Abstract
We recently reported a sugar-induced bacterial release of 13-Docosenamide and its ability to quench fluorescein. This simple handle to monitor bacterial growth is readily applicable to develop a quicker antibiotic sensitivity testing method along with a low-cost field-use optical instrumentation. Conditions were standardized to perform this new procedure in the most preferred and CLSI-recommended microdilution format in 12-well strips. A simple and portable optoelectronic prototype was used to capture the image and read the fluorescence signal of the culture medium of the 12-well strips. This new Fluorescence Quenching Method along with the device enabled the choice of the right antibiotic within 8 h of sample collection from the patient. It was compliant to the Clinical Laboratory Standard Institute’s quality control guidelines. Clinical assessment of the method using 440 urine samples from Urinary Tract Infection patients against 21 routinely used antibiotics showed a 94.3% match with the results of the Standard Disk Diffusion method. This new method saves the precious time taken for and the cost of antibiotic susceptibility testing for quicker and effective treatment with better compliance.
Similar content being viewed by others
Introduction
Among the many factors attributed to the rise of antibiotic resistance, prescription of antibiotics without susceptibility testing (AST) is the primary and the most alarming one1. In countries like India, and even in developed countries, this is a common practice, except that in the latter case, antibiotic prescriptions are restricted and required for the purchase. Lack of simple and cost-effective AST performed even in peripheral clinics to obtain results within a few hours is the sole reason for the continuation of this risky practice.
The current diagnostic tests available to determine the antibiotic sensitivity do not meet the needs of the developing countries and several pockets in the developed countries. The conventional agar-based Disk diffusion method and E-Test of AST determination can benefit from newer developments2. The current automated systems require infrastructure and skill sets that are unattainable by many laboratories. The manual method requires augmented culture techniques which are time-consuming typically returning a susceptibility profile in 48 h or longer3. The broth microdilution method, a Clinical and Laboratory Standards Institute (CLSI) reference method, which is the “gold standard” AST method, requires incubation of 16–24 hours4. After incubation, the results are read visually or by measuring OD600 nm. Even with automated system and technologies which reduce the working time to about 50%5, the current methods do not provide the relevant information in time before the initiation of the first antibiotic therapy or even by the second dose.
We had recently published that Escherichia coli and 26 other bacteria associated with human diseases when grown in glucose-containing medium, released long-chain fatty amide(s) of the type 13-docosenamide, which quenched fluorescein, a widely-used inexpensive fluorophore. This quenching of the fluorescence of fluorescein was directly proportional to the growth of bacterial culture6. Thus, an absence in the fluorescence of the growth medium indicates bacterial growth while the fluorescence of the uncultured medium remains intact. We adapted this bacterial growth monitoring technique that based on the optical shift, for developing a simpler and quicker AST (within 8 h) as reported here. This Fluorescence Quenching Method (FQM) is performed in micro-wells of commercially available 12-well strips or plates. The fluorescence of the medium is captured using an optical imager fabricated in-house for 12-well strips.
Results
Fluorescence Quenching Method (FQM) results matched CLSI quality control range for four reference ATCC bacterial strains
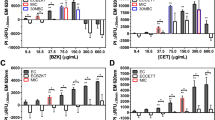
The compliance of FQM to CLSI guidelines was tested with four CLSI recommended reference ATCC bacterial strains against a panel of 10 different antibiotics, prepared according to the MIC ranges defined by CLSI (Table 1). The MIC results determined using FQM (Tryptic Soy Broth containing Glucose and Fluorescein, TSB-GF medium) for the ATCC strains were compared with that of the CLSI Broth Microdilution method (MH broth). The MIC of each antibiotic for the four ATCC strains determined by FQM and CLSI microdilution method was the same and within CLSI defined quality control range (Table 1). This absolute match was repeatable in 15 consecutive trials, each performed once daily. The fluorescence value of the medium, TSB-GF, and that of the susceptible samples did not alter significantly from 8 h to 24 h of testing indicating excellent stability of the dye in the medium even for extended incubations. The culture without antibiotic and at concentrations below MIC, the fluorescence decreased in proportion to the growth at 8 h, the cut off time for FQM, and beyond.
A simple and inexpensive fluorescence imaging device enabled field-use of FQM
The low-cost portable imager (see materials and methods) prototype generated the image of the fluorescence pattern in a 12-well test strip as given in Fig. 1a. The medium control wells appeared green owing to the intense fluorescence of fluorescein. The wells with growth were blue to bluish-green depending upon the amount of the quencher produced in proportion to the growth and the image processing software calculates the amount of green colour in each well and represents it with a number ranging from 0–256.
(a) Images of isolates cultured by FQM captured using the imager after a static incubation period of 8 h. The corresponding multimode reader values are indicated below each well. (b) The imager values for different concentrations of fluorescein compared with the corresponding fluorescence response (RFU) of a commercial fluorescence reader.
The image processing software was calibrated with standard fluorescein concentration range 0 nM–3000 nM. The values generated by the software for the different concentration of fluorescein was compared with the fluorescence reading (RFUs) values of the commercial multi-mode reader. The setting at which the linear response of the green values determined by the algorithm matched that of the commercial reader values was fixed for measuring the signal (Fig. 1b). Hence, both the imager values and the reader values were proportional to the accumulative intensity of increasing concentration of fluorescein in the solution. This setting had been verified thoroughly by routine and several repeatability tests.
FQM proved to be clinically useful AST procedure for urine samples
FQM, along with the imaging device, was evaluated for its performance as an AST tool at the Microbiological Laboratory, Coimbatore, Tamil Nadu, India, which is accredited by National Accreditation Board for Testing and Calibration Laboratories.
Among 440 urine samples tested, 20 were sterile and six contained Candida spp. Both did not quench the fluorescence of TSB-GF medium. Thirty-three samples with a colony count below 1 × 103 CFU/mL (taken as borderline and clinically insignificant) was not detected by FQM within 8 h incubation period. However, the growth and quenching could be detected conveniently on longer and overnight incubation.
Apart from the sterile samples, 381 urine samples with significant bacteriuria were tested against 21 antibiotics by FQM at three concentrations. This provided 8001 results on antibiotic susceptibility. The same samples tested by Standard Disk Diffusion (SDD) against 13 antibiotics (in discs) provided 5027 (Table 2) results for comparison with the popular clinical method. Among these, 3856 points (76.7%) in agreement, 860 (17.1%) showed a shift in MIC values categorized as inconsequential minor error (mismatch in one concentration within the range). Hence, practically, the match between the two methods was 93.8%. 311 points (6.2%) showed major and very major errors between the methods; The major errors were 165 points (3.2%) majority (1.4%) of which is due to Colistin (match was confirmed with CLSI microdilution); very major errors were 146 points (3.0%) mainly contributed by Meropenem (1.0%).
These results show FQM employing direct clinical samples of urine could give reliable antibiogram results after 8 h aerobic incubation when there is a single pathogen causing urinary tract infection, which is the case in the majority of samples. Among the infected samples, E. coli was the most prevalent, accounting for 75% of the 381 samples and 8 other bacteria together constitute the rest 25% (Table 3).
Discussion
Increasing prevalence of antibiotic resistance among community infections, time-consuming and costlier microbiological investigations demand quicker and affordable ways of knowing the right antibiotic for treatment. Survey among the Indian physicians of peripheral hospitals and private clinics catering to the upper and lower middle-class populations indicated the need for the right choice of antibiotic first within 6–8 h, even before pathogen identification, especially targeting urinary tract infections (UTI). The test results of the clinical samples reported here showing ~30% of them being resistant to 3–4 and ~12% to 9–10 types of commonly prescribed antibiotics justify such a demand. Based on the survey input, we preferred to perform AST for UTI either prior to or simultaneously with the time-consuming procedure of isolation and identification of the pathogens. Our recent discovery of growth-dependent bacterial secretion 13-Docosanamide which quenches the fluorescence of fluorescein6 offered a simple AST approach with associated inexpensive fluorescence imaging instrumentation.
After extensive standardization and optimizations to suit the field conditions, we found that the bacterial growth in the ready-to-use fluorescent medium (TSB-GF) was accompanied by significant quenching by 4 h and reached 70–90% by 8 h of incubation for a variety of clinical bacteria. Our low-cost optoelectronic-imaging device equipped with our own image processing software was comparable to a standard commercial microplate fluorescence reader in terms of quantitation and accuracy of AST results. Since fluorescein is a very stable fluorophore, quenching above 20% of its initial fluorescence was taken as bacterial growth. Hence, the wide fluorescence quenching range (from 20% to 100%) available during 8 h growth period, accommodated errors and variations typical of biological measurements and assays in reporting bacterial growth. The laboratory-level and the clinical evaluation described here have clearly borne out these advantages. The TSB-based medium prepared with fluorescein, highly fluorescent and inexpensive fluorophore was found to be stable for months7,8,9 even at room temperature when stored in dark.
In the evaluation of FQM with clinical urine samples, the discordance of 6.2% (interpreted with CLSI breakpoints) with disk diffusion was akin to those reported for the established liquid-based antibiotic susceptibility assays. Particularly, Colistin10,11 and Meropenem12,13 known for the anomalous behaviour in disk diffusion. For Colistin, CLSI broth microdilution method is recommended due to its diffusibility problem from the disks14. For meropenem, it could be due to the high instability of this β-lactam derivative in aqueous solutions as well as in solid-state15 even under the manufacturer’s recommended storage conditions. This explains that the discordance is towards susceptibility for FQM in which fresh antibiotic stocks were used versus resistance in SDD. We had also verified our results with the CLSI broth microdilution method and plating on Meropenam agar plates. The CLSI recommended antibiotic concentrations for FQM tested against the significant bacteriuria samples showed accurate AST results when compared with SDD. Candida, which can be associated with urine samples and which grows in various bacteriological media, did not quench fluorescein, apparently because of lack of production of 13-docosenamide or its analogues. Hence, the method appears to be specific to bacteria. When we tested FQM with different clinical isolates, a few Pseudomonas strains a non-fermenter of glucose, isolated from pus, blood-borne and CSF infections did not show the fluorescence quenching. However, some of these isolates showed fluorescence quenching after sub-culturing in Luria-Bertani medium The inability of Pseudomonas to catabolize glucose could have interfered in the production of the quencher compound. In such cases, the antibiogram can be determined by measuring the turbidity of the medium.
A major advantage of our method is the simplified instrumentation for quantitative optical imaging of fluorescence. Under the conditions standardized for the instrumentation, a CMOS-based camera could quantitatively record the intensity reduction of fluorescent green of the medium after the end of 8 h incubation. This is evident from the 100% correlation between the imager values and the commercial fluorescence reader values. Since the antibiogram device has the potential for several samples against many antibiotics (anywhere between 8–25 antibiotics), imaging platform for such operations need flexibility for scale-up. This design allows customization of antibiotic template to the need in terms of the number of antibiotics, number of samples and the types of antibiotics.
Though this new AST method has been tested on urine samples, a sterile biological fluid, the method can be readily adapted for positive blood cultures, cerebrospinal fluid, and other exudates. A full-fledged validation study is needed to expand such a scope. In case of a single type of bacterium present in the sample, the pattern is its antibiogram (as in this study). In case of more than one and predominantly two, the result is an aid for the right antibiotic therapy. FQM is a rapid AST compared to SDD and the testing time can be decreased (below 4 h) by mapping the fluorescence quenching pattern with hourly measurements. After completing AST by this method, identification of the pathogen, by the preferred biochemical methods can be performed with the cultured TSB-GF medium that has the high inoculum size after 6–8 h of growth.
Though the study had employed ATCC strains and clinical isolates with known identities for the sake of proving the feasibility of using in a clinical scenario, the actual use requires identification of the pathogen for deciding its breakpoints. Quicker pathogen identification methods in sterile biological fluids urine and serum are available now to identify syndrome-specific pathogens. Hence both pathogen identification and antibiogram results can be obtained within 8 h or 1 working day.
Quick spread of MDRs even among communities, especially through infections like UTIs, generation of contaminated clinical and hospital waste is an alarming concern not addressed adequately. FQM will reduce the waste by 10-folds for a typical test by using only 2.5 mL of liquid culture compared to 25 mL of contaminated agar from SDD. Based on the bill-of-material estimate of retail purchase, it is expected that FQM will drastically reduce the present cost of $5–10 per sample to <$2.50 per sample depending upon the number of antibiotics to be tested.
Materials and Methods
Antibiotics
The antibiotics for preparation of test plates were procured from the suppliers shown in parentheses: Amikacin, Ampicillin, Cefotaxime, Ceftriaxone, Ciprofloxacin, Gentamicin, Nitrofurantoin, Norfloxacin, Piperacillin/Tazobactam, Ofloxacin, (Sigma-Aldrich, USA), Amoxycillin/clavulanate (GlaxoSmithKline; Thane, India), Ceftazidime (VHB; Mumbai, India), Cefepime (Biocon; Bengaluru, India), Cefixime (GlaxoSmithKline; Thane, India), Colistin (Cipla; Pune, India), Erythromycin (Himedia; Mumbai, India), Imipenem (MSD; Mumbai, India), Levofloxacin (Alkem; Chennai, India), Meropenem (Cipla; Pune, India), Tetracycline (GlaxoSmithKline; Thane, India), Tigecycline (Natco; Chennai, India), Vancomycin (Himedia; Mumbai, India).
Strains
American Type Culture Collection (ATCC) Escherichia coli: 25922, Enterococcus faecalis: 29212, Klebsiella pneumoniae: 700603, Pseudomonas aeruginosa: 27853, Staphylococcus aureus: 29213 were purchased from Microbiologics® and maintained according to the supplier’s protocol.
Growth media
Tryptic Soy Broth containing Glucose and Fluorescein (TSB-GF) was prepared by dissolving 17 grams of Tryptone, 3 grams of Soya bean meal, 5 grams of Sodium chloride, 2.5 grams of Dipotassium phosphate, 9 grams of Glucose in 1 L of distilled water and adding 25 µL of 100 mM fluorescein solution. Mueller Hinton (MH) broth No.2 Control Cations and Mueller Hinton Agar were purchased from Himedia and prepared according to the supplier’s protocol. All the preparations were autoclaved at 121 °C at 15 psi for 20 min.
Evaluation of the fluorescence quenching method (FQM) with microdilution method using CLSI breakpoints
The antibiotic panel was prepared based on the CLSI quality control (QC) ranges for 10 antibiotics against reference ATCC cultures: E. coli, E. faecalis, P. aeruginosa and S. aureus (Table 1). Primary inoculum for each was prepared by suspending colonies of a single isolate, selected from an 18–24 h Muller Hilton agar plate culture, in MH broth. The suspension was adjusted to achieve turbidity equivalent to a 0.5 McFarland standard. This results in a suspension containing approximately 1–2 × 108 colony forming units (CFU)/mL. Working stock of 5 × 105 CFU/mL for each bacterium was prepared by diluting the primary inoculum in Mueller-Hilton medium for the CLSI broth microdilution method and TSB-GF for FQM respectively. Aliquots of 200 µL were dispensed into each of the antibiotic-coated wells of the microtitre plates and growth control wells. In the case of CLSI broth microdilution, OD600 nm was taken after 24 h static incubation at 37 °C. For FQM, after 8 h of static incubation at 37 oC, OD600 nm and fluorescence were measured using Synergy H1 Hybrid Multi-Mode Reader (commercial reader). MICs determined from the results of both the methods for ATCC organisms tested against individual antibiotic was compared with the CLSI (2017) QC range. Consecutive trials were performed for 15 days for checking the consistency of the new test.
FQM imager device
The in-house fabricated optoelectronic device housed in a black box measuring 10 cm in length 9.6 cm breadth and 20 cm in height consists of an illumination (Fig. 2a) and sample loading setup at the bottom and the CMOS-based image acquisition setup on the top (Fig. 2c(5)). The illumination setup consists of a single blue LED positioned for each well of the 12-well microtitre strip (Fig. 2c(4)) excites the medium fluorescein in the well. A strip-loading tray with a sliding mechanism at the base (Fig. 2c(3)) aligns the samples with respective blue LEDs. The camera captures the image of the strip and transfers the data to a computing device via a USB port (Fig. 2d). An in-house developed software installed in the computing device connected to the imager controls the image capture and processing operation. The software is based on an algorithm that quantifies the amount of green colour in the medium. As fluorescent medium (uncultured sample or sample sensitive to antibiotic) and the quenched medium (cultured sample or sample resistant to antibiotic) formed well-separated clusters in image processing quantification, the algorithm is coded to identify growth as antibiotic resistance and no growth as antibiotic susceptibility. Based on which the AST report is generated and securely transferred to the clinical lab/hospital/doctor.
Schematic drawings of the design and the photo of optoelectronic imager device. (a) Illumination setup with LEDs for excitation of fluorescein (b) The 12-well microtitre strip and its folded configuration within the strip holder (c) Vertical cross-sectional diagram of the imager setup (1) FQM strips with sample (2) and (3) Strip holder loaded into the Imager (4) LEDs exciting the fluorescein in the medium (5) Camera position (d) Antibiogram Imager device connected to a computing device to capture and process the image of fluorescence of TSB-GF medium within the wells.
Preparation of antibiotic coated microtitre wells
Detachable 12-well microtiter strips, which are commercially available as 8 strips (96-wells) mounted on a holding frame, were used to prepare the antibiotic coated plate for testing the clinical samples. A panel of 21 antibiotics at CLSI breakpoint concentrations for susceptible, intermediate and resistance were used. A fixed aliquot (10 µL) of the respective antibiotic working stocks prepared with sterile pyrogen-free water were delivered into A1-F3 wells of the strips according to the final concentration as shown in the template (Table 4). The prepared antibiotic strips were freeze-dried at −110 °C and stored at −80 °C until use.
Performing Fluorescence Quenching Method (FQM) with urine samples
For the FQM method, 150 µL of patient urine sample was inoculated in 15 mL TSB-GF medium. 200 µL aliquots of the inoculated medium was dispensed into A1-F3 wells (antibiotic-coated microwells) & F6, F7 (no antibiotic)-growth control wells and 200 µL of sterile medium were dispensed in F4, F5-medium control wells of the strips. The strips were covered with transparent adhesive tape to prevent spillage or contamination. After 8 h incubation, each 12-well strip of the test plate was snapped sequentially into three 4-well pieces (after cutting the adhesive tape between the segments to enable folding) and placed in the receptacle as shown in Fig. 2b (top). It was then folded into inverted U shape (Fig. 2b, bottom), placed in the tray and inserted into the device so as to align with the illumination unit (Fig. 2a–c) of the camera connected to a computing device (Fig. 2d). The software installed on the computing device captured and processed the image of the strips into susceptibility results based on the green intensity of the wells. The susceptibility report can be communicated to the clinical lab/hospital/doctor directly.
Comparison of FQM results with those of Standard Disk Diffusion (SDD) method using urine samples
The patient urine sample received for routine culture-and-susceptibility testing were selected for antibiotic susceptibility testing using SDD and FQM. The urine samples were analyzed using UROQUATTRO, HB&L™ and sterile urine sample, urine sample infected with Candida spp. and significant bacteriuria samples infected with only a single type of organism were preferred. For SDD method, after the analysis, the bacterium was isolated from the urine sample by plating on MH agar and identified by Gram staining. The isolates were then used as inoculum and hence, AST was performed 24 h after sample collection. For FQM, the corresponding urine immediately after UROQUATTRO, HB&L™ analysis was used as inoculum. For SDD method, the zone of inhibition was measured between 18–24 hours of aerobic incubation at 37 °C. The result of FQM method was generated using the fluorescence imager device after 8 h aerobic incubation at 37 °C. The antibiotic susceptibility results of both the methods were compared and the results were classified according to Table 5.
Ethical Declarations
The samples used in this study are leftover non-identified clinical urine samples thus this study is categorized under “Less than minimal risk”. Hence, this study is exempted from ethical review according to “National Ethical Guidelines for Biomedical and Health Research Involving Human Participants – Indian Council of Medical Research”. The authors Rohit Radhakrishnan, J. Rajesh, Dr N. S. Dinesh, Dr C. P. Thangavelu, and Dr K. Sankaran declare that the FQM-AST results of this study did not influence the antibiotic therapy provided to the patients.
References
English, B. K. & Gaur, A. H. The use and abuse of antibiotics and the development of antibiotic resistance. Advances in experimental medicine and biology 659, 73–82, https://doi.org/10.1007/978-1-4419-0981-7_6 (2010).
Reller, L. B., Weinstein, M., Jorgensen, J. H. & Ferraro, M. J. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clinical infectious diseases 49, 1749–1755 (2009).
Baker, C. N., Stocker, S. A., Culver, D. H. & Thornsberry, C. Comparison of the E Test to agar dilution, broth microdilution, and agar diffusion susceptibility testing techniques by using a special challenge set of bacteria. Journal of clinical microbiology 29, 533–538 (1991).
Wayne, P. Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. CLSI document M100-18. Clinical and Laboratory Standards Institute (2008).
Ligozzi, M. et al. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. Journal of clinical microbiology 40, 1681–1686 (2002).
Tamilmani, E., Radhakrishnan, R. & Sankaran, K. 13-Docosenamide release by bacteria in response to glucose during growth-fluorescein quenching and clinical application. Appl Microbiol Biotechnol, https://doi.org/10.1007/s00253-018-9127-x (2018).
Sjöback, R., Nygren, J. & Kubista, M. Absorption and fluorescence properties of fluorescein. Spectrochim Acta A 51, L7–L21, https://doi.org/10.1016/0584-8539(95)01421-P (1995).
Ueno, T. et al. Rational Principles for Modulating Fluorescence Properties of Fluorescein. J. Am. Chem. Soc. 126, 14079–14085, https://doi.org/10.1021/ja048241k (2004).
Kamiya, M. et al. Extension of the Applicable Range of Fluorescein: A Fluorescein-Based Probe for Western Blot Analysis. Angew Chem 44, 5439–5441, https://doi.org/10.1002/anie.200501542 (2005).
Dafopoulou, K. et al. Comparative Evaluation of Colistin Susceptibility Testing Methods among Carbapenem-Nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii Clinical Isolates. Antimicrobial agents and chemotherapy 59, 4625–4630, https://doi.org/10.1128/aac.00868-15 (2015).
Lo-Ten-Foe, J. R., de Smet, A. M., Diederen, B. M., Kluytmans, J. A. & van Keulen, P. H. Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrobial agents and chemotherapy 51, 3726–3730, https://doi.org/10.1128/aac.01406-06 (2007).
Bulik, C. C. et al. Comparison of meropenem MICs and susceptibilities for carbapenemase-producing Klebsiella pneumoniae isolates by various testing methods. Journal of clinical microbiology 48, 2402–2406, https://doi.org/10.1128/jcm.00267-10 (2010).
Zhang, S. X. et al. Evaluation of CLSI agar dilution method and Trek Sensititre broth microdilution panel for determining antimicrobial susceptibility of Streptococcus pneumoniae. Journal of clinical microbiology 49, 704–706, https://doi.org/10.1128/jcm.01622-10 (2011).
Satlin, M. J. The Search for a Practical Method for Colistin Susceptibility Testing: Have We Found It by Going Back to the Future? Journal of Clinical Microbiology 57, e01608–01618, https://doi.org/10.1128/JCM.01608-18 (2019).
Cielecka-Piontek, J., Zając, M. & Jelińska, A. A comparison of the stability of ertapenem and meropenem in pharmaceutical preparations in solid state. Journal of pharmaceutical and biomedical analysis 46, 52–57 (2008).
Acknowledgements
This work was carried out at National Hub for Healthcare Instrumentation Development (NHHID) with the financial support from Technology Development and Transfer Division of Department of Science and Technology, Government of India (No. IDP/MED HUB/2009-10/28.1.2011); and the University Grants Commission, Government of India under their University with Potential for Excellence scheme (F. No. 1-1/2016 (NS/PE)/Dt:07.12.2016) at Centre for Biotechnology, Anna University. We acknowledge Dr P. Kaliraj, ICMR Emeritus Scientist, Centre for Biotechnology for his valuable guidance in clinical perspective. We are grateful to Mr M. Mani, MD, Microbiological Laboratory, Coimbatore, for all the support and encouragement with guidance, and their clinical microbiologists, Dr K. N. Brahmadathan and Dr Deepa Sankari for their facilitation in the evaluation and for the discussions.
Author information
Authors and Affiliations
Contributions
Dr. K.S., Dr. C.P.T. and R.R.K. designed the study; J.R. and Dr. N.S.D. fabricated the imaging device and the software for the study; R.R.K. performed experiments under the supervision of Dr. K.S. and Dr. C.P.T.; R.R.K. consolidated the data; R.R.K. and Dr. K.S. analysed the data and wrote the manuscript. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Radhakrishnan, R., J., R., N. S., D. et al. High-throughput method for Antibiotic Susceptibility Testing based on Fluorescein Quenching by Bacteria: Application to Urinary Tract Infection. Sci Rep 10, 4058 (2020). https://doi.org/10.1038/s41598-020-60717-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-60717-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.