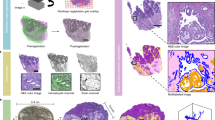
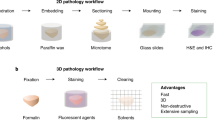
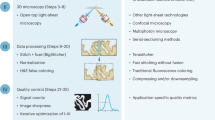
Abstract
Analysis of three-dimensional patient specimens is gaining increasing relevance for understanding the principles of tissue structure as well as the biology and mechanisms underlying disease. New technologies are improving our ability to visualize large volume of tissues with subcellular resolution. One resource often overlooked is archival tissue maintained for decades in hospitals and research archives around the world. Accessing the wealth of information stored within these samples requires the use of appropriate methods. This tutorial introduces the range of sample preparation and microscopy approaches available for three-dimensional visualization of archival tissue. We summarize key aspects of the relevant techniques and common issues encountered when using archival tissue, including registration and antibody penetration. We also discuss analysis pipelines required to process, visualize and analyze the data and criteria to guide decision-making. The methods outlined in this tutorial provide an important and sustainable avenue for validating three-dimensional tissue organization and mechanisms of disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The KESM data shown in Fig. 3 and Supplementary Video 2 have been deposited in the Figshare repository (https://doi.org/10.6084/m9.figshare.14822508) (ref. 160).
References
Gomez-Gaviro, M. V., Sanderson, D., Ripoll, J. & Desco, M. Biomedical applications of tissue clearing and three-dimensional imaging in health and disease. iScience 23, 101432 (2020).
Tian, T., Yang, Z. & Li, X. Tissue clearing technique: recent progress and biomedical applications. J. Anat. 238, 489–507 (2021).
Tomer, R., Khairy, K. & Keller, P. J. Light sheet microscopy in cell biology. Methods Mol. Biol. 931, 123–137 (2013).
Dodt, H. U. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Gritsenko, P. G. et al. p120-catenin-dependent collective brain infiltration by glioma cell networks. Nat. Cell Biol. 22, 97–107 (2020).
Ilina, O. et al. Cell–cell adhesion and 3D matrix confinement determine jamming transitions in breast cancer invasion. Nat. Cell Biol. 22, 1103–1115 (2020).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Huang, B., Jones, S. A., Brandenburg, B. & Zhuang, X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods 5, 1047–1052 (2008).
Talbot, M. J. & White, R. G. Cell surface and cell outline imaging in plant tissues using the backscattered electron detector in a variable pressure scanning electron microscope. Plant Methods 9, 40 (2013).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016).
Matsumoto, K. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537 (2019).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Murakami, T. C. et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018).
Puelles, V. G. et al. Novel 3D analysis using optical tissue clearing documents the evolution of murine rapidly progressive glomerulonephritis. Kidney Int. 96, 505–516 (2019).
Rios, A. C. et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35, 618–632 e616 (2019).
Baker, M. Biorepositories: building better biobanks. Nature 486, 141–146 (2012).
Kokkat, T. J., Patel, M. S., McGarvey, D., LiVolsi, V. A. & Baloch, Z. W. Archived formalin-fixed paraffin-embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv. Biobank 11, 101–106 (2013).
Cacciatore, S. et al. Metabolic profiling in formalin-fixed and paraffin-embedded prostate cancer tissues. Mol. Cancer Res. 15, 439–447 (2017).
Cabrita, R. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561–565 (2020).
Gao, X. H. et al. Comparison of fresh frozen tissue with formalin-fixed paraffin-embedded tissue for mutation analysis using a multi-gene panel in patients with colorectal cancer. Front. Oncol. 10, 310 (2020).
Saalfeld, S., Fetter, R., Cardona, A. & Tomancak, P. Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat. Methods 9, 717–720 (2012).
Roberts, N. et al. Toward routine use of 3D histopathology as a research tool. Am. J. Pathol. 180, 1835–1842 (2012).
Booth, M. E. et al. Three-dimensional reconstruction of ductal carcinoma in situ with virtual slides. Histopathology 66, 966–973 (2015).
Tolkach, Y., Thomann, S. & Kristiansen, G. Three-dimensional reconstruction of prostate cancer architecture with serial immunohistochemical sections: hallmarks of tumour growth, tumour compartmentalisation, and implications for grading and heterogeneity. Histopathology 72, 1051–1059 (2018).
Korehisa, S. et al. A novel histological examination with dynamic three-dimensional reconstruction from multiple immunohistochemically stained sections of a PD-L1-positive colon cancer. Histopathology 72, 697–703 (2018).
Sy, J. & Ang, L. C. Microtomy: cutting formalin-fixed, paraffin-embedded sections. Methods Mol. Biol. 1897, 269–278 (2019).
Xu, B. et al. Detection and assessment of capsular invasion, vascular invasion and lymph node metastasis volume in thyroid carcinoma using microCT scanning of paraffin tissue blocks (3D whole block imaging): a proof of concept. Mod. Pathol. 33, 2449–2457 (2020).
Yagi, Y. et al. Three-dimensional histologic, immunohistochemical, and multiplex immunofluorescence analyses of dynamic vessel co-option of spread through air spaces in lung adenocarcinoma. J. Thorac. Oncol. 15, 589–600 (2020).
Song, Y., Treanor, D., Bulpitt, A. J. & Magee, D. R. 3D reconstruction of multiple stained histology images. J. Pathol. Inform. 4, S7 (2013).
Magee, D. et al. Histopathology in 3D: from three-dimensional reconstruction to multi-stain and multi-modal analysis. J. Pathol. Inform. 6, 6 (2015).
Adler, D. H. et al. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage 84, 505–523 (2014).
Gavgiotaki, E. et al. Third Harmonic Generation microscopy distinguishes malignant cell grade in human breast tissue biopsies. Sci. Rep. 10, 11055 (2020).
Rivenson, Y. et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 3, 466–477 (2019).
van den Brand, M. et al. Sequential immunohistochemistry: a promising new tool for the pathology laboratory. Histopathology 65, 651–657 (2014).
Wentzensen, N. et al. Combined serial section-based 3D reconstruction of cervical carcinoma invasion using H&E/p16INK4a/CD3 alternate staining. Cytom. A 71, 327–333 (2007).
Jansen, I. et al. Three-dimensional histopathological reconstruction of bladder tumours. Diagn. Pathol. 14, 25 (2019).
Vasaturo, A. & Galon, J. Multiplexed immunohistochemistry for immune cell phenotyping, quantification and spatial distribution in situ. Methods Enzymol. 635, 51–66 (2020).
Viratham Pulsawatdi, A. et al. A robust multiplex immunofluorescence and digital pathology workflow for the characterisation of the tumour immune microenvironment. Mol. Oncol. 14, 2384–2402 (2020).
Hong, S. M. et al. Three-dimensional visualization of cleared human pancreas cancer reveals that sustained epithelial-to-mesenchymal transition is not required for venous invasion. Mod. Pathol. 33, 639–647 (2019).
Li, A. et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 330, 1404–1408 (2010).
Ragan, T. et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258 (2012).
Seiriki, K. et al. Whole-brain block-face serial microscopy tomography at subcellular resolution using FAST. Nat. Protoc. 14, 1509–1529 (2019).
Mayerich, D., Abbott, L. & McCormick, B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J. Microsc. 231, 134–143 (2008).
Achanta, S. et al. A comprehensive integrated anatomical and molecular atlas of rat intrinsic cardiac nervous system. iScience 23, 101140 (2020).
Weiss, S. Shattering the diffraction limit of light: a revolution in fluorescence microscopy? Proc. Natl Acad. Sci. USA 97, 8747–8749 (2000).
Jones, C. G. Scanning electron microscopy: preparation and imaging for SEM. Methods Mol. Biol. 915, 1–20 (2012).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Miyazaki, H. et al. Application of low-vacuum scanning electron microscopy for renal biopsy specimens. Pathol. Res. Pract. 208, 503–509 (2012).
Inaga, S. et al. Low vacuum scanning electron microscopy for paraffin sections utilizing the differential stainability of cells and tissues with platinum blue. Arch. Histol. Cytol. 72, 101–106 (2009).
Inaga, S. et al. Rapid three-dimensional analysis of renal biopsy sections by low vacuum scanning electron microscopy. Arch. Histol. Cytol. 73, 113–125 (2010).
Sawaguchi, A. et al. Informative three-dimensional survey of cell/tissue architectures in thick paraffin sections by simple low-vacuum scanning electron microscopy. Sci. Rep. 8, 7479 (2018).
Bates, M., Huang, B., Dempsey, G. T. & Zhuang, X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007).
Creech, M. K., Wang, J., Nan, X. & Gibbs, S. L. Superresolution imaging of clinical formalin fixed paraffin embedded breast cancer with single molecule localization microscopy. Sci. Rep. 7, 40766 (2017).
Henriques, R. et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 7, 339–340 (2010).
Ouyang, W., Aristov, A., Lelek, M., Hao, X. & Zimmer, C. Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 36, 460–468 (2018).
Xu, J., Ma, H. & Liu, Y. Stochastic optical reconstruction microscopy (STORM). Curr. Protoc. Cytom. 81, 1–27 (2017).
Andresen, V. et al. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr. Opin. Biotechnol. 20, 54–62 (2009).
Spalteholz, W. Über das Durchsichtigmachen von menschlichen und tierischen Präparaten und seine theoretischen Bedingungen, 2nd edn (n.p.; 1914).
Tainaka, K., Kuno, A., Kubota, S. I., Murakami, T. & Ueda, H. R. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell Dev. Biol. 32, 713–741 (2016).
Richardson, D. S. & Lichtman, J. W. Clarifying tissue clearing. Cell 162, 246–257 (2015).
Susaki, E. A. & Ueda, H. R. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol. 23, 137–157 (2016).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Erturk, A. et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012).
Becker, K., Jährling, N., Saghafi, S., Weiler, R. & Dodt, H.-U. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS One 7, e33916 (2012).
Hama, H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011).
Ke, M.-T., Fujimoto, S. & Imai, T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 16, 1154–1161 (2013).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Susaki, E. A. et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020).
Ueda, H. R. et al. Whole-brain profiling of cells and circuits in mammals by tissue clearing and light-sheet microscopy. Neuron 106, 369–387 (2020).
Gradinaru, V., Treweek, J., Overton, K. & Deisseroth, K. Hydrogel-tissue chemistry: principles and applications. Annu. Rev. Biophys. 20, 355–376 (2018).
van Royen, M. E. et al. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology 69, 985–992 (2016).
Noe, M. et al. Immunolabeling of cleared human pancreata provides insights into three-dimensional pancreatic anatomy and pathology. Am. J. Pathol. 188, 1530–1535 (2018).
Tanaka, N. et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat. Biomed. Eng. 1, 796–806 (2017).
Nojima, S. et al. CUBIC pathology: three-dimensional imaging for pathological diagnosis. Sci. Rep. 7, 9269 (2017).
Lai, H. M. et al. Next generation histology methods for three-dimensional imaging of fresh and archival human brain tissues. Nat. Commun. 9, 1066 (2018).
Chen, Y. et al. Three-dimensional imaging and quantitative analysis in CLARITY processed breast cancer tissues. Sci. Rep. 9, 5624 (2019).
Verhoef, E. I. et al. Three-dimensional analysis reveals two major architectural subgroups of prostate cancer growth patterns. Mod. Pathol. 32, 1032–1041 (2019).
Tanaka, N. et al. Mapping of the three-dimensional lymphatic microvasculature in bladder tumours using light-sheet microscopy. Br. J. Cancer 118, 995–999 (2018).
Yoshizawa, T. et al. Three-dimensional analysis of extrahepatic cholangiocarcinoma and tumor budding. J. Pathol. 251, 400–410 (2020).
Weiss, K. R., Voigt, F. F., Shepherd, D. P. & Huisken, J. Tutorial: practical considerations for tissue clearing and imaging. Nat. Protoc. 16, 2732–2748 (2021).
Renier, N. et al. Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016).
Lee, E. et al. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep. 6, 18631 (2016).
Tainaka, K. et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210 e2199 (2018).
Uhlen, P. & Tanaka, N. Improved pathological examination of tumors with 3D light-sheet microscopy. Trends Cancer 4, 337–341 (2018).
Tomer, R. et al. SPED light sheet microscopy: fast mapping of biological system structure and function. Cell 163, 1796–1806 (2015).
Miltenyi BioTec. UltraMicroscope Blaze. Miltenyi BioTec https://www.miltenyibiotec.com/JP-en/products/ultramicroscope-blaze.html (2021).
Chen, B. C. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014).
Chakraborty, T. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat. Methods 16, 1109–1113 (2019).
Lu, C. H. et al. Lightsheet localization microscopy enables fast, large-scale, and three-dimensional super-resolution imaging. Commun. Biol. 2, 177 (2019).
Glaser, A. K. et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017).
Glaser, A. K. et al. Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissues. Nat. Commun. 10, 2781 (2019).
Royer, L. A. et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 34, 1267–1278 (2016).
McDole, K. et al. In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 175, 859–876 e833 (2018).
Verhoef, E. I. et al. Three-dimensional architecture of common benign and precancerous prostate epithelial lesions. Histopathology 74, 1036–1044 (2019).
Korobchevskaya, K., Lagerholm, B., Colin-York, H. & Fritzsche, M. Exploring the potential of airyscan microscopy for live cell imaging. Photonics 4, 41 (2017).
Wang, H. et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019).
Jonkman, J., Brown, C. M., Wright, G. D., Anderson, K. I. & North, A. J. Tutorial: guidance for quantitative confocal microscopy. Nat. Protoc. 15, 1585–1611 (2020).
Dekkers, J. F. et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771 (2019).
Weigelin, B., Bakker, G.-J. & Friedl, P. Third harmonic generation microscopy of cells and tissue organization. J. Cell Sci. 129, 245–255 (2016).
Kalendar, W.A. Computed Tomography: Fundamentals, System Technology, Image Quality, Applications (John Wiley & Sons, 2011).
Katsamenis, O. L. et al. X-ray micro-computed tomography for nondestructive three-dimensional (3D) X-ray histology. Am. J. Pathol. 189, 1608–1620 (2019).
Virta, J. et al. X-ray microtomography is a novel method for accurate evaluation of small-bowel mucosal morphology and surface area. Sci. Rep. 10, 13164 (2020).
Ortega-Gil, A., Vaquero, J. J., Gonzalez-Arjona, M., Rullas, J. & Munoz-Barrutia, A. X-ray-based virtual slicing of TB-infected lungs. Sci. Rep. 9, 19404 (2019).
Robinson, S. K., Ramsden, J. J., Warner, J., Lackie, P. M. & Roose, T. Correlative 3D imaging and microfluidic modelling of human pulmonary lymphatics using immunohistochemistry and high-resolution muCT. Sci. Rep. 9, 6415 (2019).
Andreev, A. & Koo, D. E. S. Practical guide to storage of large amounts of microscopy data. Microsc. Today 28, 42–45 (2020).
Amat, F. et al. Efficient processing and analysis of large-scale light-sheet microscopy data. Nat. Protoc. 10, 1679–1696 (2015).
Balázs, B., Deschamps, J., Albert, M., Ries, J. & Hufnagel, L. A real-time compression library for microscopy images. Preprint at bioRxiv https://doi.org/10.1101/164624 (2017).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Ellenberg, J. et al. A call for public archives for biological image data. Nat. Methods 15, 849–854 (2018).
Allan, C. et al. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9, 245–253 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017).
Arena, E. T. et al. Quantitating the cell: turning images into numbers with ImageJ. Wiley Interdiscip. Rev. Dev. Biol. https://doi.org/10.1002/wdev.260 (2017).
Kvilekval, K., Fedorov, D., Obara, B., Singh, A. & Manjunath, B. S. Bisque: a platform for bioimage analysis and management. Bioinformatics 26, 544–552 (2010).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PloS Biol. 16, e2005970 (2018).
Sage, D. et al. Super-resolution fight club: assessment of 2D and 3D single-molecule localization microscopy software. Nat. Methods 16, 387–395 (2019).
Horl, D. et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 16, 870–874 (2019).
Bria, A. & Iannello, G. TeraStitcher—a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics 13, 316 (2012).
Pichat, J., Iglesias, J. E., Yousry, T., Ourselin, S. & Modat, M. A survey of methods for 3D histology reconstruction. Med. Image Anal. 46, 73–105 (2018).
Kartasalo, K. et al. Comparative analysis of tissue reconstruction algorithms for 3D histology. Bioinformatics 34, 3013–3021 (2018).
Swaney, J. et al. Scalable image processing technius for quantitative analysis of volumetric biological images from light-sheet microscopy. Preprint at bioRxiv https://doi.org/10.1101/576595 (2019).
Reinhard, E., Mashikhmin, M., Gooch, B. & Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 21, 34–41 (2001).
Bautista, P. A., Hashimoto, N. & Yagi, Y. Color standardization in whole slide imaging using a color calibration slide. J. Pathol. Inform. 5, 4 (2014).
Bautista, P. A. & Yagi, Y. Staining correction in digital pathology by utilizing a dye amount table. J. Digit. Imaging 28, 283–294 (2015).
Preibisch, S. et al. Efficient Bayesian-based multiview deconvolution. Nat. Methods 11, 645–648 (2014).
Becker, K. et al. Deconvolution of light sheet microscopy recordings. Sci. Rep. 9, 17625 (2019).
Khan, A. M., Rajpoot, N., Treanor, D. & Magee, D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans. Biomed. Eng. 61, 1729–1738 (2014).
Vahadane, A. et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging 35, 1962–1971 (2016).
Bejnordi, B. E. et al. Stain specific standardization of whole-slide histopathological images. IEEE Trans. Med. Imaging 35, 404–415 (2016).
Belthangady, C. & Royer, L. A. Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods 16, 1215–1225 (2019).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Borland, D. et al. Segmentor: a tool for manual refinement of 3D microscopy annotations. BMC Bioinformatics 22, 260 (2021).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Falk, T. et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019).
Arganda-Carreras, I. et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Haberl, M. G. et al. CDeep3M-Plug-and-Play cloud-based deep learning for image segmentation. Nat. Methods 15, 677–680 (2018).
Bannon, D. et al. DeepCell Kiosk: scaling deep learning-enabled cellular image analysis with Kubernetes. Nat. Methods 18, 43–45 (2021).
Kim, T., Moon, S. & Xu, K. Information-rich localization microscopy through machine learning. Nat. Commun. 10, 1996 (2019).
Speiser, A., Turaga, S. C. & Macke, J. H. Teaching deep neural networks to localize sources in super-resolution microscopy by combining simulation-based learning and unsupervised learning. Preprint at https://arxiv.org/abs/1907.00770 (2019).
van der Laak, J., Litjens, G. & Ciompi, F. Deep learning in histopathology: the path to the clinic. Nat. Med. 27, 775–784 (2021).
Yu, K. H., Beam, A. L. & Kohane, I. S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2, 719–731 (2018).
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V. & Madabhushi, A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715 (2019).
Liu, J. T. C. et al. Harnessing non-destructive 3D pathology. Nat. Biomed. Eng. 5, 203–218 (2021).
Janowczyk, A., Zuo, R., Gilmore, H., Feldman, M. & Madabhushi, A. HistoQC: an open-source quality control tool for digital pathology slides. JCO Clin. Cancer Inform. 3, 1–7 (2019).
Pietzsch, T., Saalfeld, S., Preibisch, S. & Tomancak, P. BigDataViewer: visualization and processing for large image data sets. Nat. Methods 12, 481–483 (2015).
Yushkevich, P. A. & Gerig, G. ITK-SNAP: an intractive medical image segmentation tool to meet the need for expert-guided segmentation of complex medical images. IEEE Pulse 8, 54–57 (2017).
de Chaumont, F., Dallongeville, S. & Olivo-Marin, J.-C. ICY: a new open-source community image processing software. IEEE International Symposium on Biomedical Imaging https://doi.org/10.1109/ISBI.2011.5872395 (2011).
Peng, H., Ruan, Z., Long, F., Simpson, J. H. & Myers, E. W. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 28, 348–353 (2010).
Bria, A., Iannello, G. & Peng, H. An open-source VAA3D plugin for real-time 3D visualization of terabyte-sized volumetric images. IEEE International Symposium on Biomedical Imaging https://doi.org/10.1109/ISBI.2015.7163925 (2015).
Bria, A., Iannello, G., Onofri, L. & Peng, H. TeraFly: real-time three-dimensional visualization and annotation of terabytes of multidimensional volumetric images. Nat. Methods 13, 192–194 (2016).
Peng, H., Bria, A., Zhou, Z., Iannello, G. & Long, F. Extensible visualization and analysis for multidimensional images using Vaa3D. Nat. Protoc. 9, 193–208 (2014).
Lytle, N. K., Barber, A. G. & Reya, T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 18, 669–680 (2018).
Kingston, B. R., Syed, A. M., Ngai, J., Sindhwani, S. & Chan, W. C. W. Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc. Natl Acad. Sci. USA 116, 14937–14946 (2019).
Rohé, M.-M., Datar, M., Heimann, T., Sermesant, M. & Pennec, X. SVF-Net: learning deformable image registration using shape matching. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2017, 266–274 (Springer, 2017).
Haddad, T.S. et al. Tutorial: methods for three-dimensional visualization of archival tissue material. Figshare. https://doi.org/10.6084/m9.figshare.14822508 (2021).
Falk, M., Ynnerman, A., Treanor, D. & Lundstrom, C. Interactive visualization of 3D histopathology in native resolution. IEEE Trans. Vis. Comput. Graph. https://doi.org/10.1109/TVCG.2018.2864816 (2018).
Acknowledgements
This work received funding from the Dutch Cancer Society, project number 10602/2016-2. P.F. was supported by the European Research Council (617430-DEEPINSIGHT), NIH-U54 CA210184-01, and the Cancer Genomics Center (CGC.nl). The authors thank D. Jutt for the render in Fig. 3 and Supplementary Video 2, Prof. N. Shepherd for the image slides in Fig. 2, Supplementary Video 1 and M. Falk for the renders in Supplementary Video 1, and J.-M. Bokhorst for training the deep learning network applied to the KESM dataset illustrated in Fig. 3, Supplementary Video 2.
Author information
Authors and Affiliations
Contributions
The authors contributed to the various sections of this tutorial as follows: T.H., I.N. conceptualization and draft writing; N.F., knife-edge scanning microscopy; P.F., thick sectioning without clearing.; D.T., 3D histology; I.Z., X-ray microcomputed tomography. All authors read, revised and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Strateos is a commercial entity and the affiliated author (N.F.) is a company employee. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Hiroki R. Ueda and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Video 1
3D malignant colorectal adenocarcinoma consisting of 50 slide images with an interslice distance of 20 μm digitized twice at 20x, once with an H&E stain and a second time with a Pan-cytokeratin immunohistochemical stain to reveal the epithelium in brown. Data related to Figure 2.
Supplementary Video 2
3D reconstruction of the isolated tumor class of a formalin-fixed paraffin-embedded colorectal carcinoma specimen processed using Knife-Edge Scanning Microscopy (KESM) technology and segmented using deep learning. Data related to Figure 3.
Supplementary Video 3
3D reconstruction of invasive ductal carcinoma lesion in a patient sample (30 µm Z-stack, 5 µm step). Collective strands of carcinoma cells were detected by positive E-cadherin staining; collagen bundles (SHG); nuclei (DAPI). The video is representative for 7 independent samples. Data related to Figure 5.
Supplementary Video 4
3D reconstruction of invasive lobular carcinoma lesion in a patient sample (60 µm Z-stack, 5 µm step). Collective strands of carcinoma cells were detected by positive staining for pan-cytokeratin; collagen bundles (SHG); nuclei (DAPI). The video is representative for 3 independent samples. Data related to Figure 5.
Rights and permissions
About this article
Cite this article
Haddad, T.S., Friedl, P., Farahani, N. et al. Tutorial: methods for three-dimensional visualization of archival tissue material. Nat Protoc 16, 4945–4962 (2021). https://doi.org/10.1038/s41596-021-00611-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00611-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.