batch effects of the three datasets were first removed using the sva package (version 3.38.0, http://www.bioconductor.org/packages/release/bioc/html/sva.html)18 in R3.6.1. The combined expression levels were then calculated.
Differentially expressed genes (DEGs) of low and high BMD were screened using the limma package (version 3.34.7)19 in R3.6.1. The thresholds were set as FDR < 0.05 and |log2FC|> 0.263. Directional hierarchical clustering was performed using the pheatmap package (Version 1.0.8, https://cran.r-project.org/package=pheatmap) in R3.6.1 and visualized using a heatmap.
The intersection of DEGs and EM genes was selected as differentially expressed energy metabolism genes (DE-EMGs). The methylation levels of the DE-EMGs were also determined. Finally, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis20,21 were performed using the DAVID software (version 6.8, https://david.ncifcrf.gov/)22,23. GO enrichment analysis was used to investigate the functions of the DE-EMGs, including biological processes (BP), cellular components (CC), and molecular functions (MF). P < 0.05 was defined as the threshold.
Screening of disease-related genes based on WGCNA algorithm
Weighed gene co-expression network analysis (WGCNA) is a bioinformatic algorithm for constructing co-expression networks. The network could identify modules associated with diseases and screen important pathogenic mechanisms or potential therapeutic targets. Modules related to disease status were screened based on genes detected in the merged dataset using the WGCNA package (version 1.61, https://cran.r-project.org/web/packages/WGCNA/index.html)24 in R3.6.1. WGCNA was performed by defining adjacency functions and module partitioning. The threshold for module partitioning screening was set as follows: the module set contained at least 150 genes and cutHeight = 0.995.
The selected DE-EMGs were mapped onto each WGCNA module. A hypergeometric algorithm was used to calculate the fold enrichment parameter and enrichment significance p-value of the differential genes in the module using the following formula: f (k, N, M, n) = C (k, M) × C (n-k, N-M)/C (n, N)25. N represents all genes involved in the WGCNA of the algorithms, M represents the number of genes in each module obtained by the WGCNA algorithm, n represents the number of significantly differentially expressed genes filtered in Step 2, and k represents the number of significantly differentially expressed genes mapped to the corresponding module. Modules with P < 0.05 and Fold enrichment > 1 were selected. Finally, modules significantly enriched in DEGs and DE-EMGs involved in the modules were included for further analysis.
DE-EMG-based diagnostic model
Single-factor logistic regression analysis was conducted using RMS (version 6.3-0, https://cran.r-project.org/web/packages/rms/index.html) in R3.6.1. DE-EMGs with P < 0.05 were included for further analyses. Furthermore, optimal DE-EMGs were screened using the LASSO algorithm in the Lars package (version 1.2, https://cran.r-project.org/web/packages/lars/index.html)26 from R3.6.1.
A diagnostic model based on the optimal DE-EMGs was constructed using a support vector machine in R3.6.1 e1071 (Version 1.6-8, https://cran.r-project.org/web/packages/e1071) on the merged training set. The receiver operating characteristic (ROC) curve of the merged training dataset and independent validation dataset (GSE13850) was constructed using R 3.6.1, pROC (Version 1.12.1, https://cran.r-project.org/web/packages/pROC/index.html)27 to verify the efficiency of the diagnostic model.
The alignment diagram, also known as the nomogram, integrates multiple predictive indicators using multivariate regression analysis. Here, these indicators were visualized on the same plane at a certain scale using scaled line segments to evaluate the interrelationships among the various variables in the predictive model28. The nomogram model and correct line chart were constructed using the RMS package (version 5.1-2; https://cran.r-project.org/web/packages/rms/index.html) in R3.6.1.
Evaluation of immune features based on the ssGSEA algorithm
The microenvironment comprises fibroblasts, immune cells, the extracellular matrix, growth factors, inflammatory factors, and special physicochemical characteristics. Microenvironments significantly affect the diagnosis, survival outcomes, and clinical treatment sensitivity of diseases. Cells in the microenvironment can aggregate into different categories. In addition, there are complex and significant interactions between each cell type and other cells, as well as the infiltration patterns of some robust cells. Immunological signature gene sets were downloaded from the gene set enrichment analysis database (GSEA, http://software.broadinstitute.org/gsea/index.jsp) to evaluate the immune infiltration types in the merged samples. Next, the immune infiltration types of the merged samples were evaluated using the GSVA package (Version 1.36.3)29 in R3.6.1. This was determined based on single-sample GSEA (ssGSEA, http://www.bioconductor.org/packages/release/bioc/html/GSVA.html). The proportion of immune cells in the low- and high-BMD samples was compared using the Kruskal–Wallis test. Finally, the correlation between the expression levels of the optimized DE-EMGs used to construct the diagnostic models and important immune cells was calculated using the COR function in R3.6.1.
Network construction based on DE-EMGs
Construction of TF regulatory relationships
Related TFs targeting DE-EMGs were explored using transcriptional regulatory relationships revealed by sentence-based text mining (TRRUST, https://www.grnpedia.org/trrust/)30. All the related TFs and their target genes were downloaded from the database. The DE-EMGs selected for diagnostic model construction and related TFs were screened for further studies.
Construction of the ceRNA network
The DE-EMGs selected for diagnostic model construction and related miRNAs were searched using miRWalk 3.0 (http://129.206.7.150/)31. Then, lncRNAs related with the selected miRNA were explored using DIANA-LncBasev2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-experimental)32. Similarly, lncRNAs related to osteoporosis were explored using lncRNA Disease (http://www.cuilab.cn/lncrnadisease)33. Next, an lncRNA-miRNA-mRNA network related to osteoporosis was constructed.
Construction of chemical molecules and KEGG related to osteoporosis
Chemical drug molecules and KEGG pathways were explored using the Comparative Toxicogenomics Database 2023 update (http://ctd.mdibl.org/)34. Thereafter, chemical drug molecules targeted by the DE-EMGs were selected for the diagnostic model. Finally, an integrated network of DE-EMGs was constructed for diagnostic models.
Reverse-transcription quantitative PCR (RT-qPCR)
Fresh peripheral blood was collected from eight low-BMD people (6 females, 2 males) and eight high-BMD people (4 females, 4 males). Total RNA from monocytes was extracted by Trizol reagent (GENSTAR Inc. Beijing, China) and was reverse transcribed to cDNA with Primer ScriptTM RT Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Then, real-time PCR was performed on a RT-qPCR machine (Bio-Rad, Hercules, CA, United States) with a SYBR green detection system (TargetMol Chemicals Inc. Shanghai, China). GAPDH was used as internal control. The primers used are listed in Table 1. Written and informed consent was obtained from all participants. The study was approved by the Ethics Committee of Honghui Hospital, Xi’an Jiaotong Universty.
Statistical analysis
The bioinformatics analysis was performed by corresponding packages in R3.6.1. P < 0.05 or FDR < 0.05 was regarded as threshold for statistical significance level when applicable. For the experimental validation, data were analyzed by student’s t test in GraphPad Prism 9.0 (Boston, MA, USA) with significance level of P < 0.05.
Ethics approval and consent to participate
Written and informed consent was obtained from all participants. The study was approved by the Ethics Committee of Honghui Hospital, Xi’an Jiaotong Universty.
Results
Selection of DE-EMGs
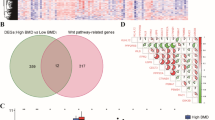
Batch effects of the three datasets were removed. Next, the datasets were merged into one. The participants were then divided into high- and low-BMD groups, which included 61 and 48 samples, respectively. A total of 427 DEGs with high vs. low BMD were obtained. A volcano plot of DEGs is shown in Fig. 2A. A heatmap of DEGs showed different expression levels of DEGs in participants with high BMD compared with those with low BMD (Fig. 2B).
Differentially expressed genes (DEGs) between high- and low-body mineral density (BMD). (A) Volcano plot of DEG selection. The blue and red dots represent significantly downregulated and upregulated DEGs, respectively. The black horizontal line represents a fold discovery rate (FDR) < 0.05, and the two vertical lines represent a | log2 fold change (FC) |> 0.263; (B) The heatmap of DEGs. Black and white bars represent the high- and low-BMD groups, respectively.
A comparison of the EM-related genes with DEGs revealed 72 overlapping genes that were identified as DE-EMGs. Furthermore, GO and KEGG pathway enrichment analysis were performed. Briefly, the identified DE-EMGs were significantly enriched in 43 GO terms (21 BPs, 10 CCs, 12 MFs), such as carbohydrate metabolic, xenobiotic metabolic, and ethanol catabolic processes (Fig. 3A). In contrast, 22 KEGG pathways, including metabolic pathways, xenobiotic metabolism by cytochrome P450, and drug metabolism by cytochrome P450, were enriched in these DE-EMGs (Fig. 3B).
Screening of osteoporosis-related genes based on the WGCNA algorithm
The value of the adjacency matrix weight parameter power was explored to satisfy the precondition of a scale-free network distribution. The range of the network construction parameters was first selected, and then the scale-free distribution topology matrix was calculated.
As shown in Fig. 4A, the value of power was selected when the square value of the correlation coefficient reached 0.9 for the first time (power = 18). The average node connectivity of the constructed co-expression network was 1, which fully conformed to the properties of small-world networks. The dissimilarity coefficient between gene points was then calculated, and a system clustering tree was obtained. The minimum number of genes for each module was set to 150, and the cut height was 0.995. Nine modules were obtained (Fig. 4B). The correlation between the BMD status of the samples and each module is shown in Fig. 4C. MEblue, MEbrown, and MEblack positively correlated with low BMD and negatively correlated with high BMD. The other five modules, namely MEturquoise, MEyellow, MEpink, MEred, and MEgrey, were positively correlated with high BMD and negatively correlated with low BMD.
Screening of osteoporosis-related genes based on the WGCNA algorithm. (A) An adjacency matrix weight parameter power selection graph and the schematic diagram of average connectivity of genes under different power parameters. The horizontal axis represents the weight parameter power, whereas the vertical axis represents the square of the correlation coefficients between log (k) and log (p (k)) in the corresponding network. The red line represents the standard line where the square value of the correlation coefficient reaches 0.9. The red line in the right graph indicates the average connectivity of network nodes under the weight parameter power of the adjacency matrix in the left figure; (B) A module division tree diagram, with each color representing different modules; (C) A module-trait related heatmap.
Here, 354 DEGs were mapped to the modules. As shown in Table 2, 60 DEGs were significantly enriched in the MEturquoise module and 12 DE-EMGs were included namely: ABCC5, ACAD11, ADH4, ADH6, B3GAT3, B4GALT2, B4GALT4, MECR, PC, PGK2, PHKG1, and PPP1R3C.
The expression levels of the 12 DE-EMGs in high- and low-BMD groups are shown in Fig. 5A. Here, the expression levels of DE-EMGs in the high-BMD group were all significantly higher than those in the low-BMD group (P < 0.05). The relationships among these 12 DE-EMGs are shown in Fig. 5B.
Construction and evaluation of the diagnostic model
Single-factor logistic regression analysis was performed to explore genes related to the survival performance of osteoporosis. Ten DE-EMGs were significantly related to the survival performance of osteoporosis (Fig. 6A; P < 0.05). The optimal DE-EMG combination was further analyzed using least absolute shrinkage and selection operator (LASSO) in the lars package in R3.6.1. The parameters of LASSO are shown in Fig. 6B. Finally, five DE-EMGs were selected as optimal for diagnosis, namely: B4GALT4, ADH4, ACAD11, B4GALT2, and PPP1R3C. The differential expression of the five DE-EMGs were further validated by qRT-PCR in our cohort of low-BMD and high-BMD patients. As shown in Fig. 6C, the expression levels of B4GALT4, ADH4, ACAD11 and PPP1R3C were significantly increased (P < 0.01), while B4GALT2 was significantly decreased in high-BMD group (P < 0.05). The experimental validation results showed a relatively high consistency rate with bioinformatics study.
A nomogram model (Fig. 7A) and corrected curves (Fig. 7B) were constructed to evaluate the efficiency of the diagnostic model based on the data from the merged training dataset. B4GALT4 made the greatest contribution to survival. Good predictive ability was observed in the model (C-index = 0.9201; P = 5.507e−14). Furthermore, the areas under the curve (AUC) of the five genes was 0.784, which had the highest predictive value, followed by ADH4 (AUC = 0.768), PPP1R3C (AUC = 0.768), B4GALT4 (AUC = 0.762), ACAD11 (AUC = 0.757), and B4GALT2 (AUC = 0.727; Fig. 7C).
A nomogram model (Fig. 8A) and corrected curves (Fig. 8B) were constructed to evaluate the efficiency of the diagnostic model based on the validation dataset. B4GALT2 made the greatest contribution to survival. The predictive ability of the model showed similar performance to the ideal model (C-index = 0.7841, P = 0.0003513). ROC analysis revealed that the AUC of B4GALT2 was 0.790. This had the highest predictive value, followed by B4GALT4 (AUC = 0.770), 5 gene (AUC = 0.760), ADH4 (AUC = 0.730), ACAD11 (AUC = 0.720), and PPP1R3C (AUC = 0.630; Fig. 8C). The specificity, sensitivity and Youden index is displayed in Table 3.
Immune characteristics based on ssGSEA
The ratios of 28 immune cells were determined using ssGSEA. A heat map of the 28 immune cell distributions between the high- and low-BMD groups is shown in Fig. 9A. Significant differences in five immune cell types were observed between the high- and low-BMD groups, including central memory, effector memory, and activated CD8 T cells, as well as regulatory T cells and activated B cells. The relationship between the five selected DE-EMGs and immune cells was further analyzed (Fig. 9B). Activated B cells and CD8 T cells were positively correlated with the five selected DE-EMGs. Central memory CD8 T cells, regulatory T cells, and effector memory CD8 + T cells were negatively correlated with the five selected DE-EMGs.
Network correlated with DE-EMGs
Seven TFs related to the five selected DE-EMGs were obtained based on the TRRUST database. Then, miRNAs targeting the five DE-EMGs were further explored based on miRWalk 3.0, and 55 DE-EMG-miRNA pairs were obtained. DANCR, a lncRNA related to the five DE-EMGs, was also investigated. Next, miRNAs related to DANCR were explored using DIANA-LncBasev2. Overlapping genes of these miRNAs and those involved in the 55 DE-EMG-miRNA pairs were explored, and seven miRNAs were identified, namely: hsa-miR-125b-2-3p, hsa-miR-146b-3p, hsa-miR-23b-3p, hsa-miR-380-5p, hsa-miR-4755-3p, hsa-miR-6852-5p, and hsa-miR-6889-3p. Furthermore, seven DE-EMG-miRNA pairs were obtained, with hsa-miR-23b-3p associated with osteoporosis.
Forty chemical drug molecules related to DE-EMGs were screened based on the CTD database. This included 17 small molecules, including bisphenol A, cadmium, dexamethasone, drugs, Chinese herbs, estradiol, ethanol, genistein, glyphosate, lipopolysaccharides, methotrexate, perfluoro-n-nonanoic acid, perfluorooctanoic acid, plant extracts, quercetin, resveratrol, streptozocin, and tretinoin.
Eight KEGG pathways related to the DE-EMGs were identified, including tyrosine metabolism, fatty acid degradation, and pyruvate metabolism pathways (Table 4). After comparing the 168 KEGG pathways involved in the CTD, two KEGG pathways were identified: hsa01100: metabolic pathways and hsa00620: pyruvate metabolism. The integrated network related to the five DE-EMGs and osteoporosis is shown in Fig. 10.
Discussion
Energy metabolism has attracted increasing attention in investigations of the pathophysiology of chronic diseases, including osteoporosis. In this study, 72 genes were selected as DE-EMGs, and a diagnostic model of five DE-EMGs were constructed: B4GALT4, ADH4, ACAD11, B4GALT2, and PPP1R3C. ROC and nomogram models confirmed good predictive ability based on the genes. Furthermore, five immune cell types showed significant differences between the high- and low-BMD groups: central memory CD8 + T cells, regulatory T cells, effector memory CD8 + T cells, activated B cells, and activated CD8 + T cells. Networks based on DE-EMGs showed that hsa-miR-23b-3p, DANCR, 17 small-molecule drugs, and two KEGG pathways, including metabolic pathways and pyruvate metabolism, might be involved in osteoporosis development.
B4GALT4, ADH4, ACAD11, B4GALT2, and PPP1R3C may be valuable biomarkers for the development of osteoporosis. ADH4, a critical member of the ADH family, is a well-known prognostic biomarker for hepatocellular carcinoma and is involved in the metabolism of ethanol and retinol. Retinol levels are associated with the occurrence of osteoporosis35,36. Osteoporosis might be caused by poor nutrition. The upregulation of PPP1R3C expression significantly increases glycogen synthesis and storage37. B4GALT family genes are associated with multiple biological processes such as cell apoptosis and proliferation38. Moreover, ACAD11 is required when cells undergo glucose starvation and is involved in fatty acid oxidation39. Thus, we propose that B4GALT4, ADH4, ACAD11, B4GALT2, and PPP1R3C are involved in the development of osteoporosis, potentially by modulating metabolic pathways.
In this study, DANCR was identified as an important lncRNA in the network based on the five selected DE-EMGs. MiR-23b-3p and DANCR act as competing endogenous RNA targeting B4GALT4, whose roles have been previously investigated. Yasuoka et al.40 showed that this gene was mainly involved in the regulation of notochord morphogenesis. Previous studies identified DANCR as an essential mediator of osteoblast differentiation. Further evidence demonstrates that blood mononuclear cells with upregulated DANCR expression could lead to osteoporosis, thereby increasing the secretion of IL-6 and TNF-α as well as bone resorbing activity41. MiR-23b-3p has been verified as a potential biomarker for osteoporosis because it participates in bone mineral density variation42. Moreover, the Wnt/β-catenin signaling pathway has been confirmed as a DANCR and miR-23b-3p targeted pathway in osteoporosis43,44. Our data predicted that metabolic pathways were enriched in the DANCR/hsa-miR-23b-3p/B4GALT4 axis. Osteoporosis is a metabolic bone disease, and its occurrence is related to various metabolic, genetic, and nutritional factors. Previous evidence has shown that a lower BMD is usually found in patients with metabolic syndrome than in those without metabolic syndrome45. Therefore, we hypothesized that the DANCR/hsa-miR-23b-3p/B4GALT4 axis may provide novel molecular insights into the development of osteoporosis.
The imbalance between bone formation and resorption is one of the causes of bone loss. Previous evidence has shown that bone resorption and formation can be modified by complex interactions among dendritic cells, B lymphocytes, and T lymphocytes46. Furthermore, several studies have confirmed the importance of T lymphocytes in the regulation of bone resorption, which might be mediated by various cytokines, such as IFN-γ, TNF-α, and IL-647,48. Patients with osteoporosis have a higher CD4 + /CD8 + ratio compared to the control group49. Our data showed that five immune cell types, central memory CD8 T, regulatory T, effector memory CD8 T, activated B, and activated CD8 + T cells, differed significantly between the high- and low-BMD groups. These data suggest that the ratio of CD8 T cells may be used to assess osteoporosis outcomes.
Our study performed a comprehensive analysis of important EMGs in osteoporosis and developed a diagnosis signature consisting 5 EMGs, including B4GALT4, ADH4, ACAD11, B4GALT2, and PPP1R3C with a relative higher AUC. The differential expression of genes in the model was validated by RT-qPCR. To the best of our knowledge, this is the first study to develop a diagnosis signature based on EMGs. However, there are some limitations in this study. First, the differential expression of the five EMGs were screened from three datasets with individual heterogeneity of background and only validated in a small sample size. Second, the diagnosis signature should still be validated in large sample-size clinical studies in future. Third, the ceRNA network established in this study is warranted for in vivo or in vitro experiments.
Overall, this study constructed a model that can predict the incidence of osteoporosis and identified B4GALT4, ADH4, ACAD11, B4GALT2, and PPP1R3C as potential biomarkers for osteoporosis development. The ratio of CD8 T cells might enable the assessment of osteoporosis outcomes. Furthermore, the DANCR/hsa-miR-23b-3p/B4GALT4 axis may provide novel molecular insights into osteoporosis development. However, further clinical studies with more participants should be conducted to verify this conclusion.
Data availability
The datasets used and/or analysed during the current study available from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/) with accession number of GSE56814, GSE62402, and GSE7158.
Abbreviations
- BMD:
-
Body mineral density
- DE-EMGs:
-
Differentially expressed energy metabolism genes
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene ontology
- BP:
-
Biological processes
- CC:
-
Cellular components
- MF:
-
Molecular functions
- WGCNA:
-
Weighed gene co-expression network analysis
- ROC:
-
Receiver operator characteristic
- GSEA:
-
Gene set enrichment analysis
- LASSO:
-
Least absolute shrinkage and selection operator
References
Sozen, T., Ozisik, L. & Basaran, N. C. An overview and management of osteoporosis. Eur. J. Rheumatol. 4(1), 46–56 (2017).
Clynes, M. A. et al. The epidemiology of osteoporosis. Br. Med. Bull. 133(1), 105–117 (2020).
Wang, J. et al. The prevalence of osteoporosis in China, a community based cohort study of osteoporosis. Front Public Health 11, 1084005 (2023).
Bougioukli, S. et al. Failure in diagnosis and under-treatment of osteoporosis in elderly patients with fragility fractures. J. Bone Miner Metab. 37(2), 327–335 (2019).
Da, W., Tao, L. & Zhu, Y. The role of osteoclast energy metabolism in the occurrence and development of osteoporosis. Front. Endocrinol. (Lausanne) 12, 675385 (2021).
Mosekilde, L., Torring, O. & Rejnmark, L. Emerging anabolic treatments in osteoporosis. Curr. Drug Saf. 6(2), 62–74 (2011).
Esen, E. et al. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 17(5), 745–755 (2013).
Maupin, K. A., Droscha, C. J. & Williams, B. O. A comprehensive overview of skeletal phenotypes associated with alterations in Wnt/beta-catenin signaling in humans and mice. Bone Res. 1(1), 27–71 (2013).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149(2), 274–293 (2012).
Kulak, C. A., Cochenski Borba, V. Z., Kulak, J. & Ribeiro Custodio, M. Osteoporosis after solid organ transplantation. Minerva Endocrinol. 37(3), 221–231 (2012).
Zheng, W., Chen, C., Yu, J., Jin, C. & Han, T. An energy metabolism-based eight-gene signature correlates with the clinical outcome of esophagus carcinoma. BMC Cancer 21(1), 345 (2021).
Tan, C. et al. Molecular signatures of tumor progression in pancreatic adenocarcinoma identified by energy metabolism characteristics. BMC Cancer 22(1), 404 (2022).
Chang, J. J. et al. Comprehensive molecular characterization and identification of prognostic signature in stomach adenocarcinoma on the basis of energy-metabolism-related genes. World J. Gastrointest. Oncol. 14(2), 478–497 (2022).
Zhou, Y. et al. Transcriptomic data identified key transcription factors for osteoporosis in Caucasian women. Calcif. Tissue Int. 103(6), 581–588 (2018).
Zhou, Y. et al. Long noncoding RNA analyses for osteoporosis risk in Caucasian women. Calcif. Tissue Int. 105(2), 183–192 (2019).
Zhou, Y. et al. A novel approach for correction of crosstalk effects in pathway analysis and its application in osteoporosis research. Sci. Rep. 8(1), 668 (2018).
Barrett, T. et al. NCBI GEO: Mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 35, D760-765 (2007).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6), 882–883 (2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7), e47 (2015).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51(D1), D587-d592 (2023).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951 (2019).
da Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4(1), 44–57 (2009).
da Huang, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37(1), 1–13 (2009).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Cao, J. & Zhang, S. A Bayesian extension of the hypergeometric test for functional enrichment analysis. Biometrics 70(1), 84–94 (2014).
Goeman, J. J. L1 penalized estimation in the Cox proportional hazards model. Biom. J. 52(1), 70–84 (2010).
Robin, X. et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Park, S. Y. Nomogram: An analogue tool to deliver digital knowledge. J. Thorac. Cardiovasc. Surg. 155(4), 1793 (2018).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Han, H. et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46(D1), D380–D386 (2018).
Sticht, C., De La Torre, C., Parveen, A. & Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 13(10), e0206239 (2018).
Paraskevopoulou, M. D. et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 44(D1), D231-238 (2016).
Bao, Z. et al. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 47(D1), D1034–D1037 (2019).
Davis, A. P. et al. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 51(D1), D1257–D1262 (2023).
Chowdhury, N. P., Moon, J. & Muller, V. Adh4, an alcohol dehydrogenase controls alcohol formation within bacterial microcompartments in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 23(1), 499–511 (2021).
Karunakara, S. H., Puttahanumantharayappa, L. D., Sannappa Gowda, N. G., Shiragannavar, V. D. & Santhekadur, P. K. Novel insights into MEG3/miR664a-3p/ADH4 axis and its possible role in hepatocellular carcinoma from an in silico perspective. Genes (Basel) 13(12), 2254 (2022).
Lee, S. K. et al. The effect of high glucose levels on the hypermethylation of protein phosphatase 1 regulatory subunit 3C (PPP1R3C) gene in colorectal cancer. J. Genet. 94(1), 75–85 (2015).
Zhou, H. et al. B4GALT family mediates the multidrug resistance of human leukemia cells by regulating the hedgehog pathway and the expression of p-glycoprotein and multidrug resistance-associated protein 1. Cell Death Dis. 4(6), e654 (2013).
Jiang, D. et al. Analysis of p53 transactivation domain mutants reveals Acad11 as a metabolic target important for p53 pro-survival function. Cell Rep. 10(7), 1096–1109 (2015).
Yasuoka, Y. Tissue-specific expression of carbohydrate sulfotransferases drives keratan sulfate biosynthesis in the notochord and otic vesicles of Xenopus embryos. Front Cell Dev. Biol. 11, 957805 (2023).
Tong, X., Gu, P. C., Xu, S. Z. & Lin, X. J. Long non-coding RNA-DANCR in human circulating monocytes: A potential biomarker associated with postmenopausal osteoporosis. Biosci. Biotechnol. Biochem. 79(5), 732–737 (2015).
Ramirez-Salazar, E. G. et al. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene 679, 19–27 (2018).
Wang, C. G., Hu, Y. H., Su, S. L. & Zhong, D. LncRNA DANCR and miR-320a suppressed osteogenic differentiation in osteoporosis by directly inhibiting the Wnt/beta-catenin signaling pathway. Exp. Mol. Med. 52(8), 1310–1325 (2020).
Li, R., Ruan, Q., Yin, F. & Zhao, K. MiR-23b-3p promotes postmenopausal osteoporosis by targeting MRC2 and regulating the Wnt/beta-catenin signaling pathway. J. Pharmacol. Sci. 145(1), 69–78 (2021).
von Muhlen, D., Safii, S., Jassal, S. K., Svartberg, J. & Barrett-Connor, E. Associations between the metabolic syndrome and bone health in older men and women: The Rancho Bernardo Study. Osteoporos. Int. 18(10), 1337–1344 (2007).
Clowes, J. A., Riggs, B. L. & Khosla, S. The role of the immune system in the pathophysiology of osteoporosis. Immunol. Rev. 208, 207–227 (2005).
Bendixen, A. C. et al. IL-4 inhibits osteoclast formation through a direct action on osteoclast precursors via peroxisome proliferator-activated receptor gamma 1. Proc. Natl. Acad. Sci. U S A 98(5), 2443–2448 (2001).
Takayanagi, H. et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature 408(6812), 600–605 (2000).
Fujita, T., Matsui, T., Nakao, Y. & Watanabe, S. T lymphocyte subsets in osteoporosis: Effect of 1-alpha hydroxyvitamin D3. Miner Electrolyte Metab. 10(6), 375–378 (1984).
Author information
Authors and Affiliations
Contributions
Q.W. and L.S. contributed to the study conception and design. Data collection and analysis were performed by W.W., Q.H., N.D., M.L. The first draft of the manuscript was written by Y.L. The manuscript was revised by K.Z. and Z.L. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, Y., Wen, W., Huang, Q. et al. Development and experimental validation of an energy metabolism-related gene signature for diagnosing of osteoporosis. Sci Rep 14, 8153 (2024). https://doi.org/10.1038/s41598-024-59062-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-59062-y
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.