Abstract
Accurate mapping and monitoring of tropical forests aboveground biomass (AGB) is crucial to design effective carbon emission reduction strategies and improving our understanding of Earth’s carbon cycle. However, existing large-scale maps of tropical forest AGB generated through combinations of Earth Observation (EO) and forest inventory data show markedly divergent estimates, even after accounting for reported uncertainties. To address this, a network of high-quality reference data is needed to calibrate and validate mapping algorithms. This study aims to generate reference AGB datasets using field inventory plots and airborne LiDAR data for eight sites in Central Africa and five sites in South Asia, two regions largely underrepresented in global reference AGB datasets. The study provides access to these reference AGB maps, including uncertainty maps, at 100 m and 40 m spatial resolutions covering a total LiDAR footprint of 1,11,650 ha [ranging from 150 to 40,000 ha at site level]. These maps serve as calibration/validation datasets to improve the accuracy and reliability of AGB mapping for current and upcoming EO missions (viz., GEDI, BIOMASS, and NISAR).
Similar content being viewed by others
Background & Summary
Tropical forests play a vital role in the Earth’s carbon cycle and contribute largely to uncertainties in the global carbon budget1. Methods to accurately map and monitor tropical forest carbon – or aboveground biomass (AGB) – are thus urgently needed to improve Earth system models and to help design carbon emission mitigation strategies in the context of Reducing Emissions from Deforestation and forest Degradation (REDD+)2,3. In the last decade, spaceborne Earth Observation (EO) data in combination with forest inventory measurements have been extensively used to generate spatially continuous AGB maps at pan-tropical scale using different modelling strategies3,4,5,6. However, existing broad-scale maps show divergent estimates among themselves and differ from field-derived forest AGB stocks at different spatial scales1,4,5,7, indicating the presence of high uncertainties in prediction maps. To improve the accuracy and reliability of AGB maps over the tropics, several ongoing and upcoming EO missions (NASA’s GEDI, ESA’s BIOMASS, NASA-ISRO’s NISAR and JAXA’s ALOS-4 missions, notably) have been specifically designed to collect satellite data sensitive to forest structure, hence to forest AGB6,8,9,10. While these new spaceborne datasets will undoubtedly revolutionise broad-scale forest AGB mapping, a network of high-quality reference data is needed to calibrate and validate the mapping algorithms11,12. Besides, using the same sets of reference data across different EO missions would vastly improve the comparability and confidence in the derived AGB maps, enabling their use in a wide range of science, policy, and management applications13.
Generating reference AGB observations over a given area is challenging since forest AGB is not directly measured through destructive sampling, but instead estimated from tree inventories and a series of statistical models propagating substantial uncertainties5. It is therefore required to reduce as far as possible and quantify the uncertainty on reference AGB predictions. In this context, the Committee on Earth Observation Satellite (CEOS) has established a good practice protocol for generating reference AGB dataset, to facilitate the production and warrant the consistency of next-generation biomass products14. The protocol suggests developing reference AGB maps over sizable areas using local forest sample plots and LiDAR data acquired using aerial platforms (hereafter airborne LiDAR). Airborne LiDAR data is currently the most informative data type for characterizing forest structure and deriving AGB maps at the landscape scale, provided they are adequately calibrated with respect to local environmental gradients and forest structural and species variability15. Besides, the high spatial resolution of airborne LiDAR data (or of derived AGB predictions) can easily be aggregated to coarser resolutions, thus bridging the scale gap between field data and the resolution of upcoming EO sensors (e.g. 100 m for NISAR, 200 m for BIOMASS).
The establishment and long-term maintenance of a network of reference forest AGB observatories across the tropics entails a myriad of challenges, particularly concerning the representativeness of the network12. Ideally, the network should be relatively evenly distributed in space and cover the main environmental gradients. While scientific discussions on site selection are on-going12, the Global Ecosystem Dynamics Investigation (GEDI) sensor on-board the International Space Station has already acquired data for a longer period than its initially projected lifetime. Data users would benefit from open-access reference AGB data, particularly in Asia where large geographic regions are not represented in the calibration/validation dataset of GEDI biomass mapping algorithm16,17. Besides the notion of spatial representativeness, hurdles related to the temporal mismatch between reference AGB and EO data should not be neglected. Rapid growth in regenerating forests or forest clearing/degradation – which notably characterise rural landscapes around central African cities, where slash-and-burn agriculture induces relatively fast dynamics – could rapidly make tens of thousands of GEDI data shots unusable. We argue that airborne LiDAR data acquired during GEDI lifetime over rapidly changing landscapes are invaluable and should be utilized to improve GEDI biomass mapping algorithms, notably on the lower-end of the forest biomass gradient to best capture forest degradation and regeneration gradients.
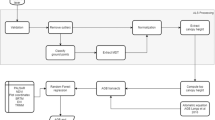
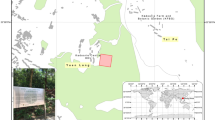
In this context, we aim to generate reference biomass datasets over the tropics for eight sites in Central Africa and five sites in South Asia (Fig. 1) by calibrating airborne LiDAR data with locally established field plots. This paper briefly describes (1) the details of the study sites and the datasets used, (2) the methodology used to generate the reference AGB maps (Fig. 2), and (3) the Monte Carlo simulation workflow used to generate uncertainty maps along with each reference AGB map. Finally, this paper provides access to these reference AGB datasets generated at 100 m and 40 m spatial resolutions over airborne LiDAR footprints ranging from 100 to 40,000 ha.
(A) Overview map showing the locations of sampling sites (n = 13) used in the current study. Outlined regions are expanded in (B): South Asian region and in (C): Central African region). Sampling site names and descriptions associated with site numbers are provided in Table 1.
Methods
Sampling sites and associated inventory and LiDAR datasets
We compiled co-located forest inventory and LiDAR datasets from 13 sampling sites in Central Africa and South Asia encompassing an array of abiotic conditions, forest types and structures (Fig. 1; Tables 1 and 2). Forest inventories were carried out at each site, and LiDAR datasets were obtained with an absolute temporal difference of 2.2 ± 1.9 years (range: 0–6.2 years) from the field measurements.
Forest inventories were conducted by different teams but followed similar protocols. In each plot, the diameter at breast height (DBH or referred to as D in this study, with D ≥ 10 cm) and the taxonomic identification of each tree were recorded. Tree relative coordinates within the plots were measured either at the individual or at the 20 × 20 m quadrat level. For a subsample of trees within the plots, tree height (H) was measured using a laser rangefinder device. Finally, plot geographic coordinates were determined using points measured every 20 m along the plot borders using a combination of differential GPS measurement system and electronic total station (in Asia) or a regular GPS system (in Africa), to warrant an accurate link between ground and remote-sensing data. The complete inventory dataset includes information on D and H measurements for respectively 97,251 and 13,303 trees, and identification rates of 89% at the species level and 92% at the genus level (8% of the trees were left unidentified). The number, size and layout of the inventory plots are uneven across sampling sites with, e.g., a single large 25-ha plot in the Forest-Geo “Rabi” site, a large 30-ha plot and smaller plots of 1-ha and 0.48-ha in the “Khao Yai” site, or a varying number of scattered 1-ha plots (ranging from 2 to 16 in the “Atout” and “Achanakmar” site, respectively). In general, the inventoried extent per site is smaller in Africa (9 ± 8 hectares) than in Asia (27 ± 13 hectares). For a breakdown of plot number, size, tree measurements and identification rates per sampling site, please refer to Table 3.
The LiDAR data at each sampling site were acquired between 2012 and 2022 using either aircraft or unmanned aerial vehicles (UAV, Table 1).
Inventory data processing: computation of reference AGB predictions
Forest inventories were first split into 1-ha (i.e. 100 × 100 m) and 0.16-ha (i.e. 40 × 40 m) plots, using information on tree location recorded in the field (i.e. either individual tree location or quadrat number). The two plot sizes correspond to the two mapping resolutions considered in this study. The 40-m resolution was chosen to account for plots where individual tree locations were only recorded at 20 × 20 m quadrat-level. In cases where the original plot size was not a multiple of the desired output size (typically when splitting 100 × 100 m plots into 40 × 40 m plots), subplots of the desired outputs size were selected at the edges of the original plot, thus leaving-out parts of the original inventory dataset (20 m wide bands in the center as per the previous example). The resulting number of 1-ha and 0.16-ha plots compiled at each sampling site is provided in Table 3.
Subsequently, the BIOMASS R package18 (version 2.1.8) within the R statistical platform (version 4.1.3) was used to compute reference AGB predictions for forest inventory plots at the two spatial resolutions (1-ha and 0.16-ha). To that end, we differentiated sites with a cumulated forest inventory area of 10 ha or more (i.e., 8 out of 13 sites, Tables 1 and 3) from those with less than 10 ha of cumulated forest inventory area (i.e., 5 sites). In the former case, we developed site-specific tree height-diameter (H-D) allometric models using second-order polynomials on log-transformed data (modelHD function in the BIOMASS package) and these models were used to predict the height of trees without H measurements in each respective site. In the latter case, which pertained to sites located in moist dense forests of Cameroon (SiteIDs 7 to 11 in Table 1), all inventory data from that country and biome were pooled into a single training dataset and the same H-D modelling procedure was applied. The resulting country- and biome-specific model was then used for predicting tree height at those sites. The H-D model coefficients for these site-level and Cameroon level model are presented in Table 4. Next, a wood density (WD) estimate was attributed to each tree based on its taxonomic identification using the getWoodDensity function.
Considering that tree AGB prediction is associated with various sources of uncertainty (including measurement errors of the independent variables such as tree diameter, height, and wood density, as well as prediction errors of the H-D models and the AGB allometric model)5,14, we used a Monte Carlo approach for uncertainty propagation. Specifically, we employed the AGBmonteCarlo function of the BIOMASS package18, which allows propagating the above-mentioned sources of uncertainty and outputs 1000 tree-level and subsequently plot-level AGB predictions. Tree AGB predictions were made using the pantropical AGB allometric model (i.e., Equation-4 in Chave et al.19). For each plot, the 1000 AGB predictions were (i) averaged to obtain a reference plot-level AGB density (hereafter AGBREF) for the development of LiDAR-AGB models and (ii) used for the propagation of uncertainties to the final AGB maps (see section “mapping forest AGB and prediction uncertainty”).
LiDAR data processing: computation of canopy height metrics
LiDAR data from African and Asian sites were processed using LAStools (version 201124) and the lidR R package (version 4.0.1), respectively. The same processing chain was applied to generate the canopy metrics in both cases. First, a digital surface model (DSM) free of pits and spikes was generated at a 1-m resolution by interpolating the highest points on a 1-m grid. Second, a ground point classification was performed on the point cloud and a digital terrain model (DTM) was interpolated from ground-points. The canopy height model (CHM) was then derived by subtracting the DTM from the DSM. Finally, the 1-m CHM was used to compute 15 canopy metrics for each plot (Table 5) as candidate predictors of forest AGB.
Specification of a general AGB model form
While LiDAR-based AGB mapping models were trained at the site or regional level (for some Cameroon sites), to minimise local bias in model predictions14,20, we privileged the use of (i) a single AGB model form across all sites to facilitate sites inter-comparison and the subsequent use of AGB predictions for spaceborne products calibration/validation and (ii) a simple, parametric modelling approach, keeping the number of predictors to a minimum to avoid overfitting and multicollinearity issues. To specify the AGB model form, we used linear mixed-effects models to identify the most predictive LiDAR-derived canopy height metrics (LCMs) on AGBREF variation while accounting for the hierarchical spatial structure of the data. In practice, we built 15 linear mixed-effects models (one for each LCM) on the log-transformed variables of AGBREF and LCM (Eq. 1):
where a and b are the model’s coefficients, LCM represents the Lidar-derived Canopy Metric, AGBREF corresponds to the field-derived AGB prediction at a given spatial resolution (i.e. 0.16- or 1-ha), REsite denotes the random site effect used in linear mixed-effects modelling and ε is the error term, assumed to follow a normally distribution with a mean of zero and a standard error σ. Based on the AIC criterion, the meanTCH metric (i.e. the mean of all CHM values in the plot area) emerged as the best predictor of AGBREF variation at both 1-ha and 0.16-ha spatial resolutions (Table 6).
A similar procedure was run on AGBREF prediction models combining each pair of LCMs rather than a single predictor. At both spatial resolutions, the best two-predictor model resulted in a modest improvement in relative RMSE (i.e., <0.2%, Table 7) compared to the model based on meanTCH only. The latter model form was thus selected for biomass mapping. In line with the H:D modelling procedure, LiDAR-based AGB mapping models were either trained at the site-level (for sites with a cumulated forest inventory area of 10 ha more) or on a pooled training dataset containing all inventory data from Cameroonian moist dense forests (for sites with a cumulated forest inventory area smaller than 10 ha), henceforth referred to as the “regional” AGB model. It is noteworthy that including sites as an additional fixed-effect covariate in the regional model did not yield significant effects for this variable at a 5% risk (neither in terms of site-level intercepts nor in terms of interactions between sites and the meanTCH predictor), suggesting a minimal site effect on the regional model’s predictions, if any.
The coefficients and calibration statistics of LiDAR-based AGB mapping models are provided in Table 8, while Fig. 3 shows scatterplots of ‘reference’ against predicted AGB values.
LiDAR-AGB models of Asian and African sites at 1-ha and 0.16-ha resolutions. The numbers for each site refer to Table 1. (7–11)* refers to the regional model established over moist dense forests of Cameroon.
Mapping forest AGB and prediction uncertainty
We mapped forest AGB and prediction uncertainty over the extent of airborne LiDAR data at each site using a Monte Carlo approach similar to that used to compute plot-level AGBREF. More specifically, we used the 1000 plot-level AGB predictions generated at the first modelling level (i.e., from tree to plot) to build 1000 LiDAR-based models per site (or at “regional” level for Cameroonian sites with less than 10 ha of cumulated forest inventory area). At the second modelling level (i.e., from plot to landscape), pixel AGB predictions derived from LiDAR-based models suffer from additional uncertainty associated to the LiDAR-based models themselves. To propagate this additional uncertainty, we mimicked the procedure used in BIOMASS to propagate the uncertainty associated to the tree-level AGB allometric model (see Appendix S1 of Réjou-Méchain et al.18 for codes and details), which entailed using a Markov chain Monte Carlo algorithm to infer the uncertainty on Lidar-based models’ parameters (i.e., models’ coefficients and associated RSE). The Markov chain outputted 1000 sets of model parameters per model. For each of the 1000 LiDAR-based model at each site, we then (1) randomly selected a set of parameters among the 1000 available sets, (2) used the model coefficient selected in (1) to predict pixels AGB and (3) added to all pixels an error term randomly drawn from a normal distribution N(0, RSEi) where RSEi is the model RSE selected in (1). This procedure led to 1000 predictions of pixels AGB embedding the prediction uncertainty from both the first and second modelling levels. Finally, reference AGB maps and associated spatial uncertainty maps were generated as the mean and standard deviation of the 1000 pixel AGB predictions, respectively. Hereafter, we refer to pixels mean AGB prediction as AGBPRED.
Additional metadata for the AGB maps
LiDAR-based AGB maps produced in the present study are intended to support calibration and validation efforts of spaceborne data. To maximise their usefulness, we provide additional information that users may require – depending on their study’s objective and methodological choices – to facilitate their integration with spaceborne data and/or develop comprehensive uncertainty propagation schemes up to the final, spaceborne-derived AGB map.
A first challenge users may face relates to the computation of the uncertainty associated with the mean AGB of arbitrary subregions of LiDAR AGB maps. Such subregions could for instance correspond to the footprints of spaceborne data unit pixels. Estimating the total mean squared error associated with a map (sub) population mean requires access to the matrix of pairwise population unit covariances, which is rarely communicated by map makers to users because of its large size. Yet, McRoberts et al.21 recently showed that pairwise population unit covariances could largely contribute to total mean squared error, and proposed an averaging and binning approach to drastically reduce the matrix size, thus facilitating its publication along with AGB maps. While we refer interested readers to McRoberts et al.21 for methodological details, we provide in Supplementary data all information recommended by the authors to allow map users to comply with IPCC good practice guidelines for greenhouse gas inventories. We note that for each pixel of the LiDAR-based AGB maps provided in this study, a bin number is available in the third map layer.
Another challenge lies in the propagation of uncertainties in multi-level hierarchical modelling, which is a likely use-case of the LiDAR-based maps we produced. These maps were generated by applying two hierarchically nested models: a tree allometric model linking field measurements to tree AGB, and a mapping model linking plot AGB to LiDAR data. LiDAR-based AGB maps users may employ a three-steps hierarchical modelling approach and add as a third step a model linking high resolution AGB predictions from the LiDAR-based maps to the coarser resolution of spaceborne data. An example of such an approach is presented in detail in Saarela et al.22 and referred to as “three-phase hierarchical model-based inference”. The uncertainty assessment in such a nested modelling approach requires information at the two first modelling steps that goes beyond the results of the Monte Carlo simulation we used to produce pixel-level uncertainty estimates. While we refer interested readers to Saarela et al.22 for methodological details, we provide in Supplementary data all information allowing users to assess uncertainty as described in Saarela et al.22. This information notably includes the variance-covariance matrix of model parameters for each sampling site as well as statistics on parameters (DBH, AGB, pixels’ height from CHM, etc.,) used at various levels in the chain of hierarchical models.
Data Records
For each site, AGB and uncertainty maps are distributed as a single GeoTiff file at the two spatial resolutions (1- and 0.16-ha) through Dataverse23. Each file comprises three individual layers. The two first layers named meanAGB and sdAGB correspond to the mean and standard deviation of AGB predictions over the 1000 Monte Carlo simulations, respectively. The file projection system is Universal Transverse Mercator. The third layer named Nbin corresponds to the bin number each map pixel is associated with in the binning approach proposed by McRoberts et al.21 to allow users reconstituting a matrix of pairwise population unit covariances estimates.
Besides data access through Rodda et al.23, data from Asian sites can be access and visualized through the Bhuvan Portal (https://bhuvan-app3.nrsc.gov.in/data/download/index.php). To access the visualisation/download through Bhuvan Portal, select the ISRO Geosphere-Biosphere Programme under the Program option and then choose the group Above Ground Biomass (AGB) Data.
In addition, we provide two supplementary data files (in excel format) that provide additional metadata details on site-level binned covariance matrices and variance-covariance matrices and summary of all the parameters used in the present study.
Technical Validation
Reference AGB maps at 1-ha resolution are shown in Fig. 4 and the density distributions of 1-ha AGB maps are represented in Fig. 5-A along with uncertainty levels in Fig. 5B expressed as a coefficient of variation (CV, in % of mean AGB) (see Fig. 6 and Fig. 7 for AGB maps and respective density distributions at 0.16-ha resolution). Figure 5-B shows that the mean uncertainty across sites is 15.4%, with site-level mean uncertainty ranging from 10.8 to 31%. It can be observed that Nachtigal and Uppangala sites have larger mean uncertainties than other sites, with 31% and 20.1%, respectively. This can be explained by larger LiDAR-AGB model uncertainties at these sites and mapping resolution (see models’ “sigma” in Table 8).
In addition to the per pixel estimates of uncertainty accompanying AGB maps, we hereafter provide (i) an assessment of mapping model predictive performances using a spatial model cross-validation technique24, to provide additional insights into the reliability of AGB predictions on each map and (ii) an assessment of mapping models extrapolation at each sampling site, which may be useful to help users for filtering-out pixels where extrapolation occurred and only retaining the highest quality AGB predictions for spaceborne products calibration/validation.
Model spatial cross-validation
Model calibration statistics in Table 8 likely overestimate model predictive performance on pixels that are not used for model training, that is, on most maps’ pixels. We performed a cross-validation (CV) of each model to provide more reliable insights into model predictive performance. Field plots at each site are iteratively split into training and test data and model CV statistics are built on the set of test data predictions. Regarding CV design, we selected a buffered leave-one-out cross-validation (LOO-CV25) where a spatial buffer around test data is used to exclude from model training dataset observations located at the neighbourhood of test data, thus avoiding inflation in CV statistics due to spatial autocorrelation in forest AGB24. As a compromise between the diversity in terms of number and spatial arrangement of field data across sites (e.g. multiple individual 1-ha plots vs. single large plot), the consistency of the CV approach across sites, as well as our expectation for a relatively weak spatial autocorrelation in forest AGB at the high resolution of the maps (<100 m2)26, we selected a LOO-CV with a 100 m buffer radius for all sites and mapping resolutions (i.e. 100 × 100 m and 40 × 40 m). This CV design notably implies that (i) when a test observation came from a large field plot (i.e. >1-ha, e.g. the 25-ha plot at Rabi), subplots at its direct neighbourhood were not used for model training (i.e., all subplots intersecting a 100 m circular buffer around the center of a test subplot were excluded from the training set, regardless of the mapping resolution), and (ii) at the 40 × 40 m mapping resolution, when a test observation came from a 1-ha field plot, the remaining three subplots of that 1-ha plot were not used for model training. The results of the buffered LOO-CV are presented in Table 9. They show that the predictive performances of mapping models developed in this study are comparable to those found in the literature (i.e. 15–20% on average for the tropical forest biome15) with relative RMSEs ranging from 10.6 to 20.1% (mean across sites: 14.1%) at 1-ha and 17.7 to 33.7% (mean across sites: 25.7%) at 0.16-ha.
Model extrapolation in the predictor space
Uncertainty maps, AGB maps, and model CV results provide insights into the reliability of AGB predictions within the calibration domain of mapping models. It is however likely that the entire gradient of forest structure sampled by LiDAR data was not fully sampled in the model’s training set, thus leading to situations of predictive extrapolation where prediction uncertainty is unknown. To investigate this issue, we compared the range of vegetation height (i.e., meanTCH) sampled by the training set of each mapping model to the full range found in the LiDAR data, restricting the analysis to pixels considered as vegetated, i.e., with meanTCH ≥ 2 m. We found that the proportion of pixels affected by predictive extrapolation strongly varied across sites and at the two mapping resolutions. Generally, the upper range of meanTCH (and thus of AGBPRED) found at a landscape scale in the LiDAR data were sampled in the training set (Fig. 8A,B), which probably is a reflection of the “majestic forest bias”27 – that is, the tendency for researchers to preferentially establish sample plots where forest stands appear the less disturbed (e.g. tallest canopy height, the highest abundance of large trees, etc.). However, a varying and often substantial proportion of maps on the lower end of the meanTCH gradient was outside the model’s calibration domain. For instance, in the Nachtigal site predictive extrapolation occurred on about 83% of the vegetated pixels on the 1-ha AGB map. This can be explained by the nature of this site, a forest-savanna mosaic, where the meanTCH of all herbaceous and shrubby savannas is lower than the height of the smallest 1-ha forest stand (ie., 16.4 m) found in model training set (Fig. 8A). However, this proportion dropped to 0% at the 0.16-ha mapping resolution thanks to the inclusion into the model training set of 18 additional 0.16-ha plots established in savannas-dominated areas (Fig. 8B, Table 3).
Proportion (in %) of map pixels outside and inside models calibration domains at 1-ha (panel A) and 0.16-ha (panel B) mapping resolutions. The proportions are computed with respect to the total number of map pixels with CHM > 2 m at the exception of the Natchigal site where a 0.4 m threshold is used so as to account for the nature of the site i.e., a forest-savanna mosaic. The proportion of map pixels within model calibration domains is represented in red. Map pixels below and above the range of model calibration domains are represented in blue and green, respectively.
We thus advise potential users of the AGB maps published here to carefully consider the bounds of the mapping model calibration domain at each site (for which we provided corresponding AGBPRED values in Fig. 8) when using these maps as reference data for larger-scale product calibration/validation.
Usage Notes
Forest AGB maps released here constitute reference estimations for the community of remote-sensing scientists interested in forest carbon stocks. For instance, we expect these maps to be of utmost usefulness for the calibration and validation of next-generation broad-scale aboveground biomass mapping models based on data from ongoing or upcoming spaceborne missions (viz., NASA”s GEDI, NASA-ISRO’s NISAR and ESA’s BIOMASS missions). These data can also be useful when assessing the accuracy of existing maps or recalibrating them (as in eg.28,29), especially since study sites presented here are located on renowned data-poor regions16 and are marked by notable uncertainties in AGB estimates17. That said, we encourage users to account for the dates of LiDAR and ground data acquisitions underlying reference AGB estimates (cf. Table 1), as temporal discrepancies with spaceborne signals or products should ideally be accounted for in any calibration or validation exercise.
More broadly, our study highlights sites of potential interest to build a network of “super-sites” (sensu11) across the tropics, that is sites combining forest inventory data over sizable areas (≥10 ha) – ideally featuring multiple forest censuses – with airborne LiDAR data. Such data have been collected thanks to the long-term vision of few organisations, to dedicated experts and to the efforts of trained labour forces in the past decades. Ongoing global changes makes the sustained monitoring of permanent forest plots in long-term study sites critical, so as to allow measuring their impacts on forest ecosystems. In spite of this crucial stake, access to funding in the tropical world for replacement of expertise, training programs and to support field data acquisition campaigns is critically limited. We thus urge National and International research and space agencies to ensure long-term funding for on-ground forest research in the tropics.
Code availability
All statistical analyses were performed in R (v.4.1.3). The BIOMASS R-package is an open source library available from the CRAN R repository. The BIOMASS vignettes and individual function helps contain detailed notes on usage to derive plot level AGB estimates with uncertainty estimates through error propagation using Monte Carlo method. The codes associated with the error propagation from plot-scale AGB to LiDAR AGB maps, generating additional metadata and binned covariance matrices as described in Methods section are available on GitHub (https://github.com/surajreddyr/LIDAR_AGB/tree/main).
References
Mitchard, E. T. A. et al. Uncertainty in the spatial distribution of tropical forest biomass: a comparison of pan-tropical maps. Carb Bal Manag., (2013).
Schimel, D. et al. Observing terrestrial ecosystems and the carbon cycle from space. Glob. Chang. Biol. 21, 1762–1776 (2015).
Herold, M. et al. The role and need for space-based forest biomass-related measurements in environmental management and policy. Surv. Geophys. 40, 757–778 (2019).
Mitchard, E. T. A. et al. Markedly divergent estimates of A mazon forest carbon density from ground plots and satellites. Glob. Ecol. Biogeogr. 23, 935–946 (2014).
Réjou-Méchain, M. et al. Upscaling Forest biomass from field to satellite measurements: sources of errors and ways to reduce them. Surv. Geophys. 40, 881–911 (2019).
Dubayah, R. et al. The Global Ecosystem Dynamics Investigation: High-resolution laser ranging of the Earth’s forests and topography. Sci. Remote Sens. 1, 100002 (2020).
Fararoda, R. et al. Improving forest above ground biomass estimates over Indian forests using multi source data sets with machine learning algorithm. Ecol. Inform. 101392 (2021).
Quegan, S. et al. The European Space Agency BIOMASS mission: Measuring forest above-ground biomass from space. Remote Sens. Environ. 227, 44–60 (2019).
Amelung, F. & others. NASA-ISRO SAR (NISAR) Mission Science Users’ Handbook. Jet Propuls. Lab. (2019).
Motohka, T., Kankaku, Y., Miura, S. & Suzuki, S. Overview of ALOS-2 and ALOS-4 L-band SAR. in 2021 IEEE Radar Conference (RadarConf21) 1–4 (2021).
Chave, J. et al. Ground data are essential for biomass remote sensing missions. Surv. Geophys. 40, 863–880 (2019).
Labrière, N. et al. Toward a forest biomass reference measurement system for remote sensing applications. Glob. Chang. Biol. 29, 827–840 (2023).
Duncanson, L. et al. The importance of consistent global forest aboveground biomass product validation. Surv. Geophys. 40, 979–999 (2019).
Duncanson, L. et al. Aboveground Woody biomass product validation good practices protocol 2021.
Zolkos, S. G., Goetz, S. J. & Dubayah, R. A meta-analysis of terrestrial aboveground biomass estimation using lidar remote sensing. Remote Sens. Environ. 128, 289–298 (2013).
Duncanson, L. et al. Aboveground biomass density models for NASA’s Global Ecosystem Dynamics Investigation (GEDI) lidar mission. Remote Sens. Environ. 270, 112845 (2022).
Rodda, S. R., Nidamanuri, R. R., Fararoda, R., Mayamanikandan, T. & Rajashekar, G. Evaluation of Height Metrics and Above-Ground Biomass Density from GEDI and ICESat-2 Over Indian Tropical Dry Forests using Airborne LiDAR Data. J. Indian Soc. Remote Sens. 1–16 (2023).
Réjou-Méchain, M., Tanguy, A., Piponiot, C., Chave, J. & Hérault, B. biomass: an r package for estimating above-ground biomass and its uncertainty in tropical forests. Methods Ecol. Evol. 8, 1163–1167 (2017).
Chave, J. et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 20, 3177–3190 (2014).
Asner, G. P. et al. A universal airborne LiDAR approach for tropical forest carbon mapping. Oecologia 168, 1147–1160 (2012).
McRoberts, R. E., Næsset, E., Saatchi, S. & Quegan, S. Statistically rigorous, model-based inferences from maps. Remote Sens. Environ. 279, 113028 (2022).
Saarela, S. et al. Three-phase hierarchical model-based and hybrid inference. MethodsX 11, 102321 (2023).
Rodda, S. R. et al. South Asian and Central African maps from: LiDAR-based reference aboveground biomass maps for tropical forests of South Asia and Central Africa. Dataverse https://doi.org/10.23708/H2MHXF (2024).
Ploton, P. et al. Spatial validation reveals poor predictive performance of large-scale ecological mapping models. Nat. Commun. 11, 4540 (2020).
Parmentier, I. et al. Predicting alpha diversity of African rain forests: models based on climate and satellite-derived data do not perform better than a purely spatial model. J. Biogeogr. 38, 1164–1176 (2011).
Réjou-Méchain, M. et al. Local spatial structure of forest biomass and its consequences for remote sensing of carbon stocks. Biogeosciences Discuss. 11, 5711 (2014).
Malhi, Y. et al. An international network to monitor the structure, composition and dynamics of Amazonian forests (RAINFOR). J. Veg. Sci. 13, 439–450 (2002).
Mitchard, E. T. A. et al. Markedly divergent estimates of Amazon forest carbon density from ground plots and satellites. Glob. Ecol. Biogeogr. 23, 935–946 (2014).
Avitabile, V. et al. An integrated pan-tropical biomass map using multiple reference datasets. Glob. Chang. Biol. 22, 1406–1420 (2016).
Acknowledgements
S.R.R. received support from French Research Institute for Development (IRD) through an International Travel grant for the execution of this study. We would like to acknowledge the assistance provided by the following entities in various aspects of this research. For the Indian (Asia) sites, we gratefully acknowledge the funding support received from ISRO-Geosphere Biosphere Program (ISRO-GBP) for LiDAR data acquisition and field work at the four Indian sites. We also express our thanks for the consistent support provided by the Indian state forest departments of Karnataka, Madhya Pradesh and Chhattisgarh, which facilitated the forest inventory measurements. We also express our gratitude to Mr. Prashant Kaiwshwar, Scientist (CCOST) for support in the Achanakmar field site in Chhattisgarh, India. Furthermore, we would like to acknowledge the assistance of Aerial Surveys and Digital Mapping Area at the National Remote Sensing Centre (ISRO) for their contributions to flight planning, LiDAR data acquisition and pre-processing. We extend our thanks to the team members who contributed to field measurements at the following locations: (A) Betul, Yellapur and Achanakmar – KV Satish, Wajeed Pasha, M. Suresh, and TR Kiran Chand and (B) Uppangala: S. Aravajy, N. Barathan, Jules Morel, Cedric Vega, A. Dharanidharan, G. Saravanan, K. Ramanujam, Jerome Feret, Ahmed Hamrouni, K. Adimoolam and T. Gopal. For African sites, research was carried out in the framework of the international joint laboratory (LMI) Dycofac, and within the One Forest Vision Initiative. At the exception of Rabi, forest inventories have been conducted thanks to recurrent funding of the IRD. We also acknowledge additional funding from EDF and Nachtigal Hydropower Company (for forest inventories and LiDAR data at Nachtigal) and from ERAMET, Maboumine and the Missouri Botanical Garden (for forest inventories and LiDAR data at Mabounie). We would like to thank Assala Gabon, Compagnie des Bois de Gabon and the Smithsonian National Zoo and Conservation Biology Institute for supporting the collection of the Rabi data in Gabon (contribution No 208 of the Gabon Biodiversity Program). Last, we warmly thank the Ministry of Forestry and Wildlife (MINFOF) of Cameroon for its continuous support to forest scientific research.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.R.R. and P.P.; data curation: R.F., S.R.R., R.G., M.M., G.D., N.B., P.P., N.J., M.R.-M.; formal analysis: S.R.R., P.P., N.J. and R.F.; writing (original draft): S.R.R., R.F., P.P. and M.R.-M.; writing (review and editing): all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rodda, S.R., Fararoda, R., Gopalakrishnan, R. et al. LiDAR-based reference aboveground biomass maps for tropical forests of South Asia and Central Africa. Sci Data 11, 334 (2024). https://doi.org/10.1038/s41597-024-03162-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03162-x
This article is cited by
-
Forest Biomass Assessment Using Multisource Earth Observation Data: Techniques, Data Sets and Applications
Journal of the Indian Society of Remote Sensing (2024)