Abstract
Synechococcus are unicellular cyanobacteria susceptible to environmental fluctuations and can be used as bioindicators of eutrophication in marine ecosystems. We examined their distribution in two Moroccan lagoons, Marchica on the Mediterranean coast and Oualidia on the Atlantic, in the summers of 2014 and 2015 using 16S rRNA amplicon oligotyping. Synechococcus representatives recruited a higher number of reads from the 16S rRNA in Marchica in comparison to Oualidia. We identified 31 Synechococcus oligotypes that clustered into 10 clades with different distribution patterns. The Synechococcus community was mainly represented by oligotype 1 (clade III) in Marchica. Cooccurring clades IV and I had an important relative abundance in Marchica in the summer of 2014, which is unusual, as these clades are widespread in cold waters. Moreover, Clades VII and subcluster “5.3” formed a sizeable percentage of the Synechococcus community in Marchica. Notably, we found low Synechococcus sequence counts in the Atlantic Lagoon. These results showed that the relative abundance of Synechococcus reads is not constant over space and time and that rare members of the Synechococcus community did not follow a consistent pattern. Further studies are required to decipher Synechococcus dynamics and the impact of environmental parameters on their spatial and temporal distributions.
Similar content being viewed by others
Introduction
Synechococcus is an important group of Cyanobacteria that contribute to global biogeochemical cycles1. They offer an attractive system to explore bacterial taxa relationships, distribution, coexistence, ecology, and evolution2. The widespread distribution of this group can be attributed to its high degree of genetic diversity3. Synechococcus comprises some of the major forms of Cyanobacteria that inhabit marine and freshwater environments3. Although their genetic diversity has been documented for these ecosystems, there is scant knowledge of the biodiversity, abundance, and distribution of this genus in coastal lagoons. The latter environments are highly productive and valued ecosystems while being morphologically and ecologically complex. Coastal lagoons are subject to more variable environmental conditions than the open sea. Due to relative isolation from the sea and their location within a hydrological catchment, these lagoons are more susceptible to changes in physicochemical parameters, leading to increasing salinity, a decrease in nutrient availability and concentration, and light spectral intensity. These factors influence the adaptation strategies of photosynthetic microorganisms, including Synechococcus species.
The emergence of next-generation sequencing approaches has granted astonishing insight into microbial biodiversity. Notably, the use of the gold standard gene marker for 16S rRNA now enables microbial diversity assessment across the globe in distinct seasons and locations depending on different environmental conditions4. Recently, the use of high-resolution methods, such as oligotyping, has allowed researchers to investigate unexplained diversity within operational taxonomic units and uncover ecologically and biologically distinct taxa5. Based on these techniques, seasonal and geographical behavioral patterns of Synechococcus strain abundances and distributions have been described in several studies6,7,8,9,10. Some studies reported higher abundances in the summer season6,7,8, whereas other studies have shown temporary blooms in spring or summer under eutrophic conditions9,10. In both cases, correlations between physicochemical parameters such as temperature, salinity, and nitrate concentration and Synechococcus abundances were reported8,10,11,12. Spatial differences were also observed; Synechococcus strains were more abundant in coastal waters than in estuaries. Surprisingly, high Synechococcus cells abundance was observed even in polar oceans, which are thought to be devoid of them13. Twenty Synechococcus clades with different patterns of distribution have been described7. Indeed, many Synechococcus clades, including Clades I and IV, have been observed in temperate or polar waters, as well as coastal and higher latitude regions. The question of consistent co-occurrence (for instance, between Clades I and IV) remains partially unanswered, and the role of environmental parameters remains poorly understood.
Synechococcus strains have been classified into three major subclusters (5.1, 5.2, and 5.3) based on 16S rRNA gene phylogeny. Marker genes used to study the diversity of marine Synechococcus were essential to describe an important number of subclades, providing more accurate resolution, such as the internally transcribed spacer (ITS), catalase-peroxidase gene (cpeA), nitrate reductase gene (narB), global nitrogen regulator gene (ntcA), ribulose bisphosphate carboxylase large chain gene (rbcL), DNA-directed RNA polymerase subunit beta gene (rpoC1), and especially petB gene coding for cytochrome b6f. This latter gene helped identify more than thirty subclades3,4. Furthermore, these markers divided the identified subclusters into more than twenty distinct genetic clades4. Marine Synechococcal clades are significantly diverse in terms of depth, temperature, and nutrient availability requirements. Clade II is found in tropical offshore environments, with 'hot spots found in the nutrient-rich coastal upwelling of Morocco and causing seasonal blooms in the Red Sea and the Gulf of Aqaba1. Clade III is predominantly found in tropical and subtropical warm waters1, whereas Clades I and IV occur in nutrient-rich, temperate, and cold environments either nearshore or offshore1. Most of these clades (I, II, III, IV, and CRD1) belong to subcluster 5.1. Marine subcluster 5.2 has been observed in nearshore, coastal, and estuarine environments1. Different clades may co-occur in similar ecological niches, with reports of as many as six clades found at once4. Co-occurrence patterns are observed in coastal waters. Clades II and III co-occurred in the Californian Current during the spring pre-bloom period, while clades I and IV predominated when the bloom itself occurred. Clades V, VI, and X coexist in the Red Sea during transitional periods between mixing and stratification8. Abundant Synechococcus clades are impacted by limiting factors such as light, nutrient availability, temperature, or viral infections8. These factors might also change across seasons and over time scales of environmental changes, leading to clade coexistence7. When less abundant, they may persist at low but stable levels14,15,16 and serve as reservoirs of genetic diversity8.
In this study, we used 16S rRNA amplicon oligotyping to investigate and compare Synechococcus population diversity and co-occurrence patterns in two Moroccan lagoons, Marchica and Oualidia, during the summers of 2014 and 2015. We hypothesized that the distribution and cooccurrence of Synechococcus oligotypes follow spatiotemporal patterns in lagoonal environments.
Results
Detection of Synechococcus oligotypes in environmental samples
A total of 535,138 16S rRNA amplicon sequence reads were generated from the microbial populations of four samples collected at two stations on the summer solstice, 21st June in 2014 and 2015. The relative Synechococcus read number compared to the total microbial community varied between both sampling sites during both timepoints (Fig. 1 and Table 1).
We identified 31 Synechococcus oligotypes, which was equivalent to 95% of all Synechococcus reads analyzed. The most abundant representative Synechococcus reads were used for downstream analyses depending on the sampling location and date. Our phylogeny confirmed that Synechococcus strains are classified into ten different clades based on representative V4–V5 16S rRNA sequences (Fig. 2).
Phylogenetic tree constructed from V4–V5 16S rRNA gene amplicon sequence representatives of Synechococcus clade and representative sequences for the most abundant oligotypes, both aligned using Muscle (version 3.8.4,17). Shown is a neighbor-joining rooted tree generated using the Geneious Prime 2021.1 software package (Biomatters Ltd, Auckland, New Zealand).
The identified clades were separated into three distinct subclusters (5.1, 5.3 and 5.2). Ten oligotypes belonged to clade III (subclade 5.1A) (O1, O15, O19, O21, O23, O24, O25, O29, O30, O31), six belonged to clade I (subclade 5.1B) (O6, O8, O9, O14, O16, O26), another six belonged to Clade 5.3 (O2, O5, O11, O20, O22, O27), two belonged to clade IV (subclade 5.1A) (O4, O12), another two belonged to clade VII (subclade 5.1B) (O3, O7), and only one oligotype belonged to clades II (subclade 5.1A) (O28), CB5 (subclade 5.2) (O10), WPC1 (subclade 5.1) (O17), VIII (subclade 5.1B)(O18), and IX (subclade 5.1B) (O13) (Table S1).
The eight abundant oligotypes in our dataset (i.e., represented by > 100 reads) shared more than 95% V4–V5 sequence similarity with each other (Table 2).
Synechococcus comprised a higher fraction of the microbial population and showed higher relative abundances in the summer of 2014 in Marchica than in 2015. We placed the eight abundant oligotypes within a phylogenetic tree that included known Synechococcus strains (Fig. 2). Table 1 shows higher read counts of Synechococcus oligotypes in the Marchica Lagoon (n = 15,447), (n = 1,667) in the summers of 2014 and 2015, respectively, compared to Oualidia (n = 26), (n = 14) during both the summers of 2014 and 2015.
Interestingly, Oligotype 1 was strongly represented in both sampling lagoons (Table 1), where it comprised a larger segment of the overall Synechococcus community. In contrast to the 2014 summer community in Marchica, a few Synechococcus oligotypes decreased in 2015; for instance, O7 changed from 177 Synechococcus reads to not detected, and O8 changed from 119 Synechococcus reads to not detected. In Oualidia, we noticed the presence of clades III (O1), IV (O4), I (O6, O14, O26), and VII (O7) in 2014 in contrast with 2015, where clade VII was absent and only 5 Synechococcus oligotypes were identified: O1, O4, O6, O9 and O14.
Distribution of Synechococcus oligotypes
Although some oligotype distribution patterns within each sampling site clearly displayed some differences over both space and time, many oligotypes were shared as well. Network analysis allowed visualization of the specificity of the oligotypes and how they were distributed in Mediterranean Marchica and Atlantic Oualidia lagoons and further investigation of which factors influenced this distribution (Fig. 3).
We identified oligotypes that were either found in one collected sample, shared by two samples, or present in all samples (Fig. 3). Oligotypes in OSD24-2014 accounted for the largest fraction (25/31), whereas oligotypes shared by the four samples made up a small portion of the total number (4/31). Most oligotypes were shared between both 2014 and 2015 samples of Mediterranean Marchica, in addition to some overlap with Atlantic Oualidia. Among all oligotypes, six were exclusively found in Marchica in 2014 (Fig. 3). Furthermore, the distribution of cooccurring oligotypes was clearly different in Marchica and Oualidia during the summers of 2014 and 2015. Cooccurring Synechococcus oligotypes tended to be well connected to each other (Table 2), showing strong correlations (e.g., O4 (clade IV) and O6 (clade I), = 0.98, O4 (clade IV) and O8 (clade I), = 0.99).
Environmental variables influencing oligotypes diversity
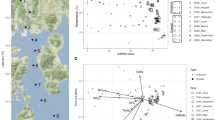
Following principle component analysis (PCA) of oligotype relative abundance, we observed that physicochemical factors in the lagoons correlated with oligotype co-occurrence patterns. The first principle component (PC1) captured 46% of the variance in oligotype relative abundance and discriminated oligotypes according to nitrate. The second principle component explained an additional 27% of the variation and discriminated oligotypes according to salinity and temperature.
The composition shift seen during the summer was supported by the statistical connections between dominating PCs and environmental variables. Notably, identified oligotypes in Marchica (O2, O3, O4, O5, O6, O7, O9, O10, O14, O15, O16, O17, O26) in 2014 and (O1, O2, O4, O5, O6, O9, O10, O14, O15, O16, O17, O26) in 2015 correlated with higher temperatures and salinities (26 Celcius, 27 Celcius, 35 ppt, 35 ppt, respectively), when oligotypes in Oualidia (O1, O4, O6, O7, O14, O26) in 2014 and (O1, O4, O6, O9, O14) in 2015 correlated with lower ones (21 Celcius, 20 Celcius, 27ppt, 29ppt respectively). Both oligotypes found in Marchica and Oualidia in 2014 correlated with a higher nitrate concentration (12 mg/l, 10 mg/l, respectively). However, those observed in 2015 correlated with a lower nitrate concentration (4 mg/l, 2 mg/l). Furthermore, Oligotype O1 in 2014 was spatially isolated, not affiliated with principal component.
Discussion
In a previous study18, we used bioinformatics tools to analyze the metagenome and the amplicon 16S sequences to gain an insight into microbial diversity in Moroccan lagoons, namely Marchica and Oualidia. 16S rRNA gene classification revealed a high percentage of bacteria in both lagoons. On average, bacteria accounted for 90% of the total prokaryotes in Marchica and ~ 70% in Oualidia. The five phyla that were the most abundant in both lagoons, Marchica and Oualidia, respectively, were Proteobacteria (53.62%, 29.18%), Bacteroidetes (16.46%, 43.49%), Cyanobacteria (0.53%, 34.35%), Verrucomicrobia (1.75%, 15.82%), and Actinobacteria (7.42%, 13.98%). At the genus level, we found that the highest assigned hits were attributed to Synechococcus, which was highly abundant in Marchica (32%) compared to Oualidia (0.07%) in 2014. This amount dropped to 22% in Marchica and 0.04% in Oualidia in 2015. Hence, in this study we performed the analysis of the Synechococcus genus community using oligotyping to investigate their dynamics and understand their co-occurrence and covariation in space and time within fragile ecosystems such as lagoons.
We may divide our results into two emerging Synechococcus communities: one dominated in 2014 and the other was less present in 2015, each composed of different cooccurring Synechococcus oligotypes. The abundant Synechococcus community in Marchica in 2014 consisted of clades I, 5.3, III, IV, and VII. These clades are typically found in either warmer or more oligotrophic environments19,20. This result is in accordance with Marchica's environmental characteristics; it is an oligotrophic ecosystem with high primary production and warmer water in summer21. The community included clades CB5 and WPC1 in Marchica 2014 and 2015 when the number of Synechococcus reads was lower. Strains belonging to the CB5 clade lack phycourobilin (PUB), contain one motile strain22,23, are present in temperate coastal waters and are prevalent in polar/subpolar waters24,25,26. WPC1 strains are observed in open-ocean and near-shore waters1,24,27. Clades IV and I usually co-occur and are more prevalent in cold coastal waters19,28,29,30. Interestingly, Clade III was prominent in Marchica. This clade is known to be motile and restricted to warm, oligotrophic water19,20,30. Although at a smaller read number, clade III was also observed in Oualidia, where the temperature is cooler compared to Marchica. Furthermore, we found that clade III growth has been shown to be severely affected at low temperatures30. Moreover, representatives of both clades I and IV were present in Oualidia in both the summers of 2014 and 2015. Some Synechococcus strains, which are known to prefer cooler water temperatures and salinities, were in higher relative abundance in the waters of Marchica. This result agrees with a previous study showing that Synechococcus isolates of clades I and IV exhibited temperature preferences31. Their growth rates were marginally lower at low temperatures in strains from clades I and IV, which were dominant in temperate regions.
Nitrate levels are typically low or undetectable in these lagoons, which allows the persistence of clades that would not typically thrive in coastal waters at other times of the year. In 2014, the nitrate concentration was higher than the average of 10 mg/l, which could be due to increased agricultural activities and wastewater treatment plant effluent21. The decreasing nitrate concentration in Marchica in 2015 could be explained by the newly installed inlet in 2010, which was designed to improve water exchange with the open sea and reduce the amount of suspended matter21. Temperature and salinity have a large effect on nitrate in marine ecosystems32; the highest nitrate degradation rates were observed at 35 °C and at increasing salinity rates. Therefore, we expected to see correlations between salinity, temperature and nitrate concentrations. Interestingly, clades CB5 in Marchica and IV in Oualidia increased in relative abundance in summer 2015 compared to 2014, when the nitrate concentration decreased. Moreover, the Synechococcus microbial community diversity and density are variables depending on the variations in the physical and chemical parameters. These parameters are strongly influenced by the marine waters passing through the artificial inlets, which have an impact on the internal hydrodynamics of both lagoons and hence the distribution and co-occurrence of Synechococcus strains. In addition, anthropogenic activities also have a great influence on Synechococcales population growth and interactions with their viruses33,34.
This study revealed some differences between Marchica and Oualidia in identified Synechococcus clades. The Marchica lagoon showed more heterogeneity (clades I, II, III, IV, VII, VIII, 5.3, WPC1, CB5, and IX) than the Oualidia lagoon, where fewer clades were identified (I, III, IV, and VII). There was a clear variation in the pattern of correlation between oligotypes of the same or different clades for both the 2014 and 2015 samplings. Furthermore, we observed complex patterns of co-occurrence among oligotypes; in 2014 (clades I, III, IV, 5.3, VII), and in 2015, we found clades CB5 and WPC1. In Oualidia, values decreased in comparison to Marchica in both 2014 and 2015 summer samplings, following a pattern of co-occurrence, especially for both clades I and IV in both sampling years. Many studies have shown that the relative proportions of cooccurring Synechococcus populations to each other at the clade and subclade levels vary in space and time based on environmental factors such as seasonal temperature fluctuations, nutrient availability and upwelling, circulation patterns, and abundance of other phytoplankton8.
We presume that the greater variability in oligotype co-occurrence behavior observed in Marchica Lagoon, especially in the summer of 2014, could be due to the higher abundance and diversity of Synechococcus oligotypes, physico-chemical parameter fluctuations or rehabilitation of the lagoon.
Less abundant oligotypes could also be considered potential bioindicators of Synechococcus genetic diversity. Their seasonal occurrence might contribute to changing ecological and biogeochemical characteristics of the marine environment35. The Synechococcus relative abundance count revealed that the Marchica Synechococcus community included the least abundant oligotypes in 2015. For instance, O7 and O8 were detected in 2014 and were absent in 2015 (Table 1). It is unclear which factors served to constrain the relative abundances of these least present oligotypes, but temperature and salinity could have an impact on their distribution in Marchica (Fig. 4) and the opposite for Oualidia, which are cooler-temperature adapted ones. We noticed that the relative abundance of cooccurring Synechococcus was not constant. For instance, oligotype 4 belonging to Clade IV showed higher values in summer 2014 (974 reads) in Marchica compared to summer 2015 (319 reads), and the opposite was observed in Oualidia, with a lower abundance compared to Marchica. Increased values of cooccurring clade I oligotypes (14, 26, and 6) were detected in the summer of 2014 in both lagoons.
Principle component analysis of Synechococcus oligotype relative abundance. The plot is generated using the relative abundance of each oligotype, T temperature, S Salinity, and NO3− Nitrate. Each point represents an oligotype. Colors represent the year of sampling; red for 2014 and blue for 2015. The shape of point indicates the sampling site; rounded points refer to Marchica lagoon, and triangles refer to Oualidia. Circles represent the normal distribution of oligotypes; the red circle refers to 2014, and the blue one refers to 2015.
In comparing our results with a study from Little Sippewissett Marsh (LSM)8 that used oligotyping to investigate the distribution of the genus Synechococcus in space and time sequencing the V4-V6 hypervariable region of the 16S rRNA gene, we found 31 oligotypes, while they identified 12. In both studies, the proportion of Synechococcus oligotypes increased in summer and in coastal waters compared to estuaries. In addition, Clades I and IV were more abundant in saline conditions, such as Marchica Lagoon. However, these clades were found in greater relative abundances at cold temperatures, in contrast to our study, where they were identified in Marchica's warm waters. Moreover, clade CB5 tended to be prominent at relatively warm temperatures (17–20 °C)6. In our work, it was not prevalent either in cooler or warmer water. Notably, the relative abundance of rare oligotypes was higher in warm hypersaline estuary waters8,18, while in our case study, they occurred in cooler moderately saline Oualidia waters.
The dominance of a certain clade could have many different ecological ramifications, especially as the clades can be incredibly diverse in their growth, loss, nutrient utilization and other attributes. The dominant clade's growth and loss patterns will set the stage for the population dynamics. For instance, if the dominant clade only blooms in a given environmental factor such as temperature, light, or salinity, it will then affect the timing of blooms, and follow-on the effects of subsequent grazing, lysis or even biogeochemical cycling. Even if the population is diverse, the dynamics as a whole will be a composite response of each individual clade's ecophysiology, making it important to understand their composition and how it changes over space and time.
While the rpoC1 gene is a higher resolution diversity marker36, 16S amplicon data can be used for exploring the entire bacterial assemblage including Synechococcus clade designations via oligotyping35. The latter has a great advantage in answering unexplained diversity contained in taxa using 16S rRNA gene sequences. Nevertheless, it has some limitations, as it acts optimally only when performed on taxa that are closely related. Regarding distantly related taxa, the high number of increased-entropy locations makes the supervision steps difficult. In addition, although oligotyping does not rely on clustering conditions or availability of existing reads within reference databases, it demands preliminary operational taxonomic unit clustering to find closely related species appropriate for the analysis. This method is under continuous improvement to better exploit the information within subtle variations in 16S rRNA gene sequences5.
In conclusion, we explored the patterns of Synechococcus diversity in space and time using an oligotyping approach to examine these populations in lagoon waters of Mediterranean Marchica and Atlantic Oualidia, in Morocco. Patterns that have been observed at the clade and subclade levels, such as Synechococcus, relative abundance and the co-occurrence of groups from different clades, were shown to occur among oligotypes. The Marchica Lagoon showed a heterogeneous Synechococcus diversity compared to Oualidia in summer 2014. Thirty-one Synechococcus oligotypes were identified. Two distinct communities emerged in the 2014 and 2015 summer samplings, abundant and rare Synechococcus species, each comprising cooccurring Synechococcus oligotypes from different clades. Network analysis showed that six oligotypes were exclusive to Marchica Lagoon. The identified clades I, III, IV, VII, and 5.3 in Marchica were in accordance with its environmental characteristics. In addition, the relative abundance of some cooccurring Synechococcus strains was not constant over time and space (e.g., clades I and IV). Using gene oligotyping, we illustrated some of the challenges associated with the identification of novel Synechococcus strains or studied their co-occurrence in space and time. Oligotyping has been instrumental in discriminating closely related Synechococcus strains. However, this study leaves open questions about how samples differ by location and whether locations differ from year to year. Do cooccurring oligotypes interact with each other and to what extent do they correlate with physicochemical parameters? What triggers the coexistence of clades I and IV with clade III in warm water or 5.3 with VII, which do not know much about. Finally, how do relative abundances change over seasons. Hence, future work needs to consider additional stations and seasons to provide better statistical support for our findings and to better understand their correlation with physical and chemical environmental parameters. Other factors were not considered in this study, such as nutrient availability, chlorophyll, irradiance, viral lysis, and greater sequencing depth, which could also influence the observed seasonal dynamics.
Methods
Sampling and sequencing
Samples were collected from Marchica Lagoon (N 35.11562, W 2.52803) and Oualidia Lagoon (N 32.74675, W 9.036667) on June 21st, 2014, and 2015, boreal summer solstice, as part of the Ocean Sampling Day (OSD) campaign (Fig. 5). Approximately 20 L was collected using a 10% acid-washed bucket and then sequentially filtered onto a 0.22 μm pore size Sterivex and frozen at − 80 °C until DNA extraction. Metadata (temperature, salinity, and nitrate) were measured and uploaded into https://github.com/MicroB3-IS/osd-analysis/wiki/Guide-to-OSD-2014-dataaccessec on 1 December 2021.
DNA was extracted using the Power Water isolation kit (MoBio, Carlsbad, CA, USA) following the manufacturer’s instructions. Amplification of the 16S rRNA gene was performed using the primer pair, designated: 515F-Y (5’-GTGYCAGCMGCCGCGGTAA-3’) and 926R (5’-CCGYCAATTYMTTTRAGTTT-3’)37. The Illumina libraries were prepared using the NuGEN Ovation Rapid DR Multiplex System 1–96. Amplicon gene sequencing (2 Å ~ 250 paired ends) was performed with Illumina MiSeq using V3 chemistry. Samples were sequenced in eight MiSeq runs (2 × 300 bp), which generated 2 × 40,000 amplicon reads per sample.
Data processing
Raw sequencing reads were preprocessed as described in the OSD workflow (github.com/MicroB3-IS/osd-analysis/wiki/Sequence-Data-Preprocessing_accessed on December 1, 2021), which produced "workable" amplicon fasta files. We used VAMPS38 to process 16S rRNA gene sequences, where taxonomy assignment was performed using Global Alignment for Sequence Taxonomy (GAST)39 and the SILVA rRNA gene reference database40. The obtained files include the reference ID, the taxonomy assigned, and the source of the taxonomy.
Oligotyping
For Synechococcus investigation, we used 16S rRNA gene oligotyping as described in5. This method is based on a supervised algorithm that identifies microdiversity using 16S rRNA gene sequences. Oligotyping is unlike regular taxonomic classification based on available reference databases available sequences or cluster analysis based on the selection of the similarity threshold. This technique tackles the taxonomic resolution limitation by finding the most information-rich nucleotide positions (i.e., oligotypes). Sequences identified as Synechococcus were extracted from the Vamps database. We aligned Synechococcus reads using PyNAST41. Of the 22,387 sequences identified as Synechococcus, 17,941 remained after quality filtration and Pynast alignment. The mean length of Synechococcus reads was 254 bp. Next, we removed the uninformative gaps in the resulting aligned sequences using the “o-trim-uninformative-columns-from-alignment” script. Subsequently, we calculated the entropy of each nucleotide position within the oligotype package. After the initial calculation of Shannon entropy using the “analyze-entropy” script, we ran 16S rRNA oligotyping for the Synechococcus genus until each oligotype had converged. Uninformative nucleotide positions were excluded. Seven nucleotide positions were used in total to define each oligotype, and to minimize the impact of sequencing errors on oligotyping results, we used a “minimum substantive abundance” criterion (M) of 5; thus, an oligotype was not included if the most common sequence for that type occurred less than five times. To reduce the noise, each oligotype was required to appear in at least one sample but was not required to comprise a certain percentage of reads or represent a minimum number of reads in all samples combined. We removed any oligotypes that did not meet these criteria from the analysis. The final number of quality-controlled oligotypes revealed by the analysis was 31 and represented 95% of the total Synechococcus reads. For each oligotype, the oligotyping pipeline chose the most abundant read as the representative sequence to be used for downstream analyses. Upon completion of oligotyping analysis, the resulting “observation matrices” are concatenated to generate a single “observation matrix” for our V4-V5 dataset. These observation matrices report counts, which are the number of reads assigned to each oligotype in each sample (Table 1). We then converted counts to percent abundances within each sample and used these normalized relative abundances for subsequent analyses. We searched the most biologically relevant representative sequence of our oligotypes using blastn version 2.2.26 to assign taxonomy for each oligotype. We kept default parameters, except ‘per. identity 100’ to have hits with 100% sequence identity reported.
Oligotype network analysis
We performed network analysis using Gephi software, version 0.9.242, to determine the distribution of all Synechococcus oligotypes from both lagoons using a force-directed graph algorithm (ForceAtlas2 in Gephi software). Every dot identifies an oligotype present in at least one sampling site, and each edge on the network connects an oligotype to one or more sampling sites.
Clade identification
We designated a clade for each oligotype’s representative sequence by matching this latter to a key reference database of 16S rRNA gene sequences from cultured Synechococcus6. Synechococcus sequences downloaded from NCBI GenBank clade classifications were obtained from the following sources4,6. We added the representative sequences for each oligotype to the Synechococcus database, and we aligned them with Muscle (version 3.8.4,17). We used exact matches between each oligotype Synechococcus sequence and the Synechococcus sequence database to infer clade designation.
Statistical analyses
To group oligotypes statistically, we computed a principal components analysis (PCA) using R package “ggfortify” with respect to a sample matrix of Synechococcus oligotype reads normalized to total Synechococcus reads for that sample. Each oligotype was projected onto the first two PCs of the matrix. To investigate environmental correlates of oligotype grouping, multiple regressions of each of the first two PCs were computed against the three environmental factors, which are the water temperature, salinity, and the concentration of nitrate.
Data availability
Datasets can be found at EBI under the Accession number ERX947554. The bacterial metagenome and 16S rRNA raw read sequence datasets were deposited in EMBL-EBI https://www.ebi.ac.uk/metagenomics/studies/MGYS00001977 accessed 1 December 2021 and OSD GitHub https://github.com/MicroB3-IS/osd-analysis/wiki accessed 1 December 2021.
References
Mazard, S., Ostrowski, M., Partensky, F. & Scanlan, D. J. Multilocus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ. Microbiol. 14(2), 372–386. https://doi.org/10.1111/j.1462-2920.2011.02514.x (2012).
Scanlan, D. J. & West, N. J. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40, 1–12 (2002).
Scanlan, D. J. et al. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299. https://doi.org/10.1128/MMBR.00035-08 (2009).
Ahlgren, N. A. & Rocap, G. Diversity and distribution of marine Synechococcus: Multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front. Microbiol. 3, 213. https://doi.org/10.3389/fmicb.2012.00213 (2012).
Eren, A. M. et al. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 4, 1111–1119. https://doi.org/10.1111/2041-210X.12114 (2013).
Xia, X., Vidyarathna, N. K., Palenik, B., Lee, P. & Liu, H. Comparison of the seasonal variations of Synechococcus assemblage structures in estuarine waters and coastal waters of Hong Kong. Appl. Environ. Microbiol. 81, 7644–7655. https://doi.org/10.1128/AEM.01895-15 (2015).
Hunter-Cevera, K. R., Post, A. F., Peacock, E. E. & Sosik, H. M. Diversity of Synechococcus at the Martha’s Vineyard Coastal observatory: Insights from culture isolations, clone libraries, and flow cytometry. Microb. Ecol. 71, 276–289. https://doi.org/10.1007/s00248-015-0644-1 (2016).
Mackey, K. R. M. et al. Seasonal succession and spatial patterns of Synechococcus Microdiversity in a salt marsh estuary revealed through 16S rRNA gene oligotyping. Front. Microbiol. 8, 1496. https://doi.org/10.3389/fmicb.2017.01496 (2017).
Nagarkar, M., Wang, M., Valencia, B. & Palenik, B. Spatial and temporal variations in Synechococcus microdiversity in the Southern California coastal ecosystem. Environ. Microbiol. 23(1), 252–266. https://doi.org/10.1111/1462-2920.15307 (2020).
Robidart, J. C. et al. Seasonal Synechococcus and Thaumarchaeal population dynamics examined with high resolution with remote in situ instrumentation. ISME J. 6, 513–523. https://doi.org/10.1038/ismej.2011.127 (2012).
Rajaneesh, K. & Mitbavkar, S. Factors controlling the temporal and spatial variations in Synechococcus abundance in a monsoonal estuary. Mar. Environ. Res. 92, 133–143. https://doi.org/10.1016/j.marenvres.2013.09.010 (2013).
Flombaum, P. et al. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. https://doi.org/10.1073/pnas.1307701110 (2013).
PaulsenML, DoréH. et al. Synechococcus in the Atlantic Gateway to the Arctic Ocean. Front. Mar. Sci. 3, 191. https://doi.org/10.3389/fmars.2016.00191 (2016).
Pedrós-Alió, C. Marine microbial diversity: Can it be determined?. Trends Microbiol. 14, 257–263. https://doi.org/10.1016/j.tim.2006.04.007 (2006).
Sogin, M. L. et al. Microbial diversity in the deep sea and theunderexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120. https://doi.org/10.1073/pnas.0605127103 (2006).
Fuhrman, J. A. Microbial community structure and its functional implications. Nature 459, 193–199. https://doi.org/10.1038/nature08058 (2009).
Edgar, R. C. MUSCLE: Multiple sequence align-ment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. https://doi.org/10.1093/nar/gkh340 (2004).
Chaouni, B. et al. Moroccan lagoon microbiomes. Water 14(11), 1715. https://doi.org/10.3390/w14111715 (2022).
Zwirglmaier, K. et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10(1), 147–161. https://doi.org/10.1111/j.1462-2920.2007.01440.x (2008).
Post, A. F. et al. Long term seasonal dynamics of Synechococcus population structure in the Gulf of Aqaba, Northern Red Sea. Front. Microbiol. 2, 131. https://doi.org/10.3389/fmicb.2011.00131 (2011).
Aknaf, A. et al. Study of the spatial and temporal variation of physical-chemical parameters characterizing the quality of surface waters of the lagoon Marchica–North-East Morocco. J. Mater. Environ. Sci. 8, 3216–3225 (2017).
Chen, F. et al. Phylogenetic diversityof Synechococcus in the Chesapeake Bayrevealed byribulose-1,5 bisphosphate carboxylase-oxygenase (RuBisCO) large subunit gene (rbcL) sequences. Aquat. Microb. Ecol. 36, 153–164. https://doi.org/10.3354/ame036153 (2004).
Waterbury, J. B., Watson, S. W., Valois, F. W. & Franks, D. G. Biological and ecological characterization of the marine unicellular cyanobacteria Synechococcus. Can. J. Fish. Aquat. Sci. 214, 71–120 (1986).
Chen, F., Wang, K., Kan, J., Suzuki, M. & Wommack, K. Diverse and unique picocyanobacterian ChesapeakeBay, revealedby16S-23S rRNAinternaltranscribedspacer sequences. Appl. Environ. Microbiol. 72, 2239–2243. https://doi.org/10.1128/AEM.72.3.2239-2243.2006 (2006).
Cai, H., Wang, K., Huang, S., Jiao, N. & Chen, F. Distinct patterns of picocyanobacterial communities in winter and summer in the Chesapeake Bay. Appl. Environ. Microbiol. 76, 2955–2960. https://doi.org/10.1128/AEM.02868-09 (2010).
Huang, S. et al. Novel lineages of Prochlorococcus and Synechococcus in theglobaloceans. ISME J. 6, 285–297 (2012).
Choi, D. & Noh, J. Phylogenetic diversity of Synechococcus strainsisolatedfromtheEastChina Sea andtheEast Sea. FEMS Microbiol. Ecol. 69, 439–448. https://doi.org/10.1111/j.1574-6941.2009.00729.x (2009).
Zwirglmaier, K. et al. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ. Microbiol. 9, 1278–1290 (2007).
Tai, V. & Palenik, B. Temporal variation of Synechococcus clades at a coastalPacific Ocean monitoring site. ISME J. 3, 903–915. https://doi.org/10.1038/ismej.2009.35 (2009).
Mella-Flores, D. et al. Is the distribution of Prochlorococcus and Synechococcus ecotypes in the Mediterranean Sea affected by global warming?. Biogeosciences 8(9), 2785–2804. https://doi.org/10.5194/bg-8-2785-2011 (2011).
Varkey, D. et al. Effects of low temperature on tropical and temperate isolates of marine Synechococcus. ISME J. 10, 1252–1263. https://doi.org/10.1038/ismej.2015.179 (2016).
Ebrahimi, S., ThiHau Nguyen, T. H. & Roberts, D. J. Effect of temperature & salt concentration on salt tolerant nitrate-perchlorate reducing bacteria: Nitrate degradation kinetics. Water Res. 83, 345–353. https://doi.org/10.1016/j.watres.2015.07.006 (2015).
Traving, S. J., Clokie, M. R. & Middelboe, M. Increased acidification has a profound effect on the interactions between the cyanobacterium Synechococcus sp. WH7803 and its viruses. FEMS Microbiol. Ecol. 87(1), 133–141. https://doi.org/10.1111/1574-6941.12199 (2014).
Gilbert, J. A. et al. Defining seasonal marine microbial community dynamics. ISME J. 6, 298–308. https://doi.org/10.1038/ismej.2011.107 (2012).
Hunter-Cevera, K. R., Hamilton, B. R., Neubert, M. G. & Sosik, H. M. Seasonal environmental variability drives microdiversity within a coastal Synechococcus population. Environ. Microbiol. 23(8), 4689–4705. https://doi.org/10.1111/1462-2920.15666 (2021).
Wang, T., Xia, X., Chen, J., Liu, H. & Jing, H. Spatio-temporal variation of Synechococcus assemblages at DNA and cDNA levels in the tropical estuarine and coastal waters. Front. Microbiol. 13, 837037. https://doi.org/10.3389/fmicb.2022.837037 (2022).
Parada, A. E., Needham, D. M. & Fuhrman, J. A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18(5), 1403–1414. https://doi.org/10.1111/1462-2920.13023 (2016).
Huse, S. M. et al. VAMPS: A website for visualization and analysis of microbial population structures. BMC Bioinform. 15, 41. https://doi.org/10.1186/1471-2105-15-41 (2014).
Huse, S. M. et al. Exploring microbial diversity and taxonomy using SSU RRNA hypervariable tag sequencing. PLoS Genet. 4, e1000255. https://doi.org/10.1371/journal.pgen.1000255 (2008).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. https://doi.org/10.1093/nar/gks1219 (2013).
Caporaso, J. G. et al. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26(2), 266–267. https://doi.org/10.1093/bioinformatics/btp636 (2010).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: An open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 3(1), 361–362 (2009).
Acknowledgements
We are grateful to Dr. Kristen Hunter-Cevera for early discussions of the dataset, providing Synechococcus database access and reviewing the manuscript, Richard Fox for his advice and assistance with the bioinformatics analysis on the MBL server, and Dr. Christiane Hassenrück for her constructive comments that significantly improved the manuscript.
Funding
This work was supported by the EU MicroB3 project, funded by the European Union’s Seventh Framework Programme (FP7; Joint Call OCEAN.2011-2: Marine microbial diversity-new insights into marine ecosystems functioning and its biotechnological potential) under grant agreement no 287589. Bouchra Chaouni is a recipient of the co-joint supervision Fulbright fellowship at the Biological Marine Laboratory, Woods Hole, MA, USA, for the 2017/18 academic year 2017/18. Hassan Ghazal is a US NIH grant recipient through the H3abionet/H3africa consortium U24HG006941.
Author information
Authors and Affiliations
Contributions
Conceptualization, H.G., L.A.-Z.; methodology, H.G., L.A.-Z., B.C.; software, B.C., A.I.A.; validation, H.G.; formal analysis, B.C.; investigation: H.G., B.C.; resources, S.R., S.A.; data curation, B.C.; writing—original draft preparation, B.C.; writing—review and editing, H.G., L.A.-Z.; visualization, B.C.; supervision, H.G., L.A.-Z., E.H.Z., N.H., F.B.; project administration H.G.; Funding acquisition, H.G., L.A.-Z., C.N. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chaouni, B., Idrissi Azami, A., Raoui, S. et al. Spatio-temporal patterns of Synechococcus oligotypes in Moroccan lagoonal environments. Sci Rep 13, 110 (2023). https://doi.org/10.1038/s41598-022-27263-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-27263-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.