Abstract
Targeted integration of large DNA cargoes (>10 kb) or genomic replacements in mammalian cells, such as human pluripotent stem cells (hPS cells), remains challenging. Here we describe a platform termed serine and tyrosine recombinase-assisted integration of genes for high-throughput investigation (STRAIGHT-IN) to circumvent this. First, a landing pad cassette is precisely inserted or used to replace specific genomic regions. The site-specific integrase Bxb1 then enables DNA constructs, including those >50 kb, to be integrated into the genome, while Cre recombinase excises auxiliary DNA sequences to prevent postintegrative silencing. Using a strategy whereby the positive selection marker is only expressed if the donor plasmid carrying the payload is correctly targeted, we can obtain 100% enrichment for cells containing the DNA payload. Procedures for expressing Cre efficiently also mean that a clonal isolation step is no longer essential to derive the required genetically modified hPS cells containing the integrated DNA, potentially reducing clonal variability. Furthermore, STRAIGHT-IN facilitates rapid and multiplexed generation of genetically matched hPS cells when multiple donor plasmids are delivered simultaneously. STRAIGHT-IN has various applications, which include integrating complex genetic circuits for synthetic biology, as well as creating panels of hPS cells lines containing, as necessary, hundreds of disease-linked variants for disease modeling and drug discovery. After establishing the hPS cell line containing the landing pad, the entire procedure, including donor plasmid synthesis, takes 1.5–3 months, depending on whether single or multiple DNA payloads are integrated. This protocol only requires the researcher to be skilled in molecular biology and standard cell culture techniques.
Key points
-
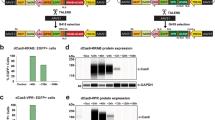
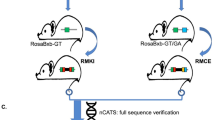
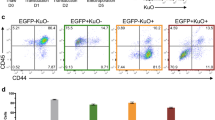
STRAIGHT-IN is a method for generating panels of genetically modified human pluripotent stem cell lines. Once a landing pad cassette is precisely inserted, the site-specific integrase Bxb1 enables DNA constructs, including those >50 kb, to be integrated into the genome, while Cre recombinase excises auxiliary DNA sequences.
-
This enables rapid and targeted integration of DNA payloads, irrespective of the size, and is achieved with minimal scarring.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The STRAIGHT-IN constructs used in this study are available from Addgene (https://www.addgene.org/Richard_Davis/). The datasets generated and analyzed in this study are provided as Source Data files. Source data are provided with this paper.
References
Kumar, M., Keller, B., Makalou, N. & Sutton, R. E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 12, 1893–1905 (2001).
Munoz-Lopez, M. & Garcia-Perez, J. DNA transposons: nature and applications in genomics. Curr. Genomics 11, 115–128 (2010).
Ellis, J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 16, 1241–1246 (2005).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Turan, S., Zehe, C., Kuehle, J., Qiao, J. & Bode, J. Recombinase-mediated cassette exchange (RMCE)–A rapidly expanding toolbox for targeted genomic modifications. Gene 515, 1–27 (2013).
Brown, W. R. A., Lee, N. C. O., Xu, Z. & Smith, M. C. M. Serine recombinases as tools for genome engineering. Methods 53, 372–379 (2011).
Duportet, X. et al. A platform for rapid prototyping of synthetic gene networks in mammalian cells. Nucleic Acids Res. 42, 13440–13451 (2014).
Davis, R. P. et al. A protocol for removal of antibiotic resistance cassettes from human embryonic stem cells genetically modified by homologous recombination or transgenesis. Nat. Protoc. 3, 1550–1558 (2008).
Li, J., Li, Y., Pawlik, K. M., Napierala, J. S. & Napierala, M. A CRISPR–Cas9, Crelox, and Flp-FRT cascade strategy for the precise and efficient integration of exogenous DNA into cellular genomes. CRISPR J. 3, 470–486 (2020).
Blanch-Asensio, A. et al. STRAIGHT-IN enables high-throughput targeting of large DNA payloads in human pluripotent stem cells. Cell Rep. Methods 2, 100300 (2022).
Blanch-Asensio, A. et al. Generation of AAVS1 and CLYBL STRAIGHT-IN v2 acceptor human iPSC lines for integrating DNA payloads. Stem Cell Res. 66, 102991 (2023).
Bulcha, J. T., Wang, Y., Ma, H., Tai, P. W. L. & Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target Ther. 6, 53 (2021).
Vijayachandran, L. S. et al. Gene gymnastics synthetic biology for baculovirus expression vector system engineering. Bioengineered 4, 279–287 (2013).
Aulicino, F. et al. Highly efficient CRISPR-mediated large DNA docking and multiplexed prime editing using a single baculovirus. Nucleic Acids Res. 50, 7783–7799 (2022).
Rostovskaya, M. et al. Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res. 40, e150 (2012).
Friedman, C. E. et al. CRaTER enrichment for on-target gene editing enables generation of variant libraries in hiPSCs. J. Mol. Cell Cardiol. 179, 60–71 (2023).
Iacovino, M. et al. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29, 1580–1588 (2011).
Zhu, F. et al. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 42, e34 (2014).
Low, B. E., Hosur, V., Lesbirel, S. & Wiles, M. V. Efficient targeted transgenesis of large donor DNA into multiple mouse genetic backgrounds using bacteriophage Bxb1 integrase. Sci. Rep. 12, 5424 (2022).
Brosh, R. et al. A versatile platform for locus-scale genome rewriting and verification. Proc. Natl Acad. Sci. USA 118, 1–11 (2021).
Tarailo-Graovac, M. et al. The genotypic and phenotypic spectrum of PIGA deficiency. Orphanet. J. Rare Dis. 10, 23 (2015).
Rosti, V. et al. Murine embryonic stem cells without pig-a gene activity are competent for hematopoiesis with the PNH phenotype but not for clonal expansion. J. Clin. Investig. 100, 1028–1036 (1997).
Yarnall, M. T. N. et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases. Nat. Biotechnol. 41, 500–512 (2023).
Anzalone, A. V. et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 40, 731–740 (2022).
Pallarès-Masmitjà, M. et al. Find and cut-and-transfer (FiCAT) mammalian genome engineering. Nat. Commun. 12, 1–9 (2021).
Xu, Z. et al. Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 13, 1–17 (2013).
Durrant, M. G. et al. Systematic discovery of recombinases for efficient integration of large DNA sequences into the human genome. Nat. Biotechnol. 41, 488–499 (2023).
Oceguera-Yanez, F. et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods 101, 43–55 (2016).
Concordet, J. P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Roberts, B. et al. Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol. Biol. Cell 28, 2854–2874 (2017).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, e16765 (2011).
Papapetrou, E. P. & Schambach, A. Gene insertion into genomic safe harbors for human gene therapy. Mol. Ther. 24, 678–684 (2016).
Cerbini, T. et al. Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines. PLoS One 10, 1–18 (2015).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Zhang, H. X., Zhang, Y. & Yin, H. Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol. Ther. 27, 735–746 (2019).
Vallone, V. F. et al. Methods for automated single cell isolation and sub-cloning of human pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 55, e123 (2020).
Jacquot, S., Chartoire, N., Piguet, F., Hérault, Y. & Pavlovic, G. Optimizing PCR for mouse genotyping: recommendations for reliable, rapid, cost effective, robust and adaptable to high-throughput genotyping protocol for any type of mutation. Curr. Protoc. Mouse Biol. 9, e65 (2019).
Fu, J., Teucher, M., Anastassiadis, K., Skarnes, W. & Stewart, A. F. A recombineering pipeline to make conditional targeting constructs. Meth. Enzymol. vol. 477, 125–144 (2010).
Wang, H. et al. Improved seamless mutagenesis by recombineering using ccdB for counterselection. Nucleic Acids Res. 42, e37 (2014).
Matreyek, K. A., Stephany, J. J., Chiasson, M. A., Hasle, N. & Fowler, D. M. An improved platform for functional assessment of large protein libraries in mammalian cells. Nucleic Acids Res. 48, e1 (2020).
Cabrera, A. et al. The sound of silence: transgene silencing in mammalian cell engineering. Cell Syst. 13, 950–973 (2022).
Johnstone, C. P. & Galloway, K. E. Supercoiling-mediated feedback rapidly couples and tunes transcription. Cell Rep. 41, 111492 (2022).
Ludwig, T. E. et al. ISSCR standards for the use of human stem cells in basic research. Stem Cell Reports 18, 1744–1752 (2023).
Capes-Davis, A. et al. Match criteria for human cell line authentication: where do we draw the line? Int. J. Cancer 132, 2510–2519 (2013).
Baker, D. et al. Detecting genetic mosaicism in cultures of human pluripotent stem cells. Stem Cell Rep. 7, 998–1012 (2016).
Assou, S. et al. Recurrent genetic abnormalities in human pluripotent stem cells: definition and routine detection in culture supernatant by targeted droplet digital PCR. Stem Cell Rep. 14, 1–8 (2020).
Miller, L. R. et al. Considering sex as a biological variable in preclinical research. FASEB J. 31, 29–34 (2017).
Crossley, B. M. et al. Guidelines for Sanger sequencing and molecular assay monitoring. J. Vet. Diagn. Investig. 32, 767–775 (2020).
McIntire, E., Taapken, S., Leonhard, K. & Larson, A. L. Genomic stability testing of pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 52, e107 (2020).
Adewumi, O. et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25, 803–816 (2007).
Allison, T. F. et al. Assessment of established techniques to determine developmental and malignant potential of human pluripotent stem cells. Nat. Commun. 9, 1925 (2018).
Bell, A. D., Usher, C. L. & McCarroll, S. A. Analyzing copy number variation with droplet digital PCR. Meth. Mol. Biol. 1768, 143–160 (2018).
Acknowledgements
We thank the LUMC hiPSC Hotel for co-ordinating the distribution of the Acceptor hiPS cell lines, N. Geijsen for providing Cas9 protein and F. Stewart for sharing plasmids and bacterial strains for recombineering. This work was supported by the Netherlands Organ-on-Chip Initiative, a Nederlandse Organisatie voor Wetenschappelijk Onderzoek Gravitation project funded by the Ministry of Education, Culture, and Science of the government of the Netherlands (024.003.001), a Starting Grant (STEMCARDIORISK; grant agreement no. 638030) from the European Research Council, a VIDI fellowship from the Netherlands Organization for Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek; ILLUMINATE; no. 91715303) and a ZonMw PSIDER consortium grant (no. 10250022120002; GREAT). The Novo Nordisk Foundation Center for Stem Cell Medicine (reNEW) is supported by a Novo Nordisk Foundation grant (NNF21CC0073729).
Author information
Authors and Affiliations
Contributions
A.B.-A. developed and optimized some of the protocols described in this manuscript, designed and performed the experiments, analyzed the data and wrote the manuscript. C.G. developed and optimized some of the protocols described in this manuscript, contributed to drafting the manuscript and revised it for important intellectual content. C.L.M. acquired some of the funding and revised the manuscript for important intellectual content. R.P.D. developed, designed and supervised the protocol, acquired some of the funding and wrote and revised the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
C.L.M. is a cofounder of Pluriomics BV (now Ncardia BV) and has advisory roles in HeartBeat.bio AG, Angios GmBH, Mogrify Limited and Sartorius AG. C.L.M. and R.P.D. declare research funding from Sartorius AG; however, this is for an unrelated study. A.B.-A. and C.G. declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Alessandro Bertero, Ran Brosh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Blanch-Asensio, A. et al. Cell Rep. Methods 2, 100300 (2022): https://doi.org/10.1016/J.CRMETH.2022.100300
Blanch-Asensio, A. et al. Stem Cell Res. 66, 102991 (2023): https://doi.org/10.1016/j.scr.2022.102991
Supplementary information
Supplementary Information
Supplementary Figs. 1–3, Note, Procedures 1–5 and Table 1.
Supplementary Data 1
ddPCR source data for Supplementary Fig. 3.
Source data
Source Data Fig. 6
Flow cytometry and ddPCR source data.
Source Data Fig. 7
ddPCR source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Blanch-Asensio, A., Grandela, C., Mummery, C.L. et al. STRAIGHT-IN: a platform for rapidly generating panels of genetically modified human pluripotent stem cell lines. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-01039-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-01039-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.