Abstract
In recent years, the global prevalence of obesity and its associated metabolic disorders has reached alarming levels, presenting a significant challenge to public health worldwide. Visfatin, also known as pre-B cell colony-enhancing factor (PBEF) or nicotinamide phosphoribosyltransferase (NAMPT), is an adipokine that has been implicated in various physiological processes, including glucose homeostasis, lipid metabolism, and inflammation. The main objective of this proposed study is to find out the association between visfatin genetic variants and metabolic syndrome. The sample size of the study consisted of 300 blood samples (150 control and 150 cases). This study found that the genotypic frequency of visfatin SNPs, including rs2302559 (OD: 18.222; 95% CI 10.228–32.466; p-value < 0.001) and rs1215113036 (OD: 129.40; 95% CI 44.576–375.693; p-value < 0.001) were significantly associated with metabolic syndrome. Moreover, the frequency of the mutant alleles of both visfatin SNPs was found to be higher in patients with metabolic syndrome as compared to controls. Results of the current study indicate that people with any genetic variation of Visfatin, such as rs2302559 and rs1215113036, are more likely to develop metabolic syndrome. Visfatin genetic variants are linked to an increased risk of metabolic syndrome, implying it’s role in disease pathophysiology.
Similar content being viewed by others
Introduction
Metabolic syndrome has been associated with an increased risk of type 2 diabetes, cardiovascular disease, cancer, and all-cause mortality. As plasma-free fatty acid levels increase, they travel to the liver and are potentiated by chronic lipolysis caused by increased adipose tissue bulk1. Diabetes, dyslipidemia, high blood pressure, and abdominal obesity are all threats to metabolic syndromes. A high-calorie, high-cholesterol diet paired with a sedentary lifestyle, smoking, drinking alcohol, and gaining weight as an adult are all risk factors for metabolic syndrome, which is linked to obesity. Surprisingly, variations in lifestyle can affect the susceptibility to metabolic syndrome due to genetic variations in insulin resistance and fat distribution2,3.
Abdominal obesity and waist circumference are major sponsors of the overall inflammatory state linked with metabolic syndrome, and even people who are minimally overweight but have a high level of abdominal obesity are at risk of unfavorable systemic effects4. The adipose tissue's release of different adipokines may be one of the primary mechanisms underlying these lifestyle-related illnesses5.
Visfatin is an adipokine released by adipose tissues, and it has been linked to obesity and inflammation in the blood. Visfatin performs various biological functions in the human body, including immunomodulation, catalyzing distinct cellular functions, and anti-apoptosis6. Visfatin may be connected to the pathophysiology of diabetes and is essential for the establishment of metabolic syndrome, according to a previous study5. The human visfatin gene is located at the 7q22.1 to 7q31.33 chromosome and encodes a protein comprised of 491 amino acids with a molecular weight of 52 kDa. The gene for visfatin has been effectively maintained throughout evolution7.
Some single nucleotide polymorphisms (SNPs) for Visfatin that were discovered have been linked to problems associated with obesity and glucose/lipid metabolism. Additionally, improvements in insulin sensitivity and glucose tolerance have both been linked to increases in visfatin concentration in obese patients who have been trained in aerobic exercise. Therefore, Visfatin may be a candidate gene for changes in glucose and obesity-related phenotypes brought on by aerobic exercise training, and its gene polymorphisms may be the reason why different reactions to the same activities are observed in different people8,9,10.
The aim of the current study was to investigate the association of visfatin SNPs, including rs2302559 and rs1215113036, with obesity-related metabolic syndrome.
Material and methods
Approval of this case–control study was given by IRB of Dow University of Health Sciences (DUHS). (IRB-1969/DUHS/Approval /2021/353). The complete study was carried out in compliance with the relevant guidelines and regulations of the mentioned authority.
This study included 300 participants of of both sexes, with age ranges between 30 to 60 years. All the participants were divided into two groups: the control group (n = 150), comprised of normal healthy individuals with BMI < 23 kg/m211, and the case group (n = 150), comprised of overweight/obese individuals (BMI ≥ 23 kg/m2)11 who are suffering from metabolic syndrome.
Metabolic syndrome was defined following the criteria provided by the modified NCEP ATP III and IDF groups. According to the modified NCEP criteria12, the presence of any three of the following five factors is required for a diagnosis of Metabolic Syndrome: abdominal obesity, hypertriglyceridaemia (triglycerides ≥ 150 mg/dL); low HDL cholesterol (HDL cholesterol ≤ 40 mg/dL for men and ≤ 50 mg/dL for women); elevated blood pressure (systolic blood pressure ≥ 130 mmHg and/or diastolic blood pressure ≥ 85 mmHg or current use of antihypertensive drugs); impaired fasting glucose (fasting plasma glucose ≥ 100 mg/dL)3.
Individuals suffering from chronic medical conditions like endocrinological problems and cardiovascular and renal diseases were excluded from the study. Written informed consent, along with clinical history, was sought from each participant. All participants of the case group were selected by non-probability purposive sampling technique.
Fasting blood of 6 ml was collected from all participants for biochemical and genetic analysis. Serum Visfatin was evaluated through ELISA techniques while fasting blood glucose analysis and lipid profiling were done by the kit method.
Genomic DNA from whole blood was isolated using a Thersmoscientific DNA isolation kit, which is GeneJET Genomic DNA Purification kit Cat no. K0721. Isolation and purification of DNA was carried out according to the manufacturer’s guidelines.
The genotype distribution of each SNP was checked for the departure from hardy Weinberg equilibrium (HWE) (p > 0.05) using the goodness of fit model among controls. HWE > 0.05 indicates that the SNPs donot differ significantly from population.however HWE < 0.05 could show that consanguinity, population structure or problem in genotyping13.
Human visfatin SNP rs2302559 anchored position is GRCh38.p14 at chromosome 7. It is a synonymous variant in which the T allele is replaced with A/C/G (NCBI; https://www.ncbi.nlm.nih.gov/snp/rs2302559). In contrast, human visfatin SNP rs1215113036 is a missense variant in which the G allele is replaced with the A allele, and it anchored at GRCh38.p14 at chromosome 7 (NCBI; https://www.ncbi.nlm.nih.gov/snp/rs1215113036). Primers for the Visfatin gene were designed using the online software Primer-1®. The specificity of all the designed primers was confirmed using the online BLAST® program/software. Genotyping of the Visfatin gene’s SNPs (i.e., rs2302559 and rs1215113036) was performed by using tARMs PCR analysis. Conditions for tARMS PCR for both visfatin SNPs are given in Table 1.
Fragments obtained after amplification were analyzed using agarose gel electrophoresis, and band sizes of selected SNPs were compared with a 100 bp DNA ladder. The confirmation of DNA sequences of both Visfatin SNPs was done using direct DNA Sequencing, and the sequencing file was analyzed with Mega11 software.
All statistical analyses were performed using SPSS software (SPSS Inc. version 20, Chicago, IL, USA). The distribution of demographic variables between the case group and controls was done by the Kolmogorov–Smirnov (KS) test. At the same time, mean differences of continuous variables between case and control groups were done by independent sample t-test. The Pearson's correlation were used for anthropometric, fasting blood glucose and lipid profile parameters with serum visfatin. Chi-square was used to determine the genotypic frequency between both groups. Relative associations between genotypes of selected visfatin SNPs and both groups (i.e., cases and controls) were assessed by calculating the odds ratios and 95% confidence intervals. P-value < 0.05 was considered statistically significant.
Results
Demographic characteristics of MetS patients and controls
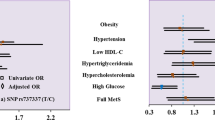
Demographic information was gathered and collected from patients with metabolic syndrome caused by obesity. MetS was significantly associated with age groups in the context of gender and age groups. In contrast, in individuals with HTN, obesity, and diabetes, MetS was found to be significantly (p-value < 0.001) linked with medical history. Positive family history were found significant in individuals with metabolic syndrome, as shown in Tables 2 and 3.
MetS association with anthropometric measurements and general physical parameters
In Table 4, it is shown that individuals with metabolic syndrome had significantly (p-value <0.001) higher levels of weight, body mass index, systolic and diastolic blood pressure, and pulse rate when anthropometric and general physical parameters were compared between the two groups.
MetS patient’s biochemical analysis
Biochemicals, including serum visfatin, serum cholesterol, triglycerides, LDL and VLDL cholesterol, and fasting blood sugar, were found to be significantly (p-value < 0.01) higher in patients with MetS as compared to controls, as shown in Table 5.
Serum Visfatin correlation with anthropometric,general physical and biochemical factors
In contrast to patients, results from Pearson correlation analysis indicated a significant (p-value < 0.001) negative correlation between MetS and diastolic blood pressure in healthy individuals. Overall findings revealed a significant (p-value < 0.001) positive correlation between visfatin and several variables, including age, weight, BMI, systolic blood pressure, pulse rate, fasting blood sugar, serum cholesterol, triglycerides, and LDL cholesterol, as shown in Table 6.
Allele and genotype frequency of visfatin SNPs (rs2302559, rs1215113036)
In patients with MetS and healthy persons, the allele and genotype frequencies of the visfatin gene and its SNPs were also determined. In patients with MetS compared to controls, the homozygous mutant genotype frequency, or CC, for the visfatin SNP rs2302559 was found to be 18-fold higher (OR = 18.222; 95%CI = 10.228–32.466). However, compared to a normal person, MetS patients had a significantly (p-value < 0.001) higher frequency of the mutant allele, C, as mentioned in Table 7. Hardy Wingeberg Equation demonstrate the consistency in the gene pool in Tables 7 and 8.
Amplification results of visfatin rs2302559 SNP showed the homozygous mutant (CC) with 383 bp and 147 bp bands and heterozygous (CT) genotypes with 383 bp, 294 bp, and 147 bp bands, as demonstrated in Fig. 1.
Additionally, it was discovered that individuals with MetS had a 129-fold higher frequency (OR = 129.40; 95% CI = 44.576–375.693) of the heterozygous mutant genotype, or GA, for the visfatin SNP rs1215113036 than did the controls. The frequency of the mutant allele, A, was significantly (p-value < 0.01) higher in MetS patients when compared to controls, as mentioned in Table 7, rs rs2302559 showed concsistency with Hardy Weinberg equation (HWE) while rs1215113036 deviated from HWE as showed in Table 8.
However, amplification results of visfatin rs1215113036 SNP showed the wild type (GG) genotypes with 258 bp and 104 bp bands and heterozygous (GA) genotypes with 258 bp, 208 bp, and 104 bp bands, as demonstrated in Fig. 2.
Sequencing results showed the complete alignment of visfatin gene sequence of selected samples with the wild-type sequence of visfatin gene. Targeted sequences of visfatin SNPs including rs2302559 and rs1215113036 along with some other variations were also confirmed through sequencing analysis as demonstrated in Fig. 3A and B respectively.
Discussion
The World Health Organization defines human obesity as an excessive buildup of adipose tissue (adiposity), which constitutes a body mass index (BMI) > 30 kg/m2. The metabolic syndrome, which includes hyperglycemia, insulin resistance, and dyslipidemia, can be thought of as a group of issues brought on by obesity1,2,3. The development of chronic diseases like type 2 diabetes (T2D) mellitus, non-alcoholic fatty liver disease (NAFLD), and cardiovascular disease (CVD) is supported by the low-grade inflammation known as metabolic syndrome. A high-fat diet is used to imitate parts of metabolic syndrome in mice, where 40–60% of caloric intake comes from dietary fat. This results in diet-induced obesity and related metabolic abnormalities, such as insulin resistance, as well as concomitant low-grade inflammation14,15,16.
The main emphasis of the present study was to investigate the association of visfatin SNPs, including rs2302559 and rs1215113036, with obesity-related metabolic syndrome in local population of Pakistan.
MetS prevalence in morbid obesity increased in both sexes after the age of 54. However, the percentage of women increased dramatically after this age, compared to a more modest increase in the same percentage of men17,18,19,20,21. Earlier studies indicated that those with a positive family history of hypertension had a considerably greater prevalence of hypertension. Additionally, the prevalence of obesity, central obesity and metabolic syndrome is linked to family history of hypertension. These investigations confirmed our findings, which point to a strong correlation between age and a positive family history in patients with metabolic syndrome22.
Insulin resistance is a significant factor in the pathophysiology of the metabolic issues associated with visceral obesity. Insulin activity is thought to be influenced by adipokines; chemical messengers generated by adipocytes. In metabolic disorders, the complex chemical visfatin circulates at higher quantities. In a prior study, it was discovered that obese MetS cases has visfatin levels that are significantly higher than those of controls and MetS people who are not obese. Additionally, visfatin serum concentration increases with increasing BMI and has a favorable correlation with lipid parameters, waist circumference, and BMI. Previous studies have also shown that MetS sufferers had higher amounts of visfatin than controls similarly in our study elevated visfatin level in MetS patients were observed23,24,25.
Several physical factors have been found to be strongly linked to metabolic syndrome, according to research. Weight and BMI (Body Mass Index) are important indicators of metabolic syndrome as excess weight is a significant risk factor for developing this condition. Biochemicals such as serum visfatin, triglycerides, fasting blood sugar, serum cholesterol, LDL (low-density lipoprotein) and VLDL (very low-density lipoprotein) cholesterol have also been found to be significant in metabolic syndrome10. Age, BMI, blood pressure, fasting blood sugar, triglycerides, serum LDL cholesterol have all been found to positively correlate with metabolic syndrome26. Another study reported Metabolic syndrome has been found to have a positive correlation with age, BMI, blood pressure, fasting blood sugar, triglycerides, serum cholesterol, and LDL cholesterol27. In some studies, visfatin found to be associated with cardiovascular metabolic disorders (31). These findings are consistent with our study which revealed a significant (p-value < 0.001) positive correlation between visfatin and several variables, including age, weight, BMI, systolic blood pressure, pulse rate, fasting blood sugar, serum cholesterol, triglycerides, and LDL cholesterol. This means that as these factors increase the likelihood of developing metabolic syndrome. Therefore, it is essential to maintain a healthy weight, manage blood pressure, and control blood sugar and lipid levels to prevent the development of metabolic syndrome and reduce the risk of developing cardiovascular disease and type 2 diabetes.
The analysis of these two SNPs, rs2302559 and rs1215113036, demonstrates different patterns of deviation from Hardy–Weinberg equilibrium. For rs2302559, there is a significant departure from HWE, indicating that some factors, such as selection or genetic drift, are likely influencing the distribution of genotypes. Further research and investigation are necessary to determine the underlying causes. On the other hand, rs1215113036 appears to be in HWE, as there is no strong evidence to suggest a departure from equilibrium. This suggests that the allele frequencies for this SNP are stable and not subject to significant evolutionary pressures in the studied population. HWE analysis is a valuable tool in population genetics to detect departures from the expected genotype frequencies28. The observed deviations from HWE for rs2302559 highlight the need for further investigation into the factors driving this departure, while the adherence to HWE for rs1215113036 suggests genetic stability for this SNP within the population under study. Understanding these genetic patterns can provide insights into evolutionary forces at play in populations and their potential health implications.
Previous investigations have proposed that visfatin may be regulated by insulin, and that visfatin and insulin are more strongly linked than visfatin and glucose. This suggests that visfatin may play a role in glucose metabolism and insulin sensitivity. It is also noted that there have been conflicting findings regarding the relationship between visfatin expression and insulin resistance. This suggests that some studies have found a strong association between visfatin and insulin resistance, while others have not found any significant association7,29,30. A potential link between serum visfatin levels and obesity was found. This implies that obesity could also impact the expression and activity of visfatin in the body, which could potentially affect insulin sensitivity and glucose metabolism10,17,31,32. One possible explanation for these inconsistent findings is the influence of gene variations involved in the production or function of visfatin and insulin. Genetic variations can impact the expression and activity of hormones in the body, and this may explain why some studies have found a strong association between visfatin and insulin resistance, while others have not.
The statement refers to several studies that have investigated the relationship between different genotypes of Visfatin and various anthropometric measurements, such as BMI, waist circumference, waist-to-height ratio, and fat mass. One study found no significant differences between the genotypes of Visfatin rs2302559 and these anthropometric measurements31. In another study, Visfatin levels were not significantly different between SNP rs2302559 genotypes. However, a different study found that carriers of the rs2302559 variant allele had lower fasting serum visfatin and fasting blood glucose levels than wild-type allele carriers in obese children33. Notwithstanding the results of the current study differ from these previous findings. Specifically, the results of the current study indicate that people with any genetic variation of Visfatin, such as rs2302559 and rs1215113036, are more likely to develop metabolic syndrome.
The study done by Javanmard et al., (2016) had investigated whether G-948T(rs2302559) gene polymorphism has an association with obesity and comorbidities such as dyslipoproteinemia in Iranian population. Their findings suggested that variations in this polymorphism were correlated with obesity, total cholesterol, and LDL-C levels in carriers of T allele. This study results were similar to our study34. Increased BMI is associated with an increased risk of developing high blood pressure, high blood sugar levels, and abnormal lipid levels, which are all components of metabolic syndrome. Blood pressure is another important physical factor that is strongly linked to metabolic syndrome34. High blood pressure can damage blood vessels and increase the risk of developing cardiovascular disease. Elevated pulse rate is also an important factor that is often associated with high blood pressure and metabolic syndrome.
The fact that genetic variations in Visfatin are associated with an increased risk of metabolic syndrome suggests that Visfatin may play a role in the development of these conditions. There have been conflicting findings in previous studies regarding the relationship between different Visfatin genotypes and various anthropometric measurements and metabolic parameters. However, the results of the current study suggest that genetic variations in Visfatin may increase the risk of developing metabolic syndrome.
Conclusion
Metabolic syndrome is characterized by multiple factors, including central obesity, dyslipidaemia, hypertension, and impaired glucose tolerance. Most significant aspect of metabolic syndrome is thought to be obesity. Influence of gene variants involved in the synthesis or action of these hormones could also explain the conflicting results when considering the relationship between serum visfatin levels and obesity. However, there is a lot of debate and very little research on these polymorphisms. Even though the SNPs for visfatin rs2302559 and rs1215113036 were found to be linked to metabolic factors in this study, further research is needed to figure out how exactly the studied SNPs affect metabolic mechanism. In order to complete the profile of these polymorphisms and confirm the association at the populational level, additional genetic studies in larger study groups are required.
Data availability
The datasets used and analysed during the current study available from the corresponding authors on reasonable request.
References
Aballay, L. R. et al. Overweight and obesity: A review of their relationship to metabolic syndrome, cardiovascular disease, and cancer in South America. Nutr. Rev. 71(3), 168–179 (2013).
Han, T. S. & Lean, M. E. A clinical perspective of obesity, metabolic syndrome and cardiovascular disease. JRSM Cardiovasc. Dis. 5, 2048004016633371 (2016).
Mendrick, D. L. et al. Metabolic syndrome and associated diseases: From the bench to the clinic. Toxicol. Sci. 162(1), 36–42 (2018).
Cojocaru, K.-A. et al. Mitochondrial dysfunction, oxidative stress, and therapeutic strategies in diabetes, obesity, and cardiovascular disease. Antioxidants 12(3), 658 (2023).
Wnuk, A. et al. Can adipokine visfatin be a novel marker of pregnancy-related disorders in women with obesity?. Obes. Rev. 21(7), e13022 (2020).
Haddad, N., Nori, E. & Hamza, S. A. J. J. J. B. S. Correlations of serum chemerin and visfatin with other biochemical parameters in Iraqi individuals with metabolic syndrome and type two diabetes mellitus. Jordan J. Biol. Sci. 11(4), 4369 (2018).
Sommer, G. et al. Visfatin/PBEF/Nampt: Structure, regulation and potential function of a novel adipokine. Clin. Sci. 115(1), 13–23 (2008).
Kaan, O. et al. Investigation of gene polymorphisms of vaspin, visfatin and chemerin in diabetic obese and non-diabetic obese patients. Sağlık Hizmetleri ve Eğitimi Dergisi 5(2), 33–38 (2021).
Ooi, D. S. Q. et al. In-vitro function of upstream visfatin polymorphisms that are associated with adverse cardiometabolic parameters in obese children. BMC Genom. 17(1), 7 (2016).
Younes, S. et al. 2021 Genetic polymorphisms associated with obesity in the Arab world: A systematic review. Int. J. Obes. 45(9), 1913 (1899).
Misra, A. Ethnic-specific criteria for classification of body mass index: a perspective for Asian Indians and American Diabetes Association position statement. Diabetes Technol. Ther. 17(9), 667–671 (2015).
Grundy, S. M. et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute scientific statement. Circulation 112(17), 2735–2752 (2005).
Wigginton, J. E., Cutler, D. J. & Abecasis, G. R. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 76(5), 887–893 (2005).
Khan, S. et al. The immune landscape of visceral adipose tissue during obesity and aging. Front. Endocrinol. 11, 267 (2020).
Crovesy, L., Masterson, D. & Rosado, E. L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 74(9), 1251–1262 (2020).
Gkastaris, K. et al. Obesity, osteoporosis and bone metabolism. J. Musculoskelet. Neuronal Interact. 20(3), 372 (2020).
Laudisio, A. et al. Metabolic syndrome and quality of life in the elderly: Age and gender differences. Eur. J. Nutr. 52, 307–316 (2013).
Araki, S. et al. Plasma visfatin concentration as a surrogate marker for visceral fat accumulation in obese children. Obesity 16(2), 384–388 (2008).
Jin, H. et al. Serum visfatin concentrations in obese adolescents and its correlation with age and high-density lipoprotein cholesterol. Diabetes Res. Clin. Pract. 79(3), 412–418 (2008).
Nourbakhsh, M. et al. Visfatin in obese children and adolescents and its association with insulin resistance and metabolic syndrome. Scand. J. Clin. Lab. Investig. 75(2), 183–188 (2015).
Misra, A. & Bhardwaj, S. Obesity and the metabolic syndrome in developing countries: focus on South Asians. International nutrition: achieving millennium goals and beyond 78, 133−140 (Karger Publishers, 2014).
Gupta, N., Shah, P., Nayyar, S. & Misra, A. Childhood obesity and the metabolic syndrome in developing countries. Indian J. Pediatr. 80, 28−37 (2013).
Berezin, A. Does visfatin predict cardiovascular complications in metabolic syndrome patients. Endocrinol. Metab. Syndr. 5(1000224), 2161–1017 (2016).
Chang, Y. H. et al. Visfatin in overweight/obesity, type 2 diabetes mellitus, insulin resistance, metabolic syndrome and cardiovascular diseases: A meta-analysis and systemic review. Diabet. Metab. Res. Rev. 27(6), 515–527 (2011).
Dakroub, A. et al. Visfatin: An emerging adipocytokine bridging the gap in the evolution of cardiovascular diseases. J. Cell. Physiol. 236(9), 6282–6296 (2021).
Zatterale, F. et al. Chronic adipose tissue inflammation linking obesity to insulin resistance and type 2 diabetes. Front. Physiol. 10, 1607 (2020).
Mohi, W., Shemran, K. & Alsaffar, Y. Comparison of visfatin and leptin levels in type 2 diabetic patients with and without atherosclerosis. Iran. J. War Public Health 15(1), 11–15 (2023).
Wesley, G., et al., Using the Hardy-Weinberg Equations: Quantifying Natural Selection and Allele Frequencies. UMBC Faculty Collection, 2023.
Sonoli, S. S. et al. Visfatin-a review. Eur. Rev. Med. Pharmacol. Sci. 15(1), 9–14 (2011).
Stastny, J. et al. Visfatin and its role in obesity development. Diabet. Metab. Syndr. Clin. Res. Rev. 6(2), 120–124 (2012).
Chen, C. C. et al. The relationship between visfatin levels and anthropometric and metabolic parameters: Association with cholesterol levels in women. Metabolism 56(9), 1216–1220 (2007).
Ezzati-Mobaser, S. et al. The up-regulation of markers of adipose tissue fibrosis by visfatin in pre-adipocytes as well as obese children and adolescents. Cytokine 134, 155193 (2020).
Vasilache, S.L. et al. Implications of visfatin genetic variants in the metabolic profile of the Romanian pediatric population. Revista Română de Medicină de Laborator 28(2), (2020).
Ugur, K. et al. Asprosin, visfatin and subfatin as new biomarkers of obesity and metabolic syndrome. Eur. Rev. Med. Pharmacol. Sci. 26(6), 2124–2133 (2022).
Author information
Authors and Affiliations
Contributions
S.H.M. conceptualize, design the study, data analysis and carry out all the experimentation. T.A.K. and A.A.B., facilities the resources related to the sampling and reviewing the manuscript S.M.H. cary out the biochemical testing Naqvi, write the manuscript, optimize the Primer and PCR, data analysis, Iqbal, Supervise the study and review the final data set and manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Masood, S.H., Khan, T.A., Baloch, A.A. et al. Association of Visfatin gene polymorphism with obesity related metabolic disorders among Pakistani population: a case control study. Sci Rep 13, 23002 (2023). https://doi.org/10.1038/s41598-023-48402-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-48402-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.