Abstract
The lack of suitable reference genomic material to enable a transparent cross-lab study of oncopanels inspired the SEQC2 Oncopanel Sequencing Working Group to develop four reference samples, sequenced with eight oncopanels at independent test laboratories with ultra-deep sequencing depth. This rich, publicly available dataset enabled performance assessment of the clinical applicability of oncopanels. In addition, this dataset present sample opportunities for developing specific and robust bioinformatics pipelines and fine-tuning parameters for more accurate variant calling, investigating ideal sequencing depth for variant calling of a given minimum VAF and variant type, and also recommending best use cases for Unique Molecular Identifier (UMI) technology.
Measurement(s) | Short Somatic Mutation |
Technology Type(s) | Tumor Panel Sequencing |
Factor Type(s) | Tumor Panel • Genomic Region |
Sample Characteristic - Organism | Homo sapiens |
Similar content being viewed by others
Background & Summary
Personalized therapy, diagnostics and prognostic prediction for cancer patients are critical components for bringing the precision medicine era from theory to practice. The advantage of emerging technologies, including the next generation sequencing, oncopanel testing, as well as the bioinformatics tools, are bringing hope to the patients and their caregivers. Physicians can guide patients to targeted therapy rather than general treatment when corresponding genomic indicators are accurately identified with these technologies. False positive variant calls from these technologies can lead to unnecessary or improper therapies as well as overestimation of biomarkers such as tumor mutational burden (TMB), a key measurement for immune checkpoint inhibitor (ICI) therapy. In recent years, the FDA has approved or cleared several cancer panels which are designed to provide physicians with clinically actionable information related to specific targets, a significant milestone for the precision medicine field. Oncopanels typically cover a small-to-large number (50–1000) of genes and can detect mutations with variant allele frequency (VAF) as low as 1% if given sufficient sequencing depth and base quality. Most pan-cancer panels are currently still for research use only. Before they can be adopted for clinical testing, a thorough understanding of their robustness, analytical characteristics, reproducibility, sensitivity, and false positive rate is greatly needed.
We proposed a multi-lab cross-oncopanel study to assess the performance and the clinical applicability of oncopanels1. Eight panel providers agreed to participant in this study: Agilent Technologies, Burning Rock Biotech, Integrated DNA Technologies, Inc., iGeneTech, Illumina Inc., QIAGEN Sciences Inc., Roche Sequencing Solutions Inc., and Thermo Fisher Scientific. In order to evaluate the performance of oncopanels in a comprehensive and fair manner, we formed the Oncopanel Sequencing Working Group under the umbrella of the Sequencing Quality Control Phase II (SEQC2) consortium. The working group contained 151 members from 100 institutes, government agencies and private companies across 14 countries. The working group mainly focused on assessing the oncopanel performance using three key metrics: sensitivity, false positive rate, and reproducibility. We developed four related reference samples with a large number of high-confident small variant positive and negatives positions to serve as known content for the performance assessment2. The four reference samples were distributed to the eight participating panel providers identified previously for library preparation, sequencing, and variant calling. We asked each panel provider to recruit at least three independent laboratories with which they were remarkably familiar to processing their panels. Each lab made four panel libraries for each reference sample following the harmonized experimental protocols developed by the panel providers accordingly. The libraries from the same panel were sequenced on various sequencing platforms, including Illumina HiSeq 2500, NovaSeq, NextSeq and ThermoFisher IonTorrent S5. Each panel provider performed the downstream analysis using their recommended or in-house bioinformatics pipelines for variant calling, but blinded to any of the known content. Variant calling results were submitted in VCF format, along with their reporting regions and allowlist/blocklist, to an independent group for performance evaluation. In addition to the primary goal of assessing the performance and the clinical applicability of oncopanels, the dataset also holds its value for: i) developing specific bioinformatics pipelines and fine-tuning parameters for more accurate variant calling for a specific panel, ii) developing robust bioinformatics pipelines that can be applied on multiple panels with promising results, iii) investigating ideal sequencing depth for variant calling of a given minimum VAF and certain variant type (i.e., SNVs, small indels and MNVs, or long indels), and iv) making the best use of UMI technology.
Methods
Study design
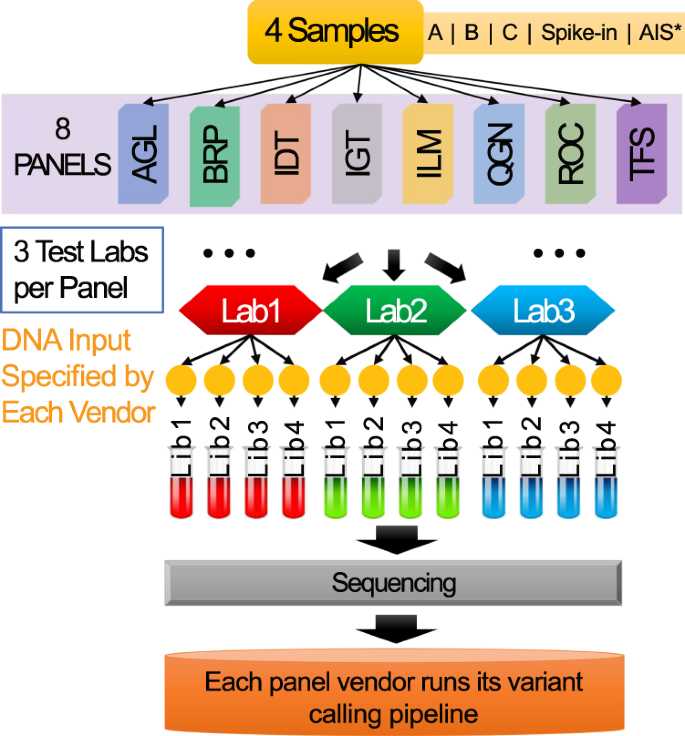
The SEQC2 Oncopanel Sequencing Working Group developed four reference samples for this study, namely: Sample A, Sample B, Sample C and Sample Spike-in (a.k.a., AC5) (Fig. 1). Sample A is the equal mass mixture of DNA from ten cancer cell-lines that are used to make the Agilent UHRR material3. Sample B is a cell line derived from a normal male individual available at Agilent. Sample C is the 1:1 dilution of Sample A and Sample B. Sample Spike-in is a mixture of Sample B with 5% AcroMetrix hotspot synthetic controls4. There were eight oncopanels included in this study: Agilent Custom Comprehensive Cancer Panel v2 (AGL), Burning Rock DX OncoScreen Plus (BRP), Integrated DNA Technologies xGen Pan-Cancer Panel (IDT), iGeneTech AIOnco-seq (IGT), Illumina TruSight Tumor 170 (ILM), QIAGEN Comprehensive cancer panel (QGN), Roche SeqCap EZ Choice custom PHC Panel (ROC), and Thermo Fisher Oncomine Comprehensive Assay v3 (TFS) (Fig. 1). Each panel provider recruited at least three independent laboratories. We distributed the four reference samples to the test laboratories and each laboratory made four DNA libraries as technical replicates. The libraries from the same panel were sequenced on the same sequencing platforms, including Illumina HiSeq 2500, NovaSeq, NextSeq and ThermoFisher IonTorrent S5. Each panel provider performed the downstream analysis using their recommended or in-house bioinformatics pipelines for variant calling. Variant calling results were submitted in VCF format, along with their reporting regions and allowlist/blocklist, to an independent group, led by Dr. Joshua Xu, for performance evaluation1.
Study design. The illustration demonstrates the workflow of sample distribution, library preparation and sequencing data processing. Four reference samples were created and distributed to each of the testing laboratories. Four technical library replicates were prepared for each sample at each laboratory. DNA libraries were then sequenced and processed by panel-specific bioinformatics pipelines for variant calling. *Sample AIS was not used in the related work. It was designed for a side study which aims to assess the usability of Accugenomics spike-in control as an orthogonal quality control in oncopanel sequencing.
Genomic DNA libraries construction of reference samples
Sample A is composed of an equal mass pooling of 10 gDNA samples prepared from Agilent’s Universal Human Reference RNA (UHRR) cancer cell lines3. Over 42 K small variants are identified with high confidence in the defined regions of over 22 million bases2.
Sample B is a gDNA sample from a normal male cell line (Agilent Human Reference DNA, Male, Agilent part #: 5190–8848).
Sample C is a 1:1 mix of Sample A and Sample B. Sample C was included to dramatically increase (4x) the number of known variants with VAF between 1% and 2.5%1.
Sample Spike-in (a.k.a., AC5) is Sample B with 5% AcroMetrix Spike hotspot synthetic controls (Thermo Fisher Scientific, Fremont, CA)4.
Samples A, B, and C were prepared by Agilent and were kindly provided to SEQC2 for study. Sample Spike-in was prepared at Thermo Fisher Scientific (Fremont, CA) after receiving Sample B from Agilent under their material transfer agreement. All four samples were stored in low-EDTA TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8.0) at 20 ng/ul concentration.
Participating panels sign-up, test sites recruitment and sample distribution
Eight oncopanel providers signed up to participate in this study, each provider recruited at least three independent laboratories which are remarkably familiar with their panels. A total of 28 testing laboratories were initially recruited. One laboratory was later excluded due to affiliation with the panel provider, one laboratory was excluded due to failing to pass quality control, and one laboratory was excluded due to an extended delay in experimental execution and data generation. We distributed the four reference samples to each laboratory. Four DNA libraries for each reference sample were then made at each laboratory as technical replicates. A total of 430 DNA libraries were prepared. Detailed information of the eight participating oncopanels are listed in Table 1. To shorten the description and file names, panel codes were used to identify panels. Laboratory codes are listed in Table 1 to identify the test laboratories for each oncopanels.
Experimental protocols
Basic information about the experimental procedures and wet-lab QC metrics are summarized in Table 1. These experimental protocols are expanded versions of descriptions in our related work1, which was approved by the panel providers accordingly.
AGL experimental protocol
Genomic DNA libraries were constructed for the reference samples according to the Agilent SureSelectXT HS Target Enrichment System for Illumina Paired-End Multiplexed Sequencing Library Protocol (Cat. No. G9702–90000 Version A1, July 2017). In brief, 30 ng of each sample’s high molecular weight genomic DNA was sonicated in a 50 μl total volume with a Covaris E220 instrument to a mean size of 350 bp (Duty Factor: 10%, Peak Incident Power: 175, Cycles per Burst: 200, Treatment Time: 2 × 30 seconds, Bath Temperature: 2° to 8 °C). DNA fragments were then end-repaired and A-tailed using a two-step cycling protocol (20 °C for 15 minutes and 72 °C for 15 minutes), followed by ligation to XTHS adaptors with UMIs for 30 minutes at 20 °C. Adapter-ligated fragments were amplified and indexed by PCR in a 50 μl total volume with Herculase II Fusion DNA Polymerase under the following conditions: 2 min at 98 °C (initial denaturation), 10 cycle amplification of 30 seconds at 98 °C, 30 seconds at 60 °C, 1 minute at 72 °C, and 5 minutes at 72 °C (final extension). Library quality control (quantity and size distribution) was then assessed using either the 2100 Bioanalyzer and DNA 1000 assay or the 2200 TapeStation and D1000 screen tape. 1 μg of prepared gDNA libraries were then hybridized to a custom Immuno-Oncology focused Comprehensive Cancer Panel (1058 targets coding regions including UTRs and 7.6 Mb in size) biotinylated RNA probes (5 minutes at 95 °C, 10 minutes at 65 °C, 1 minute at 65 °C, 60 cycles of 1 minute at 65 °C and 3 seconds at 37 °C, and 65 °C hold) and captured with Dynabeads MyOne Streptavidin T1 beads. SureSelect enriched gDNA libraries were PCR amplified using on-bead protocol in a 50 μl total volume with Herculase II Fusion DNA Polymerase under the following conditions: 2 min at 98 °C (initial denaturation), 10 cycles of 30 seconds at 98 °C, 30 seconds at 60 °C, 1 minute at 72 °C (amplification), and 5 minutes at 72 °C (final extension), followed by 4 °C hold. All DNA purifications between steps were performed with AMPure XP beads as indicated in the user manual. Post-capture library quality control was assessed using either the 2100 Bioanalyzer and a High Sensitivity DNA assay or the 2200 TapeStation and HSD1000 screen tape. Indexed samples were finally pooled and sequenced to approximately 5000X (Samples A, B, AC5, AAG) or 10,000X (Sample C) read depth on a NovaSeq 6000 instrument using a 2x150bp Paired-End protocol (Q30 scores ≥ 75%).
BRP experimental protocol
The library prep and enrichment process were performed using Burning Rock HS library preparation kit without modification. In brief, DNA shearing was performed on SEQC2 gDNA reference samples using Covaris M220 for 195 S, with Peak Incident Power = 50 W, Duty Factor: 20%, Cycle Per Burst: 200, at 6–8 °C, followed by end repair, adaptor ligation and PCR enrichment. About 1 μg of purified pre-enrichment library was hybridized to the OncoScreenPlusTM panel and further enriched following manufacturer instruction. The OncoScreenPlusTM panel is about 1.7 M bp in size and covers 520 human cancer related genes. Final DNA libraries were quantified using Qubit Fluorometer with dsDNA HS assay kit (Life Technologies, Carlsbad, CA). A LabChip GX Touch System, Agilent 2100 bioanalyzer or Agilent 4200 TapeStation D1000 ScreenTape was then performed to assess the quality and size distribution of the library. The libraries were sequenced on the NovaSeq 6000 sequencer (Illumina, Inc., California, US) with 2 × 150 bp paired-end reads with a unique dual index.
IDT experimental protocol
Libraries were constructed using 100 ng from each of the reference samples in quadruplicate. Briefly, genomic DNA was sheared to 300 bp (Covaris) followed by library construction using the NEBNext Ultra II DNA Library Prep Kit for Illumina and xGen Dual Index UMI Adapters–Tech Access. End repair and A-tailing were performed according to the manufacturer’s recommendations. Adapters were ligated using 5 µL of 15 µM stock for each reaction followed by 0.9X AMPure purification. Libraries were amplified with Illumina P5 and P7 primers using NEBNext Ultra II Q5 Master Mix using 6 cycles of amplification. Libraries were purified using 1X AMPure clean up and quantified by Qubit. Following library construction, 500 ng of each library was captured with the xGen Pan-Cancer Panel v1.5 following the manufacturer’s instructions using xGen Universal Blockers–TS Mix. Hydrophilic Streptavidin Magnetic Beads and NEBNext Ultra II Q5 Master Mix were substituted for the capture and post-capture amplification using 16 cycles of PCR. Libraries were purified, quantified, and pooled for sequencing on an Illumina NovaSeq S4 flow cell.
IGT experimental protocol
The iGeneTech AIOnco-seq gene panel was designed to target the exons of 113 cancer related genes. The total size of targeted regions was 944,153 bp. 100 ng of human genomic DNA was sheared on a Covaris S220 focused-ultrasonicator for 180 s (175 W peak incident power, 10% duty factor, and 200 cycles per burst). The resulting fragment-size distribution was about 150~250 bp. End repair, nontemplated dA-tailing, and adapter ligation were performed by using KAPA Hyper Prep Kits. The adapter was generated by annealing oligonucleotide ACACTCTTTCCCTACACGACGCTCTTCCGATC*T (the * represent a phosphorothioate bond) and oligonucleotide Pho-GATCGGAAGAGCACACGTCTGAACTCCAGTCAC. The ligation product was cleaned up and size-selected by using Beckman Ampure XP Beads (Beckman). The purified ligated product was amplified with 7 cycles of PCR to generate about 1 μg library.
A mix containing 750 ng whole genome library, 2.5 μg human Cot-1 DNA (Invitrogen), 2.5 μg salmon sperm DNA, and 2000 pmol adapter blocking oligonucleotides was concentrated to 9 μl, and then heated at 95 °C for 5 min, and held at 65 °C for 5 min. A 13 μl of 65 °C prewarmed hybridization buffer (iGeneTech) was added to the library. A 7 μl of freshly prepared, 65 °C prewarmed mixture containing 200 ng biotinylated RNA probes and 20 U SUPERase-In (Invitrogen) was added to the library and mixed by a pipette. After 16 h at 65 °C, the hybridization mix was added to 50 ul Dynabeads MyOne Streptavidin T1 (Invitrogen), which had been washed three times and resuspended in 200 μl binding buffer (iGeneTech). The binding reaction was rotated on a rotational mixer (10 rpm/min) at 25 °C for 30 min, washed once at 25 °C for 15 min with 200 μl Wash Buffer I (iGeneTech), and wash three times at 65 °C for 15 min with Wash Buffer II (iGeneTech). The beads were resuspended in 20 μl TE Buffer. The post-hybridization PCR was performed to enrich and amplify the target region library. The libraries were sequenced on an Illumina HiSeq 2500 sequencer with 125 bp paired-end sequencing reads.
ILM experimental protocol
DNA was processed according to the TruSight Tumor 170 Reference Guide5, briefly samples were sheared using Covaris to approximately 90–250 bp. Following shearing, DNA fragments were end-repaired and A-tailed in a single reaction, followed by ligation to a universal adapter. Post-ligation clean-up was performed using SPRI beads and then libraries were indexed using unique dual indexes by PCR. Target regions were captured using an overnight hybridization to biotinylated target specific oligos which covered 533Kb of genomic targets across 154 genes, followed by capture with streptavidin magnetic beads. A second hybridization and capture reaction were performed followed by PCR amplification using the universal primers compatible with Illumina’s sequencing flowcells. Libraries were normalized using bead-based normalization before being pooled in equal parts and sequenced, 10 samples per flowcell, on NextSeq v2 high-output flowcell. Sequencing reads were 2 × 101 bp with 8 bp dual indexed reads. Five independent labs were recruited as the test labs for ILM panel. One test lab was excluded from performance analysis due to an extensive delay in data generation and some deviation from the experimental protocol.
QGN experimental protocol
The Human Comprehensive Cancer QIASeq DNA Panel is a targeted enrichment panel that enriches genes using single primer extension and constructs libraries using integrated unique molecular indices (UMIs). The panel uses 11,311 primers in a single tube mix to enrich over 0.83 Mb region across full coding areas of 275 cancer genes. The selected genes harbor mutations that are commonly involved in cancer development and progression. Genomic DNA samples were first fragmented, end repaired and A-tailed within a single, controlled multi-enzyme reaction. The prepared DNA fragments were then ligated at their 5′ ends with an Illumina sequencing platform-specific adapter containing a 12-base fully random sequence of UMI and sample index. Therefore, each original DNA molecule in the sample received a unique UMI sequence during ligation prior to target enrichment and library amplification PCR. This allowed the correction of errors associated with PCR amplification and sequencing.
Target enrichment with Human Comprehensive Cancer QIASeq DNA Panel was performed post-UMI assignment to ensure that targeting regions of DNA molecules containing UMIs were sufficiently and uniformly enriched in the sequenced library. For enrichment, ligated DNA molecules were subject to six cycles of targeted PCR using one region-specific primer and one universal primer complementary to the adapter. After enrichment PCR, nineteen cycles of Universal PCR were carried out to further amplify the target enriched library and added platform specific adapter sequences and additional sample indices. Finished libraries were checked on Bioanalyzer for proper size distribution and library concentration was measured by QIAseq Library Quantification system. Libraries were normalized to 5 nM and pooled together according to quantified library concentration. Pooled libraries were sent out to Illumina for sequencing on NovaSeq with a Qiagen custom read 1 sequencing primer. Libraries within a test site were loaded on each lane of the flowcell for sequencing.
ROC experimental protocol
The panel design utilized in the study is 170907_HG38_SEQC2_PHC_EZ. This is a research panel, not intended for commercial development. The panel consists of coding sequence (CDS) from 33 cancer genes from SEQC Pan Cancer Gene list and CDS regions overlapping the Accugenomics controls. Additional targets were derived from the Roche Avenio ctDNA Panels. The panel consists of 45 genes in total (~131 kb of targets). The panel also targets 730 putative structural variant breakpoints derived from the 10X cell line sequencing (~206 kb of targets).
The ROC panel utilized an enzymatic fragmentation library prep followed by a hybridization-based workflow. The extracted DNA sample (100 ng) was fragmented using the KAPA HyperPlus Library Preparation Kit, and then ligated to Illumina-compatible, unique-dual-index (UDI) sequencing library adapters (IDT). After the ligation, the DNA library was amplified using the KAPA HiFi HotStart Ready Mix. Amplified libraries were quantified with a fluorometric method, and the maximum available amount of library (> = 0.5 ug and < = 2.5 ug) was utilized for capture. The sample was incubated overnight at 47 C with the gene panel (170907_HG38_SEQC2_PHC_EZ), which consisted of biotinylated DNA probes designed to capture the genes and regions of interest. Universal Blocking Oligos were utilized in the hybridization to prevent cross-hybridization between sequencing adapters and to increase capture specificity. The desired DNA-probe complexes were then captured on streptavidin beads, and after a series of washes, first at 47 C and then at room temperature, the samples were amplified using ligation mediated PCR (LM-PCR). The final product of the workflow was enriched libraries ready for sequencing. The final sequencing libraries were sequenced (2 × 150 bp) using the Illumina NovaSeq sequencing platform.
Four independent labs were recruited to test the ROC panel. Initial sequencing data QC analysis led to the exclusion of one test lab from performance analysis. Shorter fragments, which resulted from over fragmentation during library preparation, indicated a likely deviation from the experimental protocol.
TFS experimental protocol
The Oncomine Comprehensive Assay DNA v3C – Chef Ready Kit6 was used to generate libraries for next-generation sequencing on the Ion Torrent S5 platform. This assay enables analysis of variants across 146 genes and covers 350,350 bp of DNA target sequence. It typically is used in combination with an accompanying RNA assay bringing the gene total to 161, but the study used only the DNA assay with the RNA component omitted. The gene content of the Oncomine Comprehensive Assay was prioritized to detect relevant somatic variants in solid tumors with associations to published evidence including the indication statements of approved cancer drugs, consensus clinical treatment guidelines, and the enrolment criteria of oncology clinical trials. Purified DNA was extracted using the RecoverAll Multi-Sample RNA/DNA Isolation Workflow7. Four DNA samples were used to prepare duplicate libraries on each of two runs using the Ion Chef Instrument. An input of 10 ng per primer pool of DNA was used, and libraries were generated following the manufacturer’s instructions in the Oncomine Comprehensive Assay User Guide8 and the Ion AmpliSeq Library Preparation on the Ion Chef User Guide9. A total of 8 DNA sample libraries were combined for template preparation on the Ion Chef Instrument using the Ion 540 Kit-Chef10. Sequencing was performed with the Ion S5 XL System11 and the Ion 540 Chip12.
Sequencing, Data processing and collection
The libraries from the same panel were sequenced on the same sequencing platforms chosen by the panel providers. Even though four sequencing platforms were used in this study, including three Illumina platforms (HiSeq 2500, NovaSeq, NextSeq) and one ThermoFisher platform (IonTorrent S5), each library was sequenced on only one of the platforms. Comparison across sequencing platforms of the exact same DNA library is not available in this study. Sequencing data was required to be shared within the SEQC2 Oncopanel Working Group via Illumina BaseSpace Sequence Hub or by uploading data to the sFTP server hosted at Stanford University. Either FASTQ or BAM format was used for data sharing. All the data was collected at the National Center for Toxicological Research (NCTR), organized and renamed in a consistent manner, and then submitted to NCBI SRA data repository. The data became publicly available upon the publication of the related research manuscript1.
Bioinformatics pipelines and variant calling results
Each panel provider ran their recommended or in-house pipelines for variant calling and filtering. In this study, we considered each recommended or in-house pipeline to be part of the solution of variant detection along with the according oncopanel. The purpose of our study was not to determine the superior method for variant calling; thus, the recommended or in-house pipelines were used as they should perform reasonably at variant calling. The reproducibility, sensitivity, and false positive rate of the eight participating oncopanels were reported in the related research manuscript1. If the inquisitive reader is interested in comparing the variant calling results between the recommended or in-house pipelines, the VCF files can be downloaded at figshare13.
Data Records
The data have been deposited to NCBI SRA with accession number SRP29511314. BioSample metadata are available in the NCBI BioSample database (http://www.ncbi.nlm.nih.gov/biosample/), where Sample A is under accession number SAMN1678636015 with download size of 879 Gb, Sample B is under accession number SAMN1678636116 with download size of 801 Gb, Sample C is under accession number SAMN1678636217 with download size of 1.37 Tb, and Sample Spike-in is under accession number SAMN1678636318 with download size of 2.47 Tb. There are 400 NCBI SRA records in total for this study, 103 for Sample A, 103 for Sample B, 91 for Sample C, and 103 for Sample Spike-in. Some of the data for Panel ILM was not available due to an unrecoverable data loss, thus, these data files are not submittable to NCBI SRA. A detailed list of all 400 NCBI SRA records can be found in Online-only Table 1. An extra Sample AIS19, which contains 103 SRA records, can also be found under the BioProject PRJNA677997 umbrella. This sample was not used in the related publication1, but was designed for a side study to assess the usability of Accugenomics spike-in control as an orthogonal quality control in oncopanel sequencing. If interested in analyzing the Sample AIS, a brief description about the sample can be found in the Usage Notes. The “LibraryName” column shows the individual DNA library identifier, which combines the sample ID, panel code, testing laboratory code, DNA input amount, and library replicate together with “_”. There is a number “1” immediately after the panel code, which indicates the oncopanel study shown here. The “FileType” column show the file type of the according NCBI SRA record, where “fastq” indicates the FASTQ format and “bam” indicated the BAM format. Data for panel ROC is provided in unmapped BAM format. Data for TFS is provided in mapped BAM format, and the reference genome used for read mapping is listed in the “ReferenceGenome” column. Data for the other six panels is provided in FASTQ format. Details of read processing for all panels and read mapping for panel TFS can be found in Code Availability. Several filenames may be listed in columns “filename1” to “filename8”. These filenames are the original filenames used when uploading the data files to NCBI SRA. If retrieving the data using NCBI’s SRA ToolKit, one may see different filenames, which may be related to the SRR accession number. Filenames start with “LibraryName”, followed by part number if applicable, then the read identifier and filename extension. If the file type is “bam”, there is one file listed for each record, ending with “.bam” as the filename extension. If the file type is “fastq”, the files end with “.fastq.gz”. You will find pairs of files listed for each record, where “R1” and “R2” refer to the left and right reads. Specifically, for panel AGL, “R1” indicates the left reads, “R2” indicates the UMI barcodes, and “R3” indicates the right reads. To make the file size manageable for data transfer, we split bigger files into parts. For some records, there may be “PT1”, “PT2”, “PT3”, “PT4” in the filenames, which indicates different parts of the same data files. These parts of the same data files need to be merged in the order of 1 to 4 (or 2 if there are only two parts) before any data processing.
Data from three testing laboratories (ST25, ST11, ST17) were excluded from the study due to the clinical lab being too closely affiliated with the panel provider (ST25), an extended delay in experimental execution and data generation (ST11), and over fragmentation of DNA samples during library preparation (ST17). Due to an unexpected hard drive failure, 20 files for panel ILM were unable to be recovered, including data for ST12 (LIB3 and LIB4), ST29 (LIB1, LIB2 and LIB4).
Technical Validation
The dataset is validated in multiple aspects, (a) each of the four reference samples was prepared and sequenced four times at one testing laboratory, and is also prepared and sequenced at least 12 times (three or more testing laboratories per oncopanel) for each oncopanel; high intra- and cross-lab reproducibility was observed;1 (b) Sample Spike-in has known variants and shows very high sensitivity with a handful of readily explainable exceptions;1 (c) few false positive calls were reported with Sample B for all panels1, which indicates a low error rate of the sequencing data and high quality calling.
Usage Notes
As all eight oncopanels were designed to target certain genomic regions, variant calling should be restricted to the designed regions of each oncopanel. The corresponding regions were provided in BED format and can be downloaded from figshare20. Due to the choice of reference genome in the oncopanel design, for all panels except ROC, the regions were provided for GRCh37/hg19; and for the ROC panel, the regions were provided for GRCh38/hg38. A blocklist of AcroMetrix Oncology Hotspot Control (AOHC) variants for panel TFS is also provided, which should be used when analyzing Sample AC5 with panel TFS.
The extra Sample AIS was composed of Sample A and enzymatically fragmented Accugenomics spike-in control. As stated in our related work, the raw reads from Sample AIS should map against both the native template (NT) of reference genome (hg19 or hg38) and the modified internal standard (IS) genome template to separate the NT and IS reads21.
The 500+ enzymatically fragmented Acrometrix Synthetic Hotspot controls4 were added to Sample B to make Sample AC5. These variants can be used as positive controls for analyzing Sample AC5. The list of Acrometrix Synthetic Hotspot controls is provided in VCF format for both hg19 and hg38 and can be downloaded from figshare22.
This manuscript reflects the views of the authors and does not necessarily reflect those of the U.S. Food and Drug Administration. Any mention of commercial products is for clarification only and is not intended as approval, endorsement, or recommendation.
Code availability
The details and computational parameters of raw read processing for all panels and read mapping for panel TFS are expanded versions of descriptions in our related work1.
AGL read processing
Each sample was demultiplexed using bcl2fastq v2.20 (Illumina)23 with the base mask Y150, I8, Y10, Y150 and all default settings except for mask-short-adapter-reads, which was set to 0. Adapters were trimmed using AGeNT Trimmer (Agilent)24.
BRP read processing
After demultiplexing using bcl2fastq v2.2023, sequence data were filtered using the Trimmomatic 0.3625 with parameters “TRAILING:20 SLIDINGWINDOW:30:25 MINLEN:50”.
IDT read processing
IDT libraries were prepared with xGen Dual Index UMI Adapters—Tech Access which contain 9 bp degenerate unique molecular identifiers (UMIs) downstream of the i7 sample index. To demultiplex Illumina sequencing data containing UMIs, Illumina basecall (BCL) files were used to generate demultiplexed BAM files using Picard v2.9.0 (http://broadinstitute.github.io/picard/) IlluminaBasecallsToSam. Prior to running Picard v2.9.0 IlluminaBasecallsToSam, Picard v2.9.0 ExtractIlluminaBarcodes was used to determine the barcode for each read using the read structure 151T8B9S8B151T (T = template, B = sample barcode, M = molecular barcode, and S = skip). The UMI bases were not used for downstream analysis since there was not enough raw sequencing depth for consensus analysis. After demultiplexing, FASTQ files were generated using Picard v2.9.0 SamToFastq. FASTQ files were downsampled to an equivalent read count per sample using seqtk v1.0 (https://github.com/lh3/seqtk).
IGT read processing
Raw reads were firstly quality trimmed with Trimmomatic 0.3625, using an 8-base-pair sliding-window algorithm with a quality score cutoff of 20, clipping off ends with at least one occurrence of a quality score below 20, and discarding reads that dropped below a length of 40 base-pairs. Adaptors (a1: GATCGGAAGAGCACACGTCT, a2: AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT) were removed simultaneously.
ILM read processing
Pan Cancer sample testing analysis was performed using the standard TruSight Tumor 170 pipeline available on the BaseSpace Sequence Hub. Briefly, high level sequencing run metrics are evaluated to generate a Run QC Metrics report. Next, reads are converted into the FASTQ format using bcl2fastq v2.2023 and adapters are trimmed.
QGN read processing
Raw sequencing data were demultiplexed and converted into the FASTQ format using bcl2fastq v2.2023 in BaseSpace, with the library specific pairs of Qiagen custom read 1 primer sequences and the reverse complementary Illumina i7 primer sequences. QIAGEN’s UMI-aware variant caller smCounter226 can be used for variant calling.
ROC read processing
Two NovaSeq runs were provided by Illumina via BaseSpace (RUN1 = SeqC2_ROC1_ST_16_17_18_19; RUN2 = SeqC2_ROC1_ST16_17_18_19). BCL files were converted to unaligned BAM files using instructions provided by IDT (https://www.idtdna.com/pages/products/next-generation-sequencing/adapters/xgen-dual-index-umi-adapters-tech-access). Following the IDT tech note, Picard 2.18.3 ExtractIlluminaBarcodes and IlluminaBasecallsToSam were used to create the unaligned BAM files. The UMI for each fragment is stored in the RX tag in the BAM file.
TFS read processing and mapping
Signal processing and base calling were performed using Torrent Suite Software 5.8 (https://github.com/iontorrent/TS) using default parameters for the Oncomine Comprehensive Assay. The signal processing step consists of modeling the pH dynamics on the semiconductor surface taking into account the varying local pH in each individual sensor coming from the different reagent flows across the chip and from any nucleotide incorporation that may be happening over each sensor27. The base calling step consists of taking the estimated levels of nucleotide incorporation for each read and each nucleotide flow, and modeling the de-phasing process whereby some templates within each clonally-amplified population run ahead or behind in terms of their nucleotide incorporation. During the base calling process, sample-specific barcodes and 3′ adapters are annotated.
After completion of primary analysis with Torrent Suite Software 5.8, reads were uploaded to Ion Reporter Software 5.628 for subsequent processing. Reads were aligned with the Torrent Mapping Alignment Program (TMAP, https://github.com/iontorrent/TS/tree/master/Analysis/TMAP), which uses the BWA fastmap routine to map reads and applies post-processing of the alignments to optimize for technology-specific error patterns.
References
Gong, B. et al. Cross-oncopanel study reveals high sensitivity and accuracy with overall analytical performance depending on genomic regions. Genome Biol 22, 109 (2021).
Jones, W. et al. A verified genomic reference sample for assessing performance of cancer panels detecting small variants of low allele frequency. Genome Biol 22, 111 (2021).
Novoradovskaya, N. et al. Universal Reference RNA as a standard for microarray experiments. BMC Genomics 5, 20 (2004).
AcroMetrix Oncology Hotspot Control Package Insert. Thermo Scientific https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FCDD%2Fmanuals%2FMAN0010820-AMX-Oncology-Hotspot-Ctrl-EN.pdf&title=QWNyb01ldHJpeCBPbmNvbG9neSBIb3RzcG90IENvbnRyb2wgUGFja2FnZSBJbnNlcnQgW0VOXQ== (2020).
TruSight Tumor 170 Reference Guide. Illumina, Inc. https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/trusight/tumor-170/trusight-tumor-170-reference-guide-1000000024091-02.pdf (2019).
OncomineTM Comprehensive Assay v3C - A35806. Thermo Fisher Scientific http://www.thermofisher.com/order/catalog/product/A35806.
RecoverAllTM Multi-Sample RNA/DNA Isolation Workflow A26069. Thermo Fisher Scientific http://www.thermofisher.com/order/catalog/product/A26069.
OncomineTM Comprehensive Assay v3 User Guide - MAN0015885. Thermo Fisher Scientific https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0015885_OncomineComprehensiveAssay_v3_UG.pdf.
Ion AmpliSeqTM Library Preparation on the Ion ChefTM System User Guide MAN0013432. Thermo Fisher Scientific https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0013432_Ion_AmpliSeq_Library_Prep_on_Ion_Chef_UG.pdf.
Ion 540TM Kit-Chef A30011. Thermo Fisher Scientific http://www.thermofisher.com/order/catalog/product/A30011.
Ion S5TM XL System A27214. Thermo Fisher Scientific http://www.thermofisher.com/order/catalog/product/A27214.
Ion 540TM Chip Kit A27766. Thermo Fisher Scientific http://www.thermofisher.com/order/catalog/product/A27765.
Gong, B. & Xu, J. SEQC2 Onco-panel Sequencing Working Group - PanCancer panel Study: Variant calling results. figshare https://doi.org/10.6084/m9.figshare.c.5842112.v2 (2021).
NCBI Sequence Read Archive https://identifiers.org/insdc.sra:SRP295113 (2021).
Sample A. NCBI BioSample https://www.ncbi.nlm.nih.gov/biosample/16786360 (2021).
Sample B. NCBI BioSample https://www.ncbi.nlm.nih.gov/biosample/16786361 (2021).
Sample C. NCBI BioSample https://www.ncbi.nlm.nih.gov/biosample/16786362 (2021).
Sample Spike-in. NCBI BioSample https://www.ncbi.nlm.nih.gov/biosample/16786363 (2021).
Sample AIS. NCBI BioSample https://www.ncbi.nlm.nih.gov/biosample/16961529 (2021).
Gong, B. & Xu, J. PanCancer panels’ target regions. figshare https://doi.org/10.6084/m9.figshare.19128005 (2022).
Willey, J. C. et al. Advancing quality-control for NGS measurement of actionable mutations in circulating tumor DNA. Cell Reports Methods 1, 100106 (2021).
Gong, B. & Xu, J. Acrometrix Synthetic Hotspot controls. figshare https://doi.org/10.6084/m9.figshare.19092089 (2022).
bcl2fastq2 Conversion Software v2.20. Illumina, Inc. https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2-20.html (2017).
The Agilent Genomics NextGen Toolkit. Agilent Technologies https://www.agilent.com/en/product/next-generation-sequencing/hybridization-based-next-generation-sequencing-ngs/ngs-software/agent-232879.
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Xu, C. et al. smCounter2: an accurate low-frequency variant caller for targeted sequencing data with unique molecular identifiers. Bioinformatics 35, 1299–1309 (2019).
Rothberg, J. M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011).
Ion Reporter Software. Thermo Fisher Scientific https://www.thermofisher.com/us/en/home/life-science/sequencing/next-generation-sequencing/ion-torrent-next-generation-sequencing-workflow/ion-torrent-next-generation-sequencing-data-analysis-workflow/ion-reporter-software.html (2019).
Author information
Authors and Affiliations
Contributions
W.T. and J.X. conceived and planned the study as well as constructed the experimental design. J.X. and W.J. managed the project. B.G. managed the data deposit and wrote the manuscript with assistance from R.K., W.J. and J.X. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Agilent sample B DNA reference sample is a current product and sample A DNA and sample C DNA are potential products of Agilent Technologies, Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Table
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gong, B., Kusko, R., Jones, W. et al. Ultra-deep multi-oncopanel sequencing of benchmarking samples with a wide range of variant allele frequencies. Sci Data 9, 288 (2022). https://doi.org/10.1038/s41597-022-01359-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-022-01359-6