Abstract
The performance of ozonation for the removal of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) using Escherichia coli and Pseudomonas aeruginosa carrying ARGs from hospital wastewaters was evaluated in this study. Bacterial inactivation was determined using plate count methods and real time PCR for ARG damage (Sul1, blatem, blactx, blavim and qnrS). The reduction rate of bacterial cells and ARGs was increased by different amounts of transferred ozone dose from 11 to 45 mg/L. The concentration of 108 cfu/ml bacteria was reduced to an acceptable level by ozone treatment after a 5 min contact time, Although the removal rate was much higher for concentrations of 106 cfu/ml and 104 cfu/ml bacteria. Overall, the tendency of gene reduction by ozonation from more to less was 16S rRNA > sul1 > blatem > blactx > qnrS > blavim. Given that plasmid-borne ARGs can potentially be transferred to other bacteria even after the disinfection process, our results can provide important insights into the fate of ARGs during hospital wastewater ozonation.
Similar content being viewed by others
Introduction
Hospital wastewaters are identified as sources of nutrients, inorganic and organic pollutants and are suspected to be among the main anthropogenic sources of antibiotics, ARB (Antibiotic-resistant bacteria) and ARGs (Antibiotic resistance genes) spread into the environment. Indeed, antibiotic residues are almost enough to kill susceptible bacteria and at the same time increase the number of resistant bacteria1,2,3,4,5. Antibiotics are one of the important drugs for treating infectious diseases that are released into hospital wastewaters6,7,8. Many of these bacteria harbor acquired antibiotic resistance genes and are potential carriers for the dissemination of these genes in the environmental microbiome9,10. Antibiotic-resistant bacteria can survive in hospital wastewaters, so conventional wastewater treatment has unacceptable results in terms of ARB and ARGs removal and might even increase the concentration of certain ARGs11,12. Antibiotic resistance genes may be intrinsic or acquired. Intrinsic resistance is created in an organism whose innate chromosomal makeup predictably specifies resistance, whereas acquired resistance is due to the mutation in its genetic composition12.
In recent years, the application of new disinfection technologies, such as advanced chemical oxidation processes (AOPs) (e.g., ozonation and homogeneous and heterogeneous photocatalysis), has been paid attention for the inactivation of ARBs and the removal of ARGs in hospital wastewaters13,14. Ozonation processes have been used in hospital wastewater treatment since the 1950s, and numerous studies have shown that ozone treatment is efficient in killing many types of microorganisms, such as S. aureus, Streptococci spp., Escherichia coli, Enterococcus faecali and Pseudomonas aeruginosa15,16. Ozone is a gas molecule and allotrope of oxygen and reacts with organic and inorganic compounds due to its high oxidation potential and reactivity17,18. Ozonation is an effective process to remove organic pollutants and is used to reduce or inactivate pathogens by producing highly reactive radicals. Leakage of substances from the cell wall due to cell wall degradation and damage to nucleic acids and bonds of carbon–nitrogen of proteins are among the mechanisms of ozone disinfection that types of organism, contact time and radicals concentration affect this mechanism7,19,20,21,22,23. Ozone selectively reacts with organic compounds by reacting directly with ozone molecules, or indirectly with hydroxyl radicals (OHº) under alkaline pH conditions, resulting from the decomposition of ozone in hospital wastewater. The hydroxyl radical is considered a very strong non-selective oxidant that its reaction with most organic pollutants occurs either by hydrogen abstraction with saturated aliphatic hydrocarbons and alcohols, or electrophilic addition with unsaturated hydrocarbons13,15.
The reason for the selection of Escherichia coli and Pseudomonas aeruginosa for this study was the significant ability of these bacteria to survive on the hospital surface, the increase in antimicrobial resistance against pathogens and according to the report of the World Health Organization based on the serious threat of these bacteria to human health 24. Considering that ARGs such as blatem, blactx, blavim, Sul1 and qnrs have been found in hospital wastewaters worldwide22,25 and given that these antibiotic genes are frequently suitable tools to show the reduction efficiencies of the different treatment steps, these genes were selected26. Additionally, since the imipenem ARG (blavim) is reported among Pseudomonas aeruginosa and the spread of blavim in pathogenic bacteria is apparent, this gene was also studied19,27.
Given that it has been suggested based on the results of various studies that conventional treatment may be ineffective and chlorination was shown to increase the prevalence of antibiotic resistance28,29, methods including advanced treatment systems need to be evaluated to find a more effective and economic way to remove ARB and ARGs from hospital wastewaters30. Since according to several studies published, ozonation is effective in inactivating ARB and removing ARGs and in eliminating the potential regrowth of bacteria under optimal conditions, ozone doses and contact times, thus this method was considered to fill the gaps in knowledge about the effect of ozonation on ARB inactivation and reduction of ARGs.
Therefore, in this research, the aim of the work was to investigate and optimize the efficiency of ozonation for the removal of ARB and ARGs during treatment. Since the evaluation of antibiotic resistance in the population of natural wastewater has been studied, examining the resistance of bacteria to different groups of antibiotics and the resistance of bacteria to different concentrations of antibiotics have also been examined separately, showing the difference between this study and other studies. Additionally, regarding ARGs, it should be noted that the presence of resistance genes was examined both before exposure to antibiotics and after exposure of identified bacteria to different concentrations of antibiotics. The reason for doing this was to consider all the factors in selecting bacteria for ozone treatment and to select the most resistant bacteria in the hospital wastewater for treatment.
Materials and method
Wastewater sample collection and bacterial culture
Untreated wastewater samples were obtained from the last manhole of sewerage collecting wastewater from Tabriz University-Affiliated Hospital in Iran. Samples were taken twice during 2019 to obtain a more representative sample with a 24 h composite that was taken each period. Prior to each treatment, the pH, BOD and COD values of the samples were determined. pH values were in the neutral range (7–8), and the average BOD and COD in hospital wastewater samples were 190 ± 35 mg L−1 and 350 ± 67 mg L−1, respectively. In the first stage using the culture method and with the use of eosin methylene blue (EMB) agar and cetrimide agar, bacteria were identified for this purpose for the isolation of Escherichia coli and Pseudomonas spp. Samples were placed on agar plates in triplicate and then incubated for Escherichia coli 24 h and for Pseudomonas spp. 24–48 h at 37 °C. After identification of bacteria, different antibiotics, β-lactams (cefotaxime (Ctx), ceftriaxone (Cro), ceftazidime (Caz), cefixime (Cef), cefazolin (Cfz) imipenem (Ipm)), sulfamethoxazole (Smx) and ciprofloxacin (Cip), that were purchased from Sigma Chemical Company (Sigma Aldrich, Uk) at concentrations of 8–128 µg/mL were used to determine bacterial resistance. Bacteria grew in different concentrations of antibiotics, but considering that bacteria grown at concentration of 32 µg/mL of antibiotics compared to higher concentrations of antibiotics, identified and counted obviously, this concentration of antibiotic was studied and the number of bacteria grown in this concentration is presented.
PCR assays
PCR assays for confirmation of the identified bacteria and determination of antibiotic resistance genes
Since polymerase chain reaction (PCR) and quantitative PCR (qPCR) are very sensitive for detecting total DNA present in a sample, including DNA from live and dead cells19 and due to process-related DNA changes, PCR-based approaches allow the direct identification of DNA damage during disinfection31. After obtaining and purifying the plasmids that were extracted using a Plasmid DNA Miniprep Kit (TsingKe, Beijing, China), the concentration and the quality of the plasmids were determined by a NanoDropND-2000c spectrophotometer (Thermo Scientific, Wilmington, USA), and then isolated bacteria were confirmed by amplification of the 16S rRNA gene. PCR products were analyzed for DNA sequencing, and DNA sequences were analyzed by BLAST (Basic Local Alignment Search Tool) algorithms and databases from the National Center for Biotechnology (www.ncbi.nlm.nih.gov). After determining the identified bacteria, using standard PCR, the occurrence of genes resistant to β-lactams (blatem, blactx, blavim), fluoroquinolones (qnrS) and sulfonamides (sul1) was tested using the primers presented in SI Table 1. For this, the temperature program consisted of initial denaturing at 95 °C, followed by 37 cycles of 60 s at 94 °C, 30 s at the annealing temperature, 30 s at 72 °C and a final extension step for 5 min at 72 °C.
Determination of resistant bacteria for ozone treatment
After identifying the typical Escherichia coli and Pseudomonas aeruginosa bacteria from specific culture media, to confirm the identified colonies, the isolated bacteria were accurately identified by PCR and based on the BLAST results, five types of Pseudomonas aeruginosa and two types of Escherichia coli were confirmed, and all Pseudomonas aeruginosa and Escherichia coli were exposed to different concentrations of antibiotics (8–128 µg/ml) and examined for the presence of antibiotic resistance genes. Gene resistance to β-lactams (blatem, blactx, blavim), fluoroquinolones (qnrS) and sulfonamides (SulI) was tested using standard PCR. Finally, bacteria that were more resistant to antibiotics and had more antibiotic-resistant genes were studied for the treatment process.
Quantitative PCR
After the determination of the antibiotic resistance genes of identified Escherichia coli and Pseudomonades aeruginosa, quantitative real-time PCR (qRT-PCR) was utilized for quantification of the resistance genes in the identified bacteria. Finally, bacteria with the highest number of resistance genes were selected for the ozone disinfection process. Positive ARGs, including Sul1, blatem, blactx, blavim, and qnrS, were selected for qPCR. 16S rRNA was also employed as housekeeping gene. Plasmids carrying these target genes were prepared and used to build standard curves. qPCR reactions were performed in a LightCycler® 96 Instrument (Roche Life Science, Germany) in a 10 µL reaction mixture of 2 × SYBR Green PCR Pre-Mix (Ampliqon, Denmark), 0.2 µL of each primer, 1 µL of template DNA and 8.6μL ddH2O to a total volume of 20 μL. For this, a 10 min preheating step at 94 °C to activate the polymerase enzyme and denature the template DNA was considered. Then, amplification was carried out by 45 cycles of 94 °C for 10 s, annealing temperature for 30 s, and 72 °C for 20 s. The annealing temperature of each primer pair is presented in SI Table 1.
For generation of standard curves for Escherichia coli and Pseudomonas aeruginosa bacteria as well as for resistance genes, plasmid constructs containing 247-bp fragment of blatem, the 175-bp fragment of blactx, the 382-bp fragment of blavim, the 163-bp fragment of sul1 and the 118-bp fragment of qnrS were used which is presented in SI Figs. 1–7 and method of preparing the standard curves is explained in the SI 1.
Propidium monoazide (PMA) treatment
In this research, propidium monoazide was used to separate the surviving bacterial population from inactivated bacteria during ozone treatment. Because of loss of integrity in dead or injured bacterial membranes, PMA can enter them and intercalate with intracellular and extracellular DNA. Presence of the PMA-DNA complex blocks polymerase activity at the target sites, and consequently, no PCR product is generated32. Therefore, PMA treatment in qPCR analyses is an important step for accurate detection and quantification of ARGs31. For this treatment, after filtration of the samples, (0.2-μm Supor®-200 membrane, 47 mm diameter, Pall Life Science) filters were submerged in 300 μL of a 25 μM propidium monoazide solution and put into a 1.5 mL colorless tube. After incubation in the dark at 4 °C for 5 min, samples were exposed to the LED light of the photoactivation system at 100% intensity for 15 min. After PMA treatment, samples were prepared for DNA extraction31.
Analysis of ARB concentrations
The decrease in the number of viable Escherichia coli and Pseudomonas aeruginosa colonies following ozonation was measured using a culture-based technique. Because identified colonies were resistant to all antibiotics at a concentration of 32 µg/ml, selection plates were dosed with antibiotics at a concentration of 32 µg/mL. Samples were then serially diluted with phosphate buffer to achieve an approximate range of 30 to 300 colonies on the plate. A total of 0.1 mL of diluted sample was then placed into the agar plates in triplicate and incubated, finally were counted and calculated based on the number of dilutions.
Analysis of antibiotic resistance genes
Pseudomonas aeruginosa harbors a number of intrinsic clinically relevant ARGs and hosts a high abundance of acquired ARGs. Specific primers targeting the ecfX gene, due to its higher amplification specificity and performance in qPCR, were used to detect the abundance of Pseudomonas aeruginosa in this study before and after ozone treatment33,34. Additionally, a specific primer pair targeting the yccT gene was used to detect the abundance of Eschershia coli. After ozone treatment. For this purpose, 100 mL of the wastewater was filtered through a 0.2-μm Supor®-200 membrane (47 mm diameter, Pall Life Science) and Total DNA was extracted directly from the membranes.
Ozone treatment setup
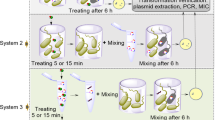
All experiments were done in a pilot plant in batch mode at ambient temperature (20 ± 2 °C) at pH 7.2–7.5. Figure 1 displays a schematic diagram of the ozonation setup. The pilot system consisted of glass columns with an inner diameter of 5 cm and a total height of 50 cm. Ozone was produced using a laboratory ozone generator (Jahad Research Center, Iran) fed by an oxygen generator (Type CFS-1, New Life Elite oxygen concentrator, AirSep corporation company, USA). Ozone gas was bubbled at constant flow rate of 0.2 L min−1 into the wastewater through a diffuser located at the bottom of the columns, and the inlet concentration of ozone was approximately 22 mg/L throughout all ozonation experiments. The off-gas was absorbed by a 2% KI solution. Due to the specific concentration of ozone at specified intervals, wastewater samples were taken from the reactor. All experiments were conducted in triplicate, and the averaged data are presented. The amount of ozone required to bacterial destruction was calculated by the dose of ozone transferred (TOD) during the ozonation. The absorbed mass of ozone transferred into the unit volume of liquid that reacts with the oxidizable compounds is defined as TOD and was calculated by Eq. (1)29,35,36.
In Eq. (1), Qgas represents the gas flow rate (0.2 L min−1), V liquid is the volume of liquid in the reactor and CO3in CO3out are the ozone concentrations in the inlet and outlet gases, respectively.
Iodometric method was used for measurement of ozone concentration in the inlet and outlet gases37 to do this, two impingers were used in series and each filled with 200 ml of 2% KI solution and gas was bubbled. Ozone-exposed KI solution was added to a solution of 10 ml of H2SO4 (2 N) and titrated with Na2S2O3 (0.005 N) solution until it turned pale yellow. Finally by adding of 1% starch indicator solution of 0.5 mL, titration continued until the blue color changed to colorless. Spectrophotometric method was used for determination of ozone concentration in aqueous solution that for this purpose, 5 ml of the sample was transferred to a test tube containing 5 ml of 2% KI solution and after approximately 30 min using cells with a 20 mm light path, the intensity of absorption was read36,38.
Ozone for the inactivation of ARB and the removal of ARGs
In this research, the abundance of ARB (P. aeruginosa and Escherichia coli) and antibiotic resistance genes in these bacteria (sul1, blatem, blactx, blavim, and qnrS) in hospital wastewater exposed to different concentrations of ozone (TOD1 = 11 mg/l, TOD2 = 25 mg/l, TOD3 = 37 mg/l, TOD4 = 45 mg/l) was evaluated. This amount of TODs that were examined is based on the amount of dissolved ozone of 1–4 mg/l. Ozone utilization efficiency (OUE) in ozonation is presented in SI Table 2 (Supplementary Materials).
Measurement of antibiotic-resistant bacteria at different concentrations of ozone
The decrease in the number of viable Escherichia coli and Pseudomonas aeruginosa colonies from the fresh cultures of these bacteria was carried out by the culture method. For this, concentrations of 104, 106 and 108 cfu/ml of bacteria were prepared according to McFarland standards, and then quantification and confirmation tests of culturable bacteria were performed. Samples were inoculated into the reactor at the start of the experiments and at different concentrations of ozone. Bacterial removal was expressed as a function of TOD values, and for this, the reduction of cultivable Escherichia coli and Pseudomonas aeruginosa bacteria with identified TOD at different times was investigated. It should be mentioned that bacterial-free wastewater was sterilized in an autoclave. To ensure the absence of bacteria in sterilized wastewater, the presence of bacteria was examined by the culture method. For each sample, triplicate plates were prepared, and the results were averaged.
Measurement of antibiotic resistance genes at different concentrations of ozone
For this purpose, DNA-free hospital wastewater was sterilized for one hour at 121 °C and 1 bar pressure. To ensure the absence of genes in sterilized wastewater, 16S rRNA bacteria were examined by PCR. At each stage of ozonation with different levels of TOD, a specific amount of DNA prepared with concentrations of 104, 106 and 108 cfu/ml bacteria was inoculated into the reactor. The amount and reduction of antibiotic resistance genes before ozonation and at different levels of TOD were examined by extracting DNA and qPCR.
Statistical analysis
The degree of log removal of specific genes was calculated using the following formula:
In this equation, j indicates specific genes (antibiotic resistance genes studied and 16S rRNA; C0j indicates the gene copy number of specific gene j in the original wastewater samples (copies per milliliter), and Cij indicates the gene copy number of specific gene j that survived the ozonation process at a dosage of i (copies per milliliter). To compare the inactivation efficiency of bacteria and genes responding to different ozone doses, one-way ANOVA was conducted. The difference was considered statistically significant at a p.value less than 0.05. All statistical analyses were performed using R software.
Results and discussion
Culture method and PCR assays for the detection of bacteria and resistance genes
In this study, all bacteria grown on EMB and cetrimide agar without antibiotics were confirmed by PCR and amplification of the 16S rRNA gene region. Based on BLAST, Escherichia coli and Pseudomonas aeruginosa were identified. Then, the resistance of the identified bacteria to different concentrations of antibiotics (8–128 μg/mL) was examined, and the presence of genes encoding antibiotic resistance determinants in all isolated bacteria was assessed by PCR. Eventually, bacteria that were more resistant to antibiotics and had more antibiotic resistance genes were studied for the treatment process, and the two bacteria determined with BLAST results are presented in Table 1.
Identified bacteria designated for ozone treatment were resistant to different classes of antibiotics, such as the resistant bacteria identified in similar studies7,39.
Inactivation of ARB by ozonation
Two identified bacteria were selected (Pseudomonas aeruginosa and Escherichia coli) having the highest resistance to most antibiotics and having the highest resistance genes to antibiotics. Three different initial concentrations of selected bacteria were treated with four different concentrations of ozone. Figures 2 and 3 show the effect of ozone disinfection on the reduction of ARB.
Ozone reacts with different compounds, such as amines, amino acids, activated aromatic compounds, reduced sulfur residues, unsaturated bonds of phospholipids, proteins, peptidoglycan and liposaccharides available in the cell envelope, and all of these substances are abundant in hospital wastewater40.
In this research, a concentration of 108 cfu/ml Escherichia coli was reduced by ozone treatment after a 5 min contact time (almost one order of magnitude in TOD1, two orders of magnitude in TOD2, four orders of magnitude in TOD3, five orders of magnitude in TOD4), while the removal rate was much higher for concentrations of 106 cfu/ml and 104 cfu/ml bacteria. Therefore, when the bacterial concentration was 106 cfu/ml, bacteria did not survive in TOD3 and TOD4 and were reduced by ozone treatment after a 5 min contact time (almost two orders of magnitude in TOD1 and five orders of magnitude in TOD2). When the Escherichia coli concentration was 104 cfu/ml, only in TOD1 and during the 5 min contact time survived and could be identified. (Reduced to 3 logs in TOD1, ozone dose = 11 mg/l). The same was true of Pseudomonas aeruginosa, and as the initial concentration of bacteria decreased (concentrations of 104 cfu/ml and 106 cfu/ml compared to 108 cfu/ml bacteria), inactivation of bacteria required a lower dose of ozone and less contact time, and this issue has been proven in various studies19,41. In general, the reduction in Pseudomonas aeruginosa was less than that in Escherichia coli, and Pseudomonas aeruginosa was more resistant than Escherichia coli and inactivated 2 log of TOD3 (ozone dose = 37 mg/l). This result was consistent with the results of other studies42,43,44.
The present study shows that ozonation of hospital wastewater is effective in the elimination of both ARB (Escherichia coli and Pseudomonas aeruginosa. Various studies have also shown that ozone has a better effect on killing bacteria and45 based on studies on the effect of ozone on multidrug resistant bacteria, even low concentrations of ozone have been shown to eliminate an acceptable percentage of these bacteria19,46.
The rate of Escherichia coli reduction was higher in the same conditions than Pseudomonas aeruginosa, but According to the statistical results and by considering equal concentrations of initial bacteria )108 cfu/ml(, only there was significant relationship between the reduction of Escherichia coli and Pseudomonas aeruginosa in TOD1 and TOD4 (p < 0.05).
Inactivation of ARGs by ozonation
The removal efficiencies of antibiotic resistance genes in the ozonation process were evaluated via the reduction of gene copy numbers. The removal efficiencies of antibiotic resistance genes in Escherichia coli and Pseudomonas aeruginosa (108 cfu/ml concentration) by ozonation are shown in Figs. 4 and 5.
The inactivation of ARGs by O3 depends on different influencing factors, such as the genome composition, ARG type, reaction time, concentration of microbial communities, ARB type, O3 dosage and wastewater parameters18. Ozonation results showed less removal of ARGs than 16S rRNA. This result can be attributed to the consumption of ozone by organic matter in wastewater and is similar to the results of studies conducted in this field47. Inactivation of selected genes increased when the ozone concentration was increased from 11 to 45 mg/L.
In this study, in accordance with other studies, O3 could acceptably reduce 16S rRNA, so that at TOD3 = 37 mg/l with a 5 min contact time, acceptable log decrease in 16S rRNA in both bacteria (Escherichia coli and Pseudomonas aeruginosa) was obtained45,48.
The results of the reduction of resistance genes by ozonation considering the concentration of genes in initial bacteria) 108 cfu/ml (, was similar to the reduction rates of qnrS, blatem and blavim being different from that of Sul1, which was acceptably reduced. Bacteria carrying the resistance genes blavim and qnrS showed more resistance against ozone treatment.
According to the statistical results, there was significant difference in the rate of gene reduction in Escherichia coli and Pseudomonas aeruginosa with different concentrations of initial bacteria. For example according to the results of one-way ANOVA, reduction of sul1 genes in both bacteria with a concentration of 108 cfu/ml was significantly different from the concentration of 104 cfu/ml. Also, gene reduction was significantly different between concentration of 106 cfu/ml and 104 cfu/ml of bacteria (p < 0.05).
In this study, with decreasing concentration of initial bacteria and consequently decreasing concentration of initial genes, the rate of gene reduction by ozonation increased so that the rate of reduction of genes with 104 cfu/ml initial bacteria was greater than the rate of reduction with 106 cfu/ml initial bacteria. The results of the decrease in the studied genes show that ARG damage requires a greater ozone dose or longer time than inactivation of the 16S rRNA gene. These results indicated that the damage to ARGs might require more energy than inactivation of the 16S rRNA gene, which is consistent with other studies conducted in this field23,45.
In this study, when the amount of ozone increases, the rate of reduction of genes also increases. One of the reasons for this is that when ozone exposure increases, protein leakage and membrane permeabilization also increase, and ozone can react too rapidly with the cell envelope and damage ARGs, which is consistent with the results of studies conducted in this field29,49. The results of this research, which was in agreement with previous studies, found that sulfonamide genes in Escherichia coli and Pseudomonas aeruginosa were removed more than other genes by ozonation, and the rate of reduction of the blactx and qnrS genes in Pseudomonas aeroginosa was very similar. At high concentrations of ozone, the removal rate of blactx was slightly greater than that of qnrS genes. In this study, the blactx gene was more difficult to remove than the blatem gene. One of the reasons for this may be that the origin of the CTX enzymes is different from TEM. TEM is generated by amino acid substitutions of their parent enzymes, but CTX is acquired by horizontal gene transfer from other bacteria using genetic apparatuses such as conjugative plasmids or transposons50,51.
The imipenem (blavim) resistant gene in Pseudomonas aeruginosa showed more resistance after ozonation and was eliminated less than other genes19,23. In research conducted by Alexander et al., which checked the efficiency of ozonation treatment to inactivate selected ARGs, the results showed that the rate of reduction genes removed through ozonation for blavim was the lowest of all genes52. Hence, Pseudomonas aeruginosa and its antibiotic resistance gene directed to imipenem are important parameters for future microbiological risk assessments.
Overall, the inactivation of genes by ozone disinfection from easy to hard was 16S rRNA > sul1 > blaem > blactx > qnrS > blavim. 16S rRNA, compared to other genes, was removed most efficiently, which can be attributed to its high original copies, which existed in most background bacteria. The results of this study on gene reduction are consistent with related studies53,54. According to the statistical results (one-way ANOVA) of different genes, significant difference was observed between the reductions of genes in different concentrations of ozone (p < 0.05).
Conclusions
In this research, multidrug-resistant Pseudomonas aeruginosa and Escherichia coli were detected in hospital wastewater, and it was also found that hospital wastewater contains ARGs that are of great clinical importance due to their resistance to different groups of antibiotics. Given that different studies are needed to provide an understanding of the ozonation effect on the removal of antibiotic resistance determinants, optimization of the ozone process (e.g., ozone dose and contact time), bacterial species and related ARGs and physicochemical properties of hospital wastewater are important factors. The findings of this study demonstrated that ozonation of hospital wastewater constitutes a promising treatment technology for the elimination of Escherichia coli and Pseudomonas aeruginosa and selected ARGs.
Because hospital wastewaters contain ARGs and ARB and given that hospital wastewaters in most countries without any restrictions and without any pretreatment are discharged into municipal sewage systems, the spread of ARB and ARGs can become a threat to human health and ecosystems. Since routine treatment can only partially remove ARB and ARGs from hospital wastewaters, so this wastewaters need advanced treatment than what is done in municipal WWTPs and must be investigated in a more useful and economic way to remove ARB and ARGs. As a result, modifications or process combinations are needed to increase the removal rate of ARB and ARGs. Additionally, other methods, including ozone catalysts, need to be investigated to find a more useful and economic way to remove ARGs from hospital wastewaters.
Since ozonation could decrease 16S rRNA more than ARGs and despite acceptable reduction of genes by ozonation, complete removal was not achieved and the genes were present even in TOD4 (45 mg/L) with a contact time of 15 min, So to achieve completely ARG removal the dosage of ozone must be much higher than that defined by common usage and more comprehensive studies should be done in this regard.
References
Lamba, M., Graham, D. W. & Ahammad, S. Z. Hospital wastewater releases of Carbapenem resistance pathogens and genes in urban India. Environ. Sci. Technol. 51, 13906–13912 (2017).
Mustapha, A. & Imir, T. Detection of Multidrug-Resistance Gram-Negative Bacteria from Hospital Sewage in North East, Nigeria. Front. Environ. Microbiol. 5, 1–7 (2019).
Khan, N. A., et al. Occurrence, Sources and Conventional Treatment Techniques for various antibiotics present in hospital wastewaters: A critical review. Trends Anal. Chem 129, 115921 (2020).
Asfaw, T., Negash, L., Kahsay, A. & Weldu, Y. Antibiotic Resistant Bacteria from Treated and Untreated Hospital Wastewater at Ayder Referral Hospital, Mekelle, North Ethiopia. Adv. Microbiol. 7, 871–886 (2017).
Khan, N. A. et al. Recent Trends in Disposal and Treatment Technologies of Emerging-Pollutants- A Critical Review. TrAC Trends Anal. Chem. 122, 115744 (2020).
O'Flaherty, E., Solimini, A. G., Pantanella, F., Giusti, M. D. & Cummins, E. Human exposure to antibiotic resistant-Escherichia coli through irrigated lettuce. Environ. Int. 122, 270–280 (2019).
Hocquet, D., Muller, A. & Bertrand, X. What happens in hospitals does not stay in hospitals:antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 93, 395–402 (2016).
Chow, L., Waldron, L. & Gillings, M. Potential impacts of aquatic pollutants: subclinical antibiotic concentrations induce genome changes and promote antibiotic resistance. Front. Microbiol. 6, 803 (2015).
Manaia, C. M. Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 25, 173–181 (2017).
Zhang, T., Kunyuan, Lv., Qingxiao, Lu., Wang, L. & Liu, X. Removal of antibiotic-resistant genes during drinking water treatment: A review. J. Environ. Sci. 104, 415–429 (2021).
Berendonk, T. et al. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317 (2015).
Thakali, O., Brooks, J.P., Shahin, S., Sherchan, S. P. & Haramoto, E. Removal of antibiotic resistance genes at two conventional wastewater treatment Plants of Louisiana, USA. Water Res. 12, 1729 (2020).
Michael-Kordatou, I., Karaloia, P. & Fatta-Kassinos, D. The role of operating parameters and oxidative damage mechanisms of advanced chemical oxidation processes in the combat against antibiotic-resistant bacteria and resistance genes present in urban wastewater. Water Res. 129, 208–230 (2018).
Kovalova, L. et al. Elimination of micropollutants during post-treatment of hospital wastewater with powdered activated carbon, ozone, and UV. Environ. Sci. Technol. 47, 7899–7908 (2013).
Heß, S. & Gallert, C. Sensitivity of antibiotic resistant and antibiotic susceptible Escherichia coli, Enterococcus and Staphylococcus strains against ozone. J Water Health 13, 1020–1028 (2015).
Azuma, T. & Hayashi, T. Disinfection of Antibiotic-resistant Bacteria in Sewage and Hospital Effluent by Ozonation. Ozone Sci. Eng. 43, 413–426 (2021).
Song, M. et al. The antibacterial effect of topical ozone on the treatment of MRSA skin infection. Mol. Med. Rep. 17, 2449–2455 (2018).
Wang, J. & Chen, X. Removal of antibiotic resistance genes (ARGs)in various wastewater treatment processes: An overview. Crit. Rev. Environ. Sci. Technol. 52, 1 (2020).
Alexander, J., Knopp, G., Dötsch, A., Wieland, A. & Schwartz, T. Ozone treatment of conditioned wastewater selects antibiotic resistance genes, opportunistic bacteria, and induce strong population shifts. Sci. Total Environ. 559, 103–112 (2016).
Ternes, T. A., Joss, A. & Öhlmann, J. Occurrence, fate, removal and assessment of emerging contaminants in water in the water cycle (from wastewater to drinking water. Water Res. 72, 1–2 (2015).
Lan, L., Kong, X., Sun, H., Li, C. & Liua, D. High removal efficiency of antibiotic resistance genes in swine wastewater via nanofiltration and reverse osmosis processes. J. Environ. Manag. 231, 439–445 (2019).
Szekeres, E. et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals.Environ. Pollut. 225, 304–315 (2017).
Hou, J. et al. Simultaneous removal of antibiotics and antibiotic resistance genes from pharmaceutical wastewater using the combinations of up-flow anaerobic sludge bed, anoxic-oxic tank, and advanced oxidation technologies. Water Res. 159, 511–520 (2019).
Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nature 543, 15 (2017).
Ng, Ch., et al. Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front. Microbiol. 8, 2200 (2017).
Jäger, T., et al. Reduction of antibiotic resistant bacteria during conventional and advanced wastewater treatment,and the disseminated loads released to the environment. Front. Microbiol. 9, 2599 (2018).
Codjoe, F. S. & Donkor, E. S. Carbapenem Resistance: A Review. Med. Sci. (Basel) 6, 1 (2018).
Lüddeke, F. et al. Removal of totaland antibiotic resistant bacteria in advanced wastewater treatment byozonation in combination with different filtering techniques. Water Res. 69, 243–251 (2015).
Marce, M., Domenjoud, B., Esplugas, S. & Baig. S. Ozonation treatment of urban primary and biotreated wastewaters: Impacts and modeling. Chem. Eng. J. 283, 768–777 (2016).
Zhuang, Y. et al. Inactivation of antibiotic resistance genes inmunicipal wastewater by chlorination, ultraviolet, and ozonation disinfection. Environ. Sci. Pollut. Res. 22, 7037–7044 (2015).
Süss, J., Volz, S., Obst, U. & Schwartz, T. Application of a molecular biology concept for thedetection of DNA damage and repair during UV disinfection. Water Res. 43, 3705–3716 (2009).
Hembach, N. et al. Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in germany. Front. Microbiol. 8, 1282 (2017).
Alexander, J., Hembach, N. & Schwartz, T. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 10, 8952 (2020).
Moremi, N. et al. Extended-spectrum β-lactamase bla CTX-M-1 group in gram-negative bacteria colonizing patients admitted at Mazimbu hospital and Morogoro Regional hospital in Morogoro, Tanzania. BMC Res. Not. 14, 77 (2021).
Shokouhi, S. B., Dehghanzadeh, R., Aslani, H. & Shahmahdi, N. Activated carbon catalyzed ozonation (ACCO) of Reactive Blue 194 azo dye in aqueous saline solution: Experimental parameters, kinetic and analysis of activated carbon properties. J. Water Process. Eng. 35, 101188 (2020).
Shahmahdi, N., Dehghanzadeh, R., Aslani, H. & Shokouhi, S. B. Performance evaluation of waste iron shavings (Fe0) for catalytic ozonation in removal of sulfamethoxazole from municipal wastewater treatment plant effluent in a batch mode pilot plant. Chem. Eng. J. 383, 123093 (2020).
American Public Health Association, A.W.W.A., WaterEnvironment Federation. Standard methods for examination of water and wastewater. 23rd Ed (2017).
Shechter, H., Spectrophotometric method for determination of ozone in aqueous solutions. Water Res. 7, 729–739 (1973).
Fuentefria, D. B., Ferreira, A. E. & Corção, G. Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water: are they genetically related? J. Environ. Manag. 92, 250–255 (2011).
Stangea, C., Sidhu, J. P. S., Toze, S. & Tiehm, A. Comparative removal of antibiotic resistance genes during chlorination,ozonation, and UV treatment. Int. J. Hyg. Environ. Health 222, 541–548 (2019).
Selma, M. V., Beltran, D., Allende, A., Chacon-Vera, E. & Gil, M. I. Elimination by ozone of Shigella sonnei in shredded lettuce and water. Food Microbiol. 24, 492–499 (2007).
Lezcano, I., Perez Rey, R., Baluja, C. & Sánchez, E. Ozone inactivation of Pseudomonas aeruginosa, Escherichia coli, Shigella sonnei and Salmonella typhimurium in Water. Ozone Sci. Eng. 21, 293-300 (2008).
Restaino, L., Frampton, E., Hemphill, J. & Palnikar, P. Efficacy of ozonated water against various food-related microorganisms. Appl. Environ. Microbiol. 61, 3471–3475 (1995).
Fontes, B. et al. Effect of low-dose gaseous ozone on pathogenic bacteria. BMC Infect. Dis. 12, 358 (2012).
Zhuang, Y., et al. Inactivation of antibiotic resistance genes inmunicipal wastewater by chlorination, ultraviolet, and ozonation disinfection. Environ. Sci. Pollut. Res. Int. 22, 7037–7044 (2015).
Oh, J., Salcedo, D. E., Medriano, C. A. & Kim, S. Comparison of different disinfection processes in the effective removal of antibiotic-resistant bacteria and genes. J. Environ. Sci. 26, 1238–1242 (2014).
Macauley, J., Qiang, Z., Adams, C., Surampalli, R. & Mormile, M. Disinfection of swine wastewater using chlorine, ultraviolet light and ozone. Water Res. 40, 2017–2026 (2006).
Sousa, J. M. et al. Ozonation and UV 254nm radiation for the removal of micro organisms and antibiotic resistance genes from urban wastewater. J. Hazard. Mater. 323, 434–441 (2017).
Rekhate, C. V. & Srivastava, J. K. Recent advances in ozone-based advanced oxidation processes for treatment of wastewater- A review. Chem. Eng. J. Adv. 3, 100031 (2020).
Ehlers, M., Veldsman, Ch., Makgotlho, E. P., Dove, M. G., Hoosen, A. & Kock, M. .Detection of blaSHV, blaTEM and blaCTX-M antibiotic resistance genes in randomly selected bacterial pathogens from the Steve Biko Academic Hospital. FEMS Immunol. Med. Microbiol. 56, 191–196 (2009).
Estrada, A., Wright, D. & Anderson, A. C. Antibacterial antifolates:from development through resistance to the next generation. Cold Spring Harb. Perspect. Med. 6, a028324 (2016).
Alexander, J., Knopp, G., Dotsch, A., Wieland, A., & Schwartz, T. Ozone treatment of conditioned wastewater selects antibiotic resistance genes, opportunistic bacteria, and induce strong population shifts. Sci. Total Environ. 559, 103–112 (2016).
Colomer-Lluch, M. et al. Antibiotic resistance genes in bacterial and bacteriophage fractions of Tunisian and Spanish wastewaters as markers to compare the antibiotic resistance patterns in each population. Environ. Int. 73, 167–175 (2014).
Tiirik, K. et al. The Effect of the Effluent from a Small-Scale Conventional Wastewater Treatment Plant Treating Municipal Wastewater on the Composition and Abundance of the Microbial Community, Antibiotic Resistome, and Pathogens in the Sediment and Water of a Receiving Stream. Water 13, 865 (2021).
Acknowledgements
This research has been supported by the Tehran University of Medical Sciences, Institute for Environmental Research, Center for Water Quality Research, Tehran University of Medical Sciences, Tehran, Iran (Grant No. 97-03-46-41302) as part of a PhD dissertation (IR.TUMS.VCR.REC.1398.241).
Author information
Authors and Affiliations
Contributions
F.B.A.; Investigation/Writing Original draft, review, editing. M.H.D.; Review, Editing, Supervision. R.D.; Review, Editing, Supervision. D.F.; Review& Editing. D.S.; Review& Editing, Supervision. A.H.M.; Review, Editing. K.Y.; Review, Editing. A.R.; Investigation/Review, Editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baghal Asghari, F., Dehghani, M.H., Dehghanzadeh, R. et al. Performance evaluation of ozonation for removal of antibiotic-resistant Escherichia coli and Pseudomonas aeruginosa and genes from hospital wastewater. Sci Rep 11, 24519 (2021). https://doi.org/10.1038/s41598-021-04254-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-04254-z
This article is cited by
-
A database on the abundance of environmental antibiotic resistance genes
Scientific Data (2024)
-
Review of hospital effluents: special emphasis on characterization, impact, and treatment of pollutants and antibiotic resistance
Environmental Monitoring and Assessment (2023)
-
Optimizing ozone dose and contact time for removal of antibiotic-resistant P. aeruginosa, A. baumannii, E. coli, and associated resistant genes in effluent of an activated sludge process in a municipal WWTP
Environmental Science and Pollution Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.