Abstract
All tropically adapted humped cattle (Bos indicus or “zebu”), descend from a domestication process that took place >8,000 years ago in South Asia. Here we present an intercontinental survey of Y-chromosome diversity and a comprehensive reconstruction of male-lineage zebu cattle history and diversity patterns. Phylogenetic analysis revealed that all the zebu Y-chromosome haplotypes in our dataset group within three different lineages: Y3A, the most predominant and cosmopolitan lineage; Y3B, only observed in West Africa; and Y3C, predominant in South and Northeast India. The divergence times estimated for these three Zebu-specific lineages predate domestication. Coalescent demographic models support either de novo domestication of genetically divergent paternal lineages or more complex process including gene flow between wild and domestic animals. Our data suggest export of varied zebu lineages from domestication centres through time. The almost exclusive presence of Y3A haplotypes in East Africa is consistent with recent cattle restocking in this area. The cryptic presence of Y3B haplotypes in West Africa, found nowhere else, suggests that these haplotypes might represent the oldest zebu lineage introduced to Africa ca. 3,000 B.P. and subsequently replaced in most of the world. The informative ability of Interspersed Multilocus Microsatellites and Y-specific microsatellites to identify genetic structuring in cattle populations is confirmed.
Similar content being viewed by others
Introduction
The last two decades of genetic studies on the origin of livestock species have unveiled a remarkable set of new questions regarding the origin and spread of domesticated animals. Research on single versus multiple domestications, interbreeding of wild and domestic animals, and cultural and faunal exchanges is transforming current perspectives on domestication trajectories and biodiversity1,2,3. Genetic studies of cattle domestication have pointed out that modern cattle (Bos taurus and B. indicus) resulted from at least two genetically distinct auroch populations (B. primigenius and B. nomadicus)4,5. The domestication of B. taurus in the Near East and expansion of taurine cattle into Europe has received particular attention6,7,8. However, studies of the domestication and spread of the zebu cattle lineage, B. indicus, are relatively scant.
Archaeological findings suggest that zebu cattle were domesticated 8,000–9,000 years ago (B.P.) and dispersed throughout northwestern South Asia by 6,000 years before present9,10. South India may have been an additional centre for cattle domestication11,12, as may have been Gujarat in western India9,11,12,13. Several areas of domestication or wild capture are consistent with genetic studies, which have identified two major mtDNA haplogroups (or lineages) in zebu cattle: I1 and I24. An extensive survey of modern cattle suggests that the maternal zebu cattle lineage I1 likely originated from the domestication of local wild cattle (Bos namadicus) in northwestern South Asia4. Animals of this lineage may have spread through South Asia (≈5,500–4,000 years B.P.) and beyond the Subcontinent eastwards to Southeast Asia and southern China (by ca. 2,500 B.P.)4. After this initial spread, additional genetic diversity was recruited to domestic herds from South Asian wild cattle populations carrying the haplogroup I2.
In comparison with mtDNA4 and genome-wide14 information studies conducted on B. indicus analyses of Y-chromosome diversity are rare. Seminal studies using Single Nucleotide Polymorphisms (SNPs) identified a single zebu Y-chromosome lineage (Y3)15. Admixture between B. taurus and B. indicus in Africa and differences in diversity parameters assessed using Y-chromosome markers16,17,18 have contributed to understand the complexity of domestication processes, the early spread of male zebu cattle in South Asia and the introduction of zebu to Africa through Indian Ocean trade.
Humped zebu-like cattle were introduced to Egypt from the Levant in small numbers and depicted in tombs and temples 3,400–3,000 B.P.19 but present African zebu cattle populations are the result of multiple introductions. Archaeology suggests that most animals came through eastern rather than northern Africa. Pre-Aksumites, Aksumites and their trading partners in Yemen and the Red Sea brought zebu cattle to the Horn of Africa 2,000–1,600 B.P.20,21 following well known sea trade routes from South Asia to the Gulf of Oman22,23. Humped cattle followed Sahelian routes, appearing in West Africa by 1,000 B.P.24. The rinderpest panzootics of 1889–1896 are estimated to have annihilated up to 80% of herds in many regions or over 5.2 million African taurine cattle25 and resulted in zebu being massively reintroduced along the eastern coastline of Africa, largely replacing African taurine bulls.
Genetic data have demonstrated that African cattle mainly carry the maternal T1 taurine mtDNA lineage26. However, the African zebu and sanga populations (crosses between African taurine and zebu cattle) mainly carry Y-chromosomes of zebu origin17. The absence of zebu mitochondria in African cattle implies that introgression of the zebu lineage into Africa was mostly male mediated. A number of microsatellite-based studies have also shown that zebu introgression into Africa declines from East to West26. Moreover, a study using Y-chromosome markers, found that the zebu-specific Y-chromosome sub-lineage Y3B27 was absent from the extant zebu population from the Indian subcontinent. Very recently, the X-degenerate region within the male-specific part of the bovine Y-Chromosome was resequenced28 allowing to identify two sub-haplogroups within the B. indicus Y3 chromosome: the Y3a sub-haplogroup identified in Chinese cattle and the Y3b sub-haplogroup mainly carried out by Indian zebu sires. However, African zebu was not included in that analysis28. Here, we report on the genetic analysis of a large number of West African and South Asian zebu Y-chromosomes to shed light on the origin and spread of zebu cattle within south Asia and from south Asia to Africa.
Methods
Sampling
Samples from 248 B. indicus males belonging to 22 cattle populations from Asia and Africa were analysed (Supplementary Table S1). The Asian dataset totalled 100 made up of three Indian native populations (from northeastern, Central and southern Indian areas; totalling 26 samples) plus unrelated samples from major zebu breeds kept in Argentina and Brazil that descend from pure zebu sires from India: Brahman (6), Gir (10), Guzerat (5), Indubrazil (2), Nelore (46) and Tabapuã (5). When necessary, samples from Brahman, Indubrasil and Tabapuã breeds were pooled into a single population due to their historical admixed origin (which included the use of Guzerat, Nelore and Gir individuals). We consider this pooled Indian population to originate from a geographical location that averages those of parental populations. We also analysed samples from 5 sires (obtained in 3 different countries) from Central Asia and 8 sires from Yemen. The African dataset totalled 135 samples, from East Africa (3 populations and 38 samples) and West Africa (6 populations, including Central African M’Bororo cattle, and 97 samples).
Genotyping
The two Y-specific Interspersed Multilocus Microsatellites (IMMs; UMN2405 and UMN2303) and the six Y-chromosome specific microsatellites (INRA189, UMN0103, UMN0307, BM861 and BYM1)16, with two loci typed for microsatellite UMN010329 were genotyped following a protocol described previously27. We have adopted the terminology recommended by the Human Y-Chromosome Consortium30.
IMM bands and microsatellite alleles were combined into haplotypes. Observed haplotypes were analysed as follows: (a) Analysis of correspondence was performed using the “Proc Corresp” of the SAS/STAT package (SAS Institute Inc., Cary, USA); (b) applying the Bayesian procedures implemented in the program MrBayes31,32. Two MCMC runs starting from different random trees were completed. Each run consisted of 20 million replications and four chains. All sample points prior to reaching convergence were discarded as burn-in samples. The remaining samples were used to generate a majority rule consensus tree, where the percentage of samples recovering any particular clade represented the clades posterior probability32; and (c) a MJ network connecting different haplotypes was constructed using the program Network 4.5.233 (available at http://www.fluxus-engineering.com/). To avoid reticulation, a reduced median algorithm34 was used to generate an.rmf file to which the median joining network method33 was applied to this file. Following the recommendations of35, the “frequency >1” option was applied to discard singly occurring Y-types. The same weights were assigned to each polymorphism.
Phylogenetic and population genetic analyses
An unbiased estimate of haplotype diversity, h, and its variance, V(h), were calculated according to Nei36 (formulas 8.5 and 8.13 therein). The standard error of h, SE(h), was calculated by taking the \(\sqrt{V(h)}\). Within population gene diversity adjusted for sampling size was also computed following Nei36. The between-populations genetic identity matrix was computed as the average across loci of the term to \(\sum _{ij}\,{x}_{ij}{y}_{ij}\) 36,where xij and yij are the frequencies of the ith allele at the jth locus within the populations x and y, using the program MolKin37. The information provided by the between-population genetic identity was summarized, from the complementary of this matrix, using a PCA. The PC scores and the canonical dimensions computed for each analysed population were used to construct interpolation maps drawn using the Spatial Analyst Extension of ArcView, available at http://www.esri.com/software/arcview/. The Inverse Distance Weighted (IDW) option with a power of two was selected for the interpolation of the surface. IDW assumes that each input point has a local influence that decreases with distance. The sampling area of each population was used as geographic coordinates and interpolation surfaces were divided into eight equal classes.
Demographic scenarios
Three historical scenarios for domestication and the spread of zebu cattle were modelled (Fig. 1) using the coalescent sampler implemented in SIMCOAL 2.1.238. We refer only to the initial capture of males as domestication (≈9,000 years ago1,2) and refer to later instances of capture and recruitment of wild bulls to herds as introgression event or gene flow. Model 1 assumed an ancestral Y-chromosome population undergoing a bottleneck associated with a single domestication and subsequent population split at the first introduction of zebu into Africa (ca. 3,500 B.P.19,20) into three different populations, two of them being merged at 2,000 B.P.; Model 2 assumed three different ancestral Y-chromosome domesticated populations evolving separately during 250,000 years39, with two of them being merged at 2,000 B.P.; Model 3 assumed two ancestral Y-chromosome domesticated populations one of them splitting into two subpopulations at 3,500 B.P. (which, in turn, merge at 2,000 B.P. like in the other two Models) and received migrants in an introgression event at 3,000 B.P. from the other population of the ancestral lineage. The scenarios modelled represent simple (Model 1) or complex (Models 2 and 3) domestication processes that originated the present admixed zebu Y-chromosome population. Model 3 represents the more complex, and sustained in time, domestication scenario including a dominant ancestral Y-chromosome population which acts as a genetic source of the others. The merging of two of the ancestral populations at 2,000 B.P. represents a secondary, less intense, recruitment of wild oxen, probably out of the Indus Valley, suggested by mtDNA structure4. Current effective population sizes (103–105 individuals), IMMs mutation rate (ranging from 2 × 10−5 to 2 × 10−4 mutation/generation) and the size of the initial domesticated populations (1–200 individuals) were modelled assuming uniform distributed priors. Divergence times were measured in years and transformed to generations for simulations using a generation time of 4.84 years40. For Y-chromosome microsatellites, rapidly mutating markers mutation rate41 (0.008 mutations/generation) was used under a strict stepwise mutation model. The two IMMs, which are considered slow mutation dominant markers27, were simulated as independent markers. Both the IMMs and the six different microsatellite loci typed were simulated using SIMCOAL for 180 individuals. This latter figure coincides with the number of available non-American zebu Y-chromosomes plus those haplotypes that were only observed in American zebu.
Schematic view of the demographic models compared in the ABC model choice. Model 1, a single domestication followed by the diversification in three descendant lineages. Model 2, three independent domestications or early recruitment from three differentiated ancestral populations. Model 3, a wild oxen admixture (grey arrow) between a single domesticated lineage and an ancestral differentiated population (with a proportion of admixture input into the domesticated lineage of 0.7950. In all cases, the complete admixture of two of the descendant lineages occurred 2,000 B.P. The “b” within the grey square illustrates the bottleneck associated with domestication. Arrows represent the direction of the events backwards in time (coalescent simulations). The last admixture at time 0 (present) was included in the coalescent simulations to account for the artificial mixture caused by the unique sample. Details of the parameters used for simulations are provided in the Materials and Methods section.
The posterior probability of each proposed scenario was then calculated using the software ABCtoolbox42 with 106 simulation replicates generated under each scenario and using four summary statistics, the standard deviation over loci of the number of alleles (Ksd), the mean heterozygosity over loci (\(\bar{H}\)) and standard deviation over loci of the heterozygosity (H) and the mean total heterozygosity (\(\overline{Ht}\)) calculated with the software alrsumstat43. Half a percent (0.5%) of the simulations matching closest the empirical data based on distances between observed and simulated statistics was retained for the estimation of the marginal densities under the General Linear Model (GLM)44 for each evolutionary scenario. These were also used for the assessment of the posterior odds (Bayes factors)45 for each model given the observed data. The ABC model choice was validated using simulated replicates under the three models as pseudo-observed datasets to estimate the power to distinguish between models, the percentage of model misclassification and the posterior probabilities supporting wrong choices43.
Divergence times estimation
Divergence times were estimated via the ρ statistic (i.e. the average number of mutations from derived haplotypes) to a haplotype designated as ancestral for the haplogroup18,46. The average distance to the node of interest (ρ was transformed to absolute time estimates by multiplication (ρ × years per 1 mutation). As a phylogeny-based statistic, ρ offers the advantage of being unbiased by demographic processes. The sampling error of ρ was approximated as \(\sqrt{\frac{{\boldsymbol{\rho }}}{n}}\), where n denotes the sample size. Due to the fact that our dataset included IMMs, that are likely to be dominant slowly-mutating markers27 and rapid mutating Y-chromosome microsatellites, divergence times were computed assuming the average mutation rate of 0.0008/generation reported in39 for intermediate- rate mutating Y-chromosome markers.
Additional resequencing
The very recent identification of two sub-haplogroups (Y3a and Y3b) within the zebu Y3 Chromosomes by Chen et al.28 made necessary carrying out additional analyses to ascertain their consistency with the variability identified in the current analysis. A 443pb fragment (from position 3631054 to 3631601; reference assembly Btau_5.0.1 GCF_000003205.7) of the X-degenerate region within the male-specific part of the bovine Y-Chromosome was resequenced for diagnostic purposes. The amplified fragment included three mutations (g.3631254a > g, g. 3631400t > c and g. 3631401t > g) separating the Chen et al.’s28 B. indicus Y-Chromosome sub-haplogroups Y3a and Y3b. A total of 51 sequences were obtained. A complete description of the laboratory methods used is in Supplementary Table S2.
Results
Sixteen of the bands (seven for UMN2405 and nine for UMN2303) described for the IMMs used in Pérez-Pardal et al.27 were polymorphic (in terms of presence/absence) in zebu cattle. The number of alleles per microsatellite varied from one (BM861) to four (smaller loci of the marker UMN0103, BYM1 and INRA189), revealing a total of 19 alleles across the six loci here considered (Supplementary Table S3).
We identified 47 different haplotype combinations resulting from the genotypes of two IMMs and six microsatellites markers. Of these, 29 are unique. The three most frequent haplotypes (H19, 24 samples; H20, 37; and H21, 55) were identified in 18 different populations. However, these three haplotypes were neither found in non-Indian Asian samples nor in sires from northeastern India. Haplotype H24 was identified in 23 samples from the six West African populations analyzed (Supplementary Table S3). Within-population haplotypic diversity ranged from 0.089 ± 0.015 in Nelore cattle to 0.800 ± 0.213 in Ethiopian Raya-Azebo and Nigerian Goudali cattle (Supplementary Table S1). Within-population gene diversity varied from 0.014 in Nelore cattle to 0.181 in Malian Bororo cattle. Regarding geographic areas, (not considering the strongly bottlenecked American zebu samples) the highest haplotypic and gene diversity values were found in non-Indian Asian samples (0.583 ± 0.101 and 0.202, respectively) or Indian (0.500 ± 0.008 and 0.209, respectively) sires. Overall gene diversity and haplotype diversity were 0.152 and 0.190, respectively (Supplementary Table S1).
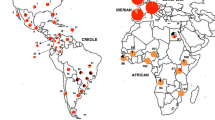
All the Bayesian phylogeny, median-joining (MJ) network and the correspondence analyses concur in the identification of three Y3 haplotypic families (Fig. 2). The main one, Y3A, included 26 haplotypes and 186 samples (55% and 75% of the total haplotypes and samples, respectively). The Y3B included 12 (25%) haplotypes and 24 (10%) samples, while the Y3C included 9 (20%) haplotypes and 38 (15%) samples (Supplementary Table S1). When compared with haplotypic family Y3A, the Y3B is mainly defined by the presence of three bands (126, 127 and 128) on the IMM UMN2303, while family Y3C is defined by the absence of the allele 149 on microsatellite UMN0307 and the predominant presence of band 124 on IMM UMN2303 (Supplementary Table S3). The Bayesian analysis provided a statistically significant confidence (0.94) for the separation between haplotypic families Y3A and Y3B while that between families Y3A and Y3C was slightly lower (0.87). Correspondence analysis separated the haplotypic families Y3B and Y3C on Dimension 1 (X-axis) while the main haplotypic family (Y3A) was differentiated on Dimension 2 (Y-axis). The network obtained using the whole dataset was highly reticulated (Supplementary Fig. S1). However, after removal of singly occurring haplotypes35 the network obtained was highly consistent with the other analyses (Fig. 2C) identifying H20, H24 and H44 as the central haplotypes within families Y3A, Y3B and Y3C, respectively. Observed haplotype families also showed differential geographic frequencies (Fig. 2D). The Y3A was predominant in India and was present in all populations sampled except for Yemen and The Central African Republic. Haplotypic family Y3B was only present in West Africa while Y3C was mainly present in South India and Yemen and only has traces in Northeast India, Central Asia and East Africa.
Graphical representation of genetic diversity and phylogeny of the zebu specific Y-chromosome lineages. Plot (A) shows the phylogenetic tree constructed from the 146 identified haplotypes using the Bayesian procedures implemented in the program MrBayes 3.1.Plot (B) shows the two dimensions calculated on the polymorphism of the 47 Y-chromosome haplotypes identified via correspondence analysis; and Plot (C) shows a network tree constructed using the program Network 4.5. The three identified haplotypic families (Y3A, Y3B and Y3C) are, respectively, in orange, blue and green. Plot (D) shows a geographic map and the frequency of the haplotypic families identified. Here, size of the circles does not correspond with sample size.
Between-population genetic relationships were assessed via Principal Components Analysis (PCA; Supplementary Table S4) and graphically visualized on synthetic maps (Supplementary Fig. S2). The first three principal components (PC) with eigenvalue >1, explain 65.5, 16.4 and 7.4% of the total variability. The first PC differentiates the genetic variability of most northern Indian subcontinent and Central Asian samples (mainly carrying Y3A haplotypes) from those of Southern India and Yemen (where family Y3C haplotypes are present). The second PC contrasts the genetic variability of the Yemen, and southern and northeastern India populations with those samples from West Africa in which haplotypes belonging to family Y3B are frequent (Supplementary Fig. S2). Consistency of this geographically-related analysis was evaluated by a second PCA from which zebu individuals of Argentinean and Brazilian origin were excluded. This latter analysis allowed identification of two PCs with eigenvalue >1, explaining 64.7 and 17.1% of the total variability (not shown).
Three historical scenarios (Fig. 1) for the domestication and spread of zebu cattle were modelled using the obtained data. Two of the three models compared under the ABC model choice (Model 1 and Model 2) had high relative posterior probabilities (PP), with Bayes factors (BF) of 8.9 and 6.4 with Model 3. Therefore, Model 3 was clearly disposable. Although Model 2 was the most probable model, its probability is only slightly higher than that of Model 1 (BF = 1.4), which is insufficient statistical evidence to draw clear conclusions about a possible separate domestication of different zebu Y-chromosome populations. Nevertheless, our cross-validation study of the statistical properties of the ABC model choice indicated that this slight difference in probabilities might be enough to seriously consider Model 2 as the best candidate. This claim is supported on the fact that, when comparing the three models tested based on pseudo-observed data sets (PODS; Supplementary Fig. S3), the estimated posterior probability favouring Model 2 given real data (PP = 0.548) was never reached in false positives (i.e., when the true model that generated the data was Model 1).
Moreover, Model 2 was additionally run considering the admixture of the three descendant populations at 2,000 B.P. This scenario was less supported by the observed data (results not shown), indicating that two of the descendants of these domesticated populations would share recent history but a third population would remain isolated (or partially isolated) until the present. This fact leaded us to retain the hypothesis underlying Model 2 as more plausible.
Resequencing (Supplementary Table S2) informed that sequences assigned to the haplotypic family Y3A and to the “African” haplotypic family Y3B identified here can be included into the Chen et al.’s28 B. indicus Y-Chromosome sub-haplogroup Y3b, identified in zebu sires of Indian origin, while the sequences assigned to the current the haplotypic family Y3C carried mutations g.3631254a > g, g. 3631400t > c and g. 3631401t > g and, therefore, are consistent with the definition of the Chen et al.’s28 B. indicus Y-Chromosome sub-haplogroup Y3a, mainly identified in Chinese zebu sires.
Discussion
The diversity observed in the zebu cattle lineage was lower than that for taurine cattle at the same Y-chromosome loci16,27. In keeping with the early history of movement of the lineage, B. taurus Y-chromosomes belong to two different haplogroups (Y1 and Y2)15. All zebu cattle Y-chromosome haplotypes fall within a distinct Y3 haplogroup. More specifically, zebu cattle Y-chromosomes can be classified into three different Y3 haplotypic families (Fig. 2). However, the low genetic variability found is consistent with a short divergence between these haplotypic families (Table 1) indicating a short divergence between the wild populations involved in the domestication process. The ABC model choice used to compare the support of the data to three possible domestication histories points towards a scenario of separated domestication for each the three Y-chromosome families identified or multiple early and regionally diverse events of gene flow as likely, with no simple discrimination among them possible. With the revelation of how common gene flow is among domestic animals and their wild relatives today researchers are leaning towards requiring a high bar for identification of multiple domestication1,2,47. In the light of the validation study carried out, however, the results of the ABC model choice must be interpreted as additional evidence to be considered together with other information, such as projection of zebu cattle Y-chromosome genetic variation on geographical maps, to clarify to the history of the species.
The comparison of the findings of the current study with recently reported by Chen et al.28 give insights of general interest as well:
-
(a)
The consistency of our majority haplotypic families Y3A and Y3B with the Chen et al.’s28 B. indicus Y-Chromosome sub-haplogroup Y3b is in agreement with the fact that haplotypic families Y3A and Y3B had the lower divergence times (6,957 ± 4,017 years; Table 1). Therefore, it can be hypothesized that sires carrying Y3A and Y3B haplotypes could belong to the same or closely related domesticated populations. Furthermore, the Indian origin of our “African” Y3B haplotypic family is confirmed.
-
(b)
The consistency of our South and Northeast India haplotypic family Y3C with the Chen et al.’s28 B. indicus Y-Chromosome sub-haplogroup Y3a clarifies that this “Chinese” sub-habplogroup is likely to have an Indian subcontinent origin. Therefore, the significance of the Indian subcontinent in the domestication of B. indicus4 is confirmed again.
History of zebu cattle
Despite the fact that the Indian subcontinent was the centre of domestication of zebu cattle9,10, the haplotypic family Y3B was only observed in African zebu. These data most likely reflect the effect of waves of introduction of zebu to Africa. The total absence of Y3B haplotypes in South Asia suggests interbreeding between local African and domestic Asian cattle, the traces of which were swamped by subsequent population shifts in South Asia, after the linage was introduced to Africa.
Currently available archaeological data indicates that humped cattle were not introduced to Africa earlier than ≈3,500 B.P.19,20 and that humped cattle may have reached these areas from abroad in small numbers19,21,24,48. The range of breeding choices made by the varied owners of zebu cattle are unknown49, but only zebu sires have provided a genetic signal traceable to the present50.
Considering that all three zebu-specific Y3 sub-lineages discovered in this research originated in the domestication centre, the exclusivity of the Y3B haplotypes in West Africa zebu sires can only be explained by the history of zebu movements and population demographic events. It is likely that this Y3B haplotypic family derived from an ancient South Asian process of domestication and was later replaced by herder’s recruitment of new male lineages from wild stock into the zebu gene pool.
The origin of West African zebu
The exclusivity of the Y3B haplotypic family in West African zebu cattle is significant as a reservoir of male biodiversity27. Given the significance of males for breeding programs we think that these populations should be further explored and seriously considered in any cattle biodiversity conservation program. Within the Indian subcontinent, the presence of the Y3C family in southern and southeastern India (Fig. 1) is also notable, particularly, when considering that the zebu mtDNA lineage (I2), is projected to date around ≈3,500 B.P.4. An early expansion of cattle-oriented Neolithic cultures carrying recent domesticated zebu cattle from the Indus Valley eastward into the Indo-Gangetic plains, and southward into the southern tip of the Indian subcontinent4,9,11, may have resulted in introgression of local wild stock into domesticated herds. In fact, population size estimates for B. indicus can only be explained if one assumes a substantial and posterior admixture event between the bottlenecked early domestic zebu population and wild animals39. Our estimates can support the posterior recruitment of I2 females as well as introgression of Y3C sires into early zebu herds. In the light of archaeological and ethnographic data it is likely that a range of practices in different regions of the continent resulted in wild-domestic gene flow, including extensive herding, herders intentional turning out domestic cows to breed with wild bulls, and wild capture to maintain hardiness and herd sizes2,9,10,11,51.
The elevated frequency of the haplotypic family Y3A in our African dataset may represent recent zebu introductions over the past two centuries, which have increased the frequency of Y3A sires in East Africa. The restocking of African herds with animals carrying Y3A haplotypes suggests that this lineage was already the most frequent in South Asia. Samples are smaller from Yemen, but given the pivotal role of the Gulf of Oman trade routes from India in ancient plant exchanges and traditional maritime coastal trading routes to the Horn of Africa, the absence of the Y3A lineage raises the question of whether nineteenth century introductions of East African zebu followed southerly Indian Ocean routes22 (Y3C; Supplementary Table S1).
The observed geographic patterns have led us to speculate that the first zebu males transported to Africa were mostly from Y3B and Y3C lineages and that recent introductions brought the Y3A lineage to Africa. As the eastern coast of Africa was more exposed to the influence of zebu introductions, the older Y3B and Y3C were replaced by the Y3A lineage, especially in the last 200 years. The Atlantic coast of Africa, on the other hand, was less exposed to the influence of these maritime routes, and the distance from eastern Africa was a factor in the persistence of the older Y3B male zebu lineage in western Africa cattle today. These findings of our research demonstrate how a livestock species such as cattle, has expanded through time in ways in which one wave of expansion might erase the former one, with persistence of unexpected biodiversity linked to ancient histories of trade and exchange.
Ethics Statement
Blood and hair root samples were collected by veterinary practitioners with the permission and in presence of the owners. For this reason, permission from the Ethics Committee for Health Research in Burkina Faso (Joint Order 2004-147/MS/MESSE of May 11, 2004) was not required. In all instances, veterinarians followed standard procedures and relevant national guidelines to ensure appropriate animal care. Semen doses were routinely collected by different companies not directly related to our research project.
Data Availability Statement
All genotypes, haplotypes and sequences obtained are provided as Supplementary Table S3 (xlsx file).
References
Larson, G. et al. Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. USA 111, 6139–6146, https://doi.org/10.1073/pnas.1323964111 (2014).
Marshall, F. B., Dobney, K., Denham, T. & Capriles, J. M. Evaluating the roles of directed breeding and gene flow in animal domestication. Proc. Natl. Acad. Sci. USA 111, 6153–6158, https://doi.org/10.1073/pnas.1312984110 (2014).
Zeder, M. A. Core questions in domestication research. Proc. Natl. Acad. Sci. USA 112, 3191–3198, https://doi.org/10.1073/pnas.1501711112 (2015).
Chen, S. et al. Zebu cattle are an exclusive legacy of the South Asia neolithic. Mol. Biol. Evol. 27, 1–6, https://doi.org/10.1093/molbev/msp213 (2010).
Bradley, D. G., MacHugh, D. E., Cunningham, P. & Loftus, R. T. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl. Acad. Sci. USA 93, 5131–5135 (1996).
Park, S. D. et al. Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biol. 16, 234, https://doi.org/10.1186/s13059-015-0790-2 (2015).
Decker, J. E. et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 10, e1004254, https://doi.org/10.1371/journal.pgen.1004254 (2014).
Beja-Pereira, A. et al. The origin of European cattle: evidence from modern and ancient DNA. Proc. Natl. Acad. Sci. USA 103, 8113–8118, https://doi.org/10.1073/pnas.0509210103 (2006).
Patel, A. Occupational histories, settlements, and subsistence in Western India: What bones and genes can tell us about the origins and spread of pastoralism. Anthropozoologica 44, 173–188, https://doi.org/10.5252/az2009n1a8 (2009).
Meadow, R. The Origins and Spread of Pastoralism in Northwestern South Asia in The Origins and Spread of Agriculture and Pastoralism in Eurasia (ed. D. Harris), 390–412 (London, University College London Press, 1996).
Allchin, F. & Allchin, B. Some New Thoughts on Indian Cattle in South Asian Archaeology (eds J. van Lohuizen-de Leeuw & J. Ubaghs), 71–77 (Leiden, Germany, Brill, 1973).
Fuller, D. Q. Agricultural origins and frontiers in SouthAsia: A working synthesis. J World Prehist. 20, 1–86, https://doi.org/10.1007/s10963-006-9006-8 (2006).
García-Granero, J., Lancelotti, C., Madella, M. & Ajithprasad, P. Millets and Herders: The Origins of Plant Cultivation in Semiarid North Gujarat (India). Curr. Anthrop. 57, 149–173, https://doi.org/10.1086/685775 (2016).
Porto-Neto, L. R. et al. Genomic divergence of zebu and taurine cattle identified through high-density SNP genotyping. BMC Genomics 14, 876, https://doi.org/10.1186/1471-2164-14-876 (2013).
Gotherstrom, A. et al. Cattle domestication in the Near East was followed by hybridization with aurochs bulls in Europe. Proc. Biol. Sci. B 272, 2345–2350, https://doi.org/10.1098/rspb.2005.3243 (2005).
Perez-Pardal, L. et al. Y-specific microsatellites reveal an African subfamily in taurine (Bos taurus) cattle. Anim. Genet. 41, 232–241, https://doi.org/10.1111/j.1365-2052.2009.01988.x (2010).
Hanotte, O. et al. Geographic distribution and frequency of a taurine Bos taurus and an indicine Bos indicus Y specific allele amongst sub-saharan African cattle breeds. Mol. Ecol. 9, 387–396, https://doi.org/10.1046/j.1365-294x.2000.00858.x (2000).
Anderung, C. et al. Investigation of X- and Y-specific single nucleotide polymorphisms in taurine (Bos taurus) and indicine (Bos indicus) cattle. Anim. Genet. 38, 595–600, https://doi.org/10.1111/j.1365-2052.2007.01663.x (2007).
Nicolotti, M. & Guerin, C. Le zebu (Bos indicus) dans l’Egypte ancieenee. Archaeozoologia 5, 87–108 (1992).
Marshall, F. The Origins and Spread of Domestic Animals in East Africa in Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography (eds R. Blench & K. MacDonald), 181–221 (London, University College London Press, 2000).
Chaix, L. The Fauna from the Uno/Bu Excavations at Bieta Giyorgis (Aksum) in Tigray, Northern Ethiopia: Campaigns 1995–2003; Pre-Aksumite, 700–400 Bc to Late Aksumite, Ad 800–1200. J. Afr. Archaeol. 11, 211–241, https://doi.org/10.3213/2191-5784-10244 (2013).
Boivin, N., Crowther, A., Prendergast, M. & Fuller, D. Q. Indian Ocean Food Globalisation andAfrica. Afr. Archaeol. Rev. 31, 547–581, https://doi.org/10.1007/s10437-014-9173-4 (2014).
Woldekiros, H. S. & D’Andrea, A. C. Early Evidence for Domestic Chickens (Gallus gallus domesticus) in the Horn of Africa. Int. J. Osteoarchaeol. 27, 329–341, https://doi.org/10.1002/oa.2540 (2017).
Linseele, V. From first stock keepers to specialised pastoralists in the West African savannah. In Pastoralism in Africa: Past, Present and Future (eds M. Bollig, M. Schnegg & H.-P. Wotzka), pp. 145–170. New York, Berghan Books.
Spinage, C. 2003 Cattle Plague. A History. (New York, Kluwer Academic/Plenum, 2013).
Hanotte, O. et al. African pastoralism: genetic imprints of origins and migrations. Science 296, 336–339, https://doi.org/10.1126/science.1069878 (2002).
Perez-Pardal, L. et al. Multiple paternal origins of domestic cattle revealed by Y-specific interspersed multilocus microsatellites. Heredity 105, 511–519, https://doi.org/10.1038/hdy.2010.30 (2010).
Chen, N. et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 9, 2337, https://doi.org/10.1038/s41467-018-04737-0 (2018).
Perez-Pardal, L. et al. Genetic structure of the bovine Y-specific microsatellite UMN0103 reflects the genetic history of the species. Anim. Genet. 42, 566–567, https://doi.org/10.1111/j.1365-2052.2011.02177.x (2011).
The Y-Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12, 338–348, https://doi.org/10.1101/gr.217602 (2002).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574, https://doi.org/10.1093/bioinformatics/btg180 (2003).
Huelsenbeck, J. & Ronquist, F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755, https://doi.org/10.1093/bioinformatics/17.8.754 (2001).
Bandelt, H. J., Forster, P. & Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48, https://doi.org/10.1093/oxfordjournals.molbev.a026036 (1999).
Bandelt, H. J., Forster, P., Sykes, B. C. & Richards, M. B. Mitochondrial portraits of human populations using median networks. Genetics 141, 743–753 (1995).
Forster, P. et al. A short tandem repeat-based phylogeny for the human Y chromosome. Am. J. Hum. Genet. 67, 182–196, https://doi.org/10.1086/302953 (2000).
Nei, M. Molecular Evolutionary Genetics (New York, Columbia University Press, 1987).
Gutierrez, J. P., Royo, L. J., Alvarez, I. & Goyache, F. MolKinv2.0: A computer program for genetic analysis of populations using molecular coancestry information. J. Hered. 96, 718–721, https://doi.org/10.1093/jhered/esi118 (2005).
Laval, G. & Excoffier, L. SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics 20, 2485–2487, https://doi.org/10.1093/bioinformatics/bth264 (2004).
Murray, C., Huerta-Sanchez, E., Casey, F. & Bradley, D. G. Cattle demographic history modeled from autosomal sequence variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2531–2539, https://doi.org/10.1098/rstb.2010.0103 (2010).
Gutierrez, J. P. et al. Pedigree analysis of eight Spanish beef cattle breeds. Genet. Sel. Evol. 35, 43–63, https://doi.org/10.1051/gse:2002035 (2003).
Zhivotovsky, L. A. et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 74, 50–61, https://doi.org/10.1086/380911 (2004).
Wegmann, D., Leuenberger, C., Neuenschwander, S. & Excoffier, L. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics 11, 116, https://doi.org/10.1186/1471-2105-11-116 (2010).
Peter, B. M., Wegmann, D. & Excoffier, L. Distinguishing between population bottleneck and population subdivision by a Bayesian model choice procedure. Mol. Ecol. 19, 4648–4660, https://doi.org/10.1111/j.1365-294X.2010.04783.x (2010).
Leuenberger, C. & Wegmann, D. Bayesian Computation and Model Selection Without Likelihoods. Genetics 184, 243–252, https://doi.org/10.1534/genetics.109.109058 (2010).
Kass, R. & Raftery, A. Bayes Factor. J. Am. Stat. Assoc. 90, 773–795 (1995).
Forster, P., Harding, R., Torroni, A. & Bandelt, H. J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 59, 935–945 (1996).
Frantz, L. A. F. et al. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet. 47, 1141–1148, https://doi.org/10.1038/ng.3394 (2015).
Magnavita, C. Ancient Humped Cattle inAfrica: A View from the Chad Basin. Afr. Archaeol. Rev. 23, 55–84, https://doi.org/10.1007/s10437-006-9008-z (2006).
Stock, F. & Gifford-Gonzalez, D. Genetics and African Cattle Domestication. Afr. Archaeol. Rev. 30, 51–72, https://doi.org/10.1007/s10437-013-9131-6 (2013).
Loftus, R. & Cunningham, P. Molecular genetic analysis of African zeboid populations. In The Origins and Development of African Livestock - Archaeology, Genetics, Linguistics and Ethnography (eds R. Blench & K. MacDonald), 251–258. (London, University College of London Press, 2000).
Choudhury, A. Wild water buffalo: Bhubalus arnee (Kerr, 1792). In Ecology, Evolution and Behavior of Wild Cattle(eds M. Melletti & J. Burton), 255–301. (Cambridge, Cambridge University Press, 2014).
Acknowledgements
This work was partially supported by the projects AGL2011-27585, MICIIN-FEDER AGL2016-77813-R. CompGen and the FCT grant PTDC/CVT/117851/2010 (COMPETE: FCOMP-01-0124-FEDER-020141). LP-P is supported by grant SFRH/BPD/94518/2013. ASG was supported by a grant under the program Beatriu de Pinós (2010BP-B 00175). SC is partially supported by National Natural Science Foundation of China (No. 31460281) and VC by an FCT grants SFRH/BD/88129/2012, respectively. Authors thank Claudio C. Fioretti, Morné de la Rey and Hiver Boussini for their support and help with sampling and Alba Rodríguez Saavedra for editing this paper.
Author information
Authors and Affiliations
Contributions
F.G., L.P.-P., A.B.-P. and I.A. conceived and planned the project; A.B.-P., F.G. and L.P.-P. wrote the paper; A.S.-G., L.P.-P., F.G. and I.F. did the data analyses; A.S.-G. run and interpreted ABC models; A.T., J.B.S.F., V.C., S.C., M.T. and R.J.C.C. undertook sampling and discussed and interpreted genetic data in light of the statistical and breeding evidence; A.P., R.H.M. and F.B.M. discussed and interpreted genetic data in light of the statistical and historical evidence; L.P.-P. and I.A. did the laboratory work. F.G. and A.B.-P. are the senior authors of this paper. All authors gave final approval for publication.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pérez-Pardal, L., Sánchez-Gracia, A., Álvarez, I. et al. Legacies of domestication, trade and herder mobility shape extant male zebu cattle diversity in South Asia and Africa. Sci Rep 8, 18027 (2018). https://doi.org/10.1038/s41598-018-36444-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-36444-7
This article is cited by
-
Global dispersal and adaptive evolution of domestic cattle: a genomic perspective
Stress Biology (2023)
-
Genome-wide investigations reveal the population structure and selection signatures of Nigerian cattle adaptation in the sub-Saharan tropics
BMC Genomics (2022)
-
Y-chromosome genetic diversity of Bos indicus cattle in close proximity to the centre of domestication
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.