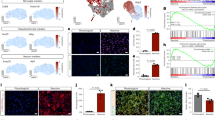
Abstract
Astrocytes become reactive in response to insults to the central nervous system by adopting context-specific cellular signatures and outputs, but a systematic understanding of the underlying molecular mechanisms is lacking. In this study, we developed CRISPR interference screening in human induced pluripotent stem cell-derived astrocytes coupled to single-cell transcriptomics to systematically interrogate cytokine-induced inflammatory astrocyte reactivity. We found that autocrine–paracrine IL-6 and interferon signaling downstream of canonical NF-κB activation drove two distinct inflammatory reactive signatures, one promoted by STAT3 and the other inhibited by STAT3. These signatures overlapped with those observed in other experimental contexts, including mouse models, and their markers were upregulated in human brains in Alzheimer’s disease and hypoxic-ischemic encephalopathy. Furthermore, we validated that markers of these signatures were regulated by STAT3 in vivo using a mouse model of neuroinflammation. These results and the platform that we established have the potential to guide the development of therapeutics to selectively modulate different aspects of inflammatory astrocyte reactivity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Bulk RNA-seq data of hiPSC-derived astrocytes generated in this study shown in Extended Data Fig. 3 are available on the Gene Expression Omnibus (GEO) under accession code GSE182307. The raw single-cell RNA-seq data and UMI count matrices from the CROP-seq experiment are available on the GEO under accession code GSE182308. Processed data from the CRISPRi screens can also be interactively explored on CRISPRbrain (https://www.crisprbrain.org/screens/) by selecting ‘Astrocyte’ as the screening cell type. The public databases used in this study include Enrichr (https://maayanlab.cloud/Enrichr/), GENCODE (https://www.gencodegenes.org/human/), GEO (https://www.ncbi.nlm.nih.gov/geo/) and Synapse (https://www.synapse.org/). Previously published datasets available on the GEO that were reanalyzed in this study include GSE143598, GSE120411, GSE76097, GSE148611 and GSE130119; previously published datasets available on Synapse and reanalyzed in this study include syn21861229.
Code availability
The full analysis pipeline (including code and processed data objects) used for master regulator analysis, analysis of CROP-seq data and integration with previously published single-cell RNA-seq datasets is available at https://kampmannlab.ucsf.edu/inflammatory-reactive-astrocyte-analysis.
References
Escartin, C. et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325 (2021).
Sofroniew, M. V. & Vinters, H. V. Astrocytes: biology and pathology. Acta Neuropathol. 119, 7–35 (2010).
Burda, J. E. et al. Divergent transcriptional regulation of astrocyte reactivity across disorders. Nature 606, 557–564 (2022).
Wang, Q., Tang, X. N. & Yenari, M. A. The inflammatory response in stroke. J. Neuroimmunol. 184, 53–68 (2007).
Hausmann, O. N. Post-traumatic inflammation following spinal cord injury. Spinal Cord 41, 369–378 (2003).
Ponath, G., Park, C. & Pitt, D. The role of astrocytes in multiple sclerosis. Front. Immunol. 9, 217 (2018).
Heneka, M. T. et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405 (2015).
Han, R. T., Kim, R. D., Molofsky, A. V. & Liddelow, S. A. Astrocyte–immune cell interactions in physiology and pathology. Immunity 54, 211–224 (2021).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Guttenplan, K. A. et al. Knockout of reactive astrocyte activating factors slows disease progression in an ALS mouse model. Nat. Commun. 11, 3753 (2020).
Guttenplan, K. A. et al. Neurotoxic reactive astrocytes drive neuronal death after retinal injury. Cell Rep. 31, 107776 (2020).
Yun, S. P. et al. Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat. Med. 24, 931–938 (2018).
Clarke, L. E. et al. Normal aging induces A1-like astrocyte reactivity. Proc. Natl Acad. Sci. USA 115, E1896–E1905 (2018).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 16, 465–480 (2020).
TCW, J. et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Rep. 9, 600–614 (2017).
Li, X. et al. Fast generation of functional subtype astrocytes from human pluripotent stem cells. Stem Cell Rep. 11, 998–1008 (2018).
Williams, J. L. et al. Astrocyte–T cell crosstalk regulates region-specific neuroinflammation. Glia 68, 1361–1374 (2020).
Gimenez, M. A., Sim, J. E. & Russell, J. H. TNFR1-dependent VCAM-1 expression by astrocytes exposes the CNS to destructive inflammation. J. Neuroimmunol. 151, 116–125 (2004).
Rosenman, S. J., Shrikant, P., Dubb, L., Benveniste, E. N. & Ransohoff, R. M. Cytokine-induced expression of vascular cell adhesion molecule-1 (VCAM-1) by astrocytes and astrocytoma cell lines. J. Immunol. 154, 1888–1899 (1995).
Rubio, N., Sanz-Rodriguez, F. & Arevalo, M. A. Up-regulation of the vascular cell adhesion molecule-1 (VCAM-1) induced by Theilerʼs murine encephalomyelitis virus infection of murine brain astrocytes. Cell Commun. Adhes. 17, 57–68 (2010).
Labib, D. et al. Proteomic alterations and novel markers of neurotoxic reactive astrocytes in human induced pluripotent stem cell models. Front. Mol. Neurosci. 15, 870085 (2022).
Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Kampmann, M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 13, 406–416 (2018).
Castro, M. A. et al. Regulators of genetic risk of breast cancer identified by integrative network analysis. Nat. Genet. 48, 12–21 (2016).
Fletcher, M. N. et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat. Commun. 4, 2464 (2013).
Campbell, T. M. et al. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis 37, 741–750 (2016).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Liberti, S. et al. HuPho: the human phosphatase portal. FEBS J. 280, 379–387 (2013).
Horlbeck, M. A. et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 (2016).
Shih, V. F., Tsui, R., Caldwell, A. & Hoffmann, A. A single NFκB system for both canonical and non-canonical signaling. Cell Res. 21, 86–102 (2011).
Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2, 17023 (2017).
Cardinaux, J. R., Allaman, I. & Magistretti, P. J. Pro-inflammatory cytokines induce the transcription factors C/EBPbeta and C/EBPdelta in astrocytes. Glia 29, 91–97 (2000).
Alonzi, T. et al. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol. Cell. Biol. 21, 1621–1632 (2001).
Herrmann, J. E. et al. STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J. Neurosci. 28, 7231–7243 (2008).
Anderson, M. A. et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200 (2016).
Ben Haim, L. et al. The JAK/STAT3 pathway is a common inducer of astrocyte reactivity in Alzheimer’s and Huntington’s diseases. J. Neurosci. 35, 2817–2829 (2015).
Wang, Z. H. et al. C/EBPβ regulates delta-secretase expression and mediates pathogenesis in mouse models of Alzheimer’s disease. Nat. Commun. 9, 1784 (2018).
Strohmeyer, R., Shelton, J., Lougheed, C. & Breitkopf, T. CCAAT-enhancer binding protein-β expression and elevation in Alzheimer’s disease and microglial cell cultures. PLoS ONE 9, e86617 (2014).
Feng, H., Zhang, Y. B., Gui, J. F., Lemon, S. M. & Yamane, D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 17, e1009220 (2021).
Lehtonen, A., Matikainen, S. & Julkunen, I. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J. Immunol. 159, 794–803 (1997).
Ng, S. L. et al. IκB kinase ε (IKKε) regulates the balance between type I and type II interferon responses. Proc. Natl Acad. Sci. USA 108, 21170–21175 (2011).
Hasel, P., Rose, I. V. L., Sadick, J. S., Kim, R. D. & Liddelow, S. A. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 24, 1475–1487 (2021).
Mayer-Barber, K. D. & Yan, B. Clash of the cytokine titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell. Mol. Immunol. 14, 22–35 (2017).
Gan, W. et al. LATS suppresses mTORC1 activity to directly coordinate Hippo and mTORC1 pathways in growth control. Nat. Cell Biol. 22, 246–256 (2020).
He, L. et al. mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol. Cell 70, 949–960 (2018).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Banks, T. A. et al. A lymphotoxin-IFN-β axis essential for lymphocyte survival revealed during cytomegalovirus infection. J. Immunol. 174, 7217–7225 (2005).
Weichhart, T., Hengstschläger, M. & Linke, M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 15, 599–614 (2015).
Balamurugan, K. et al. The tumour suppressor C/EBPδ inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 29, 4106–4117 (2010).
Heinrich, P. C., Castell, J. V. & Andus, T. Interleukin-6 and the acute phase response. Biochem. J. 265, 621–636 (1990).
Sonderegger, I. et al. GM-CSF mediates autoimmunity by enhancing IL-6-dependent Th17 cell development and survival. J. Exp. Med. 205, 2281–2294 (2008).
Metzemaekers, M., Vanheule, V., Janssens, R., Struyf, S. & Proost, P. Overview of the mechanisms that may contribute to the non-redundant activities of interferon-inducible CXC chemokine receptor 3 ligands. Front. Immunol. 8, 1970 (2017).
Wang, Y., van Boxel-Dezaire, A. H., Cheon, H., Yang, J. & Stark, G. R. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc. Natl Acad. Sci. USA 110, 16975–16980 (2013).
Au-Yeung, N., Mandhana, R. & Horvath, C. M. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2, e23931 (2013).
Hungness, E. S. et al. Transcription factors C/EBP-β and -δ regulate IL-6 production in IL-1β-stimulated human enterocytes. J. Cell. Physiol. 192, 64–70 (2002).
Tsai, M. H., Pai, L. M. & Lee, C. K. Fine-tuning of type I interferon response by STAT3. Front Immunol. 10, 1448 (2019).
Michalska, A., Blaszczyk, K., Wesoly, J. & Bluyssen, H. A. R. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls type I and type II IFN responses. Front. Immunol. 9, 1135 (2018).
Metwally, H. et al. Noncanonical STAT1 phosphorylation expands its transcriptional activity into promoting LPS-induced IL-6 and IL-12p40 production. Sci. Signal 13, eaay0574 (2020).
Nan, J., Wang, Y., Yang, J. & Stark, G. R. IRF9 and unphosphorylated STAT2 cooperate with NF-κB to drive IL6 expression. Proc. Natl Acad. Sci. USA 115, 3906–3911 (2018).
Haan, S., Keller, J. F., Behrmann, I., Heinrich, P. C. & Haan, C. Multiple reasons for an inefficient STAT1 response upon IL-6-type cytokine stimulation. Cell. Signal. 17, 1542–1550 (2005).
Qing, Y. & Stark, G. R. Alternative activation of STAT1 and STAT3 in response to interferon-γ. J. Biol. Chem. 279, 41679–41685 (2004).
van Boxel-Dezaire, A. H. et al. Major differences in the responses of primary human leukocyte subsets to IFN-β. J. Immunol. 185, 5888–5899 (2010).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Heppner, F. L., Ransohoff, R. M. & Becher, B. Immune attack: the role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 16, 358–372 (2015).
Lyra, E. et al. Pro-inflammatory interleukin-6 signaling links cognitive impairments and peripheral metabolic alterations in Alzheimer’s disease. Transl. Psychiatry 11, 251 (2021).
Roy, E. R. et al. Concerted type I interferon signaling in microglia and neural cells promotes memory impairment associated with amyloid β plaques. Immunity 55, 879–894 (2022).
Eikelenboom, P., Hack, C. E., Rozemuller, J. M. & Stam, F. C. Complement activation in amyloid plaques in Alzheimer’s dementia. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 56, 259–262 (1989).
Abraham, C. R., Selkoe, D. J. & Potter, H. Immunochemical identification of the serine protease inhibitor α1-antichymotrypsin in the brain amyloid deposits of Alzheimer’s disease. Cell 52, 487–501 (1988).
Habib, N. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020).
Orzyłowska, O., Oderfeld-Nowak, B., Zaremba, M., Januszewski, S. & Mossakowski, M. Prolonged and concomitant induction of astroglial immunoreactivity of interleukin-1beta and interleukin-6 in the rat hippocampus after transient global ischemia. Neurosci. Lett. 263, 72–76 (1999).
Deng, Y., Lu, J., Sivakumar, V., Ling, E. A. & Kaur, C. Amoeboid microglia in the periventricular white matter induce oligodendrocyte damage through expression of proinflammatory cytokines via MAP kinase signaling pathway in hypoxic neonatal rats. Brain Pathol. 18, 387–400 (2008).
Taher, N. A. B. et al. Altered distributions and functions of natural killer T cells and γδ T cells in neonates with neonatal encephalopathy, in school-age children at follow-up, and in children with cerebral palsy. J. Neuroimmunol. 356, 577597 (2021).
Liu, F. & McCullough, L. D. Inflammatory responses in hypoxic ischemic encephalopathy. Acta Pharmacol. Sin. 34, 1121–1130 (2013).
Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016).
Sloan, S. A. et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779–790 (2017).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Han, H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386 (2018).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Barbar, L. et al. CD49f is a novel marker of functional and reactive human iPSC-derived astrocytes. Neuron 107, 436–453 (2020).
Wheeler, M. A. et al. MAFG-driven astrocytes promote CNS inflammation. Nature 578, 593–599 (2020).
Miyaoka, Y. et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods 11, 291–293 (2014).
TCW, J. et al. Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell 185, 2213–2233 (2022).
Krencik, R. et al. Dysregulation of astrocyte extracellular signaling in Costello syndrome. Sci. Transl. Med. 7, 286ra266 (2015).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron 104, 239–255 (2019).
Li, X. L. et al. Highly efficient genome editing via CRISPR–Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 46, 10195–10215 (2018).
Cheng, C., Fass, D. M., Folz-Donahue, K., MacDonald, M. E. & Haggarty, S. J. Highly expandable human iPS cell-derived neural progenitor cells (NPC) and neurons for central nervous system disease modeling and high-throughput screening. Curr. Protoc. Hum. Genet. 92, 21.28.21 (2017).
Krencik, R. & Zhang, S. C. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat. Protoc. 6, 1710–1717 (2011).
Bowles, K. R., TCW, J., Qian, L., Jadow, B. M. & Goate, A. M. Reduced variability of neural progenitor cells and improved purity of neuronal cultures using magnetic activated cell sorting. PLoS ONE 14, e0213374 (2019).
Wolf, J., Rose-John, S. & Garbers, C. Interleukin-6 and its receptors: a highly regulated and dynamic system. Cytokine 70, 11–20 (2014).
Neal, E. H. et al. A simplified, fully defined differentiation scheme for producing blood–brain barrier endothelial cells from human iPSCs. Stem Cell Rep. 12, 1380–1388 (2019).
Fernandopulle, M. S. et al. Transcription factor-mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 79, e51 (2018).
Niu, J. et al. Oligodendroglial ring finger protein Rnf43 is an essential injury-specific regulator of oligodendrocyte maturation. Neuron 109, 3104–3118 (2021).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Lachmann, A. et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 9, 1366 (2018).
Lachmann, A., Xie, Z. & Ma’ayan, A. Elysium: RNA-seq alignment in the cloud. Preprint at https://www.biorxiv.org/content/10.1101/382937v1 (2018).
Krawczyk, M. C. et al. Human astrocytes exhibit tumor microenvironment-, age-, and sex-related transcriptomic signatures. J. Neurosci. 42, 1587–1603 (2022).
Zhang, Y., Parmigiani, G. & Johnson, W. E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2, lqaa078 (2020).
Hill, A. J. et al. On the design of CRISPR-based single-cell molecular screens. Nat. Methods 15, 271–274 (2018).
Gaublomme, J. T. et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat. Commun. 10, 2907 (2019).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2020).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Stephens, M. False discovery rates: a new deal. Biostatistics 18, 275–294 (2017).
Torre, D., Lachmann, A. & Ma’ayan, A. BioJupies: automated generation of interactive notebooks for RNA-seq data analysis in the cloud. Cell Syst. 7, 556–561 (2018).
Perriot, S. et al. Human induced pluripotent stem cell-derived astrocytes are differentially activated by multiple sclerosis-associated cytokines. Stem Cell Rep. 11, 1199–1210 (2018).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Smithson, M. & Verkuilen, J. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol. Methods 11, 54–71 (2006).
Cribari-Neto, F. & Zeileis, A. Beta regression in R. J. Stat. Softw. 34, 1–24 (2010).
Kelley, K. W., Nakao-Inoue, H., Molofsky, A. V. & Oldham, M. C. Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci. 21, 1171–1184 (2018).
Huang, R. et al. The NCATS BioPlanet—an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front. Pharm. 10, 445 (2019).
Cao, J. et al. A human cell atlas of fetal gene expression. Science 370, eaba7721 (2020).
Acknowledgements
We thank B. Desousa, V. Jovanovic, Z. Krejciova and N. Sun for contributions to preliminary studies and discussions. We thank A. Molofsky, A. Kao and M. Oldham for serving on K.L.ʼs thesis committee. We thank members of the Kampmann laboratory (G. Mohl, S. Sattler and O. Teter) for discussions and feedback on the manuscript. We thank B. Woo for cloning the transcription factors sgRNA library and B. Ramani for help with obtaining primary mouse astrocytes. We thank the Conklin laboratory for the gift of the WTC11 hiPSC line. This research was supported by National Institutes of Health (NIH) grant F30 AG066418 to K.L.; California Institute for Regenerative Medicine grant EDUC4-12812 and NIH grant T32 NS115706 to I.V.L.R.; Chan Zuckerberg Initiative Ben Barres Early Career Acceleration Awards to E.S.L. and M. Kampmann; NIH/NIND grants (R01NS097551, P01NS083513 and R21NS119954) to S.F.; and NIH grants P30 EY02162-39 and R03AG063157 to E.M.U. S.F. is a Harry Weaver Neuroscience Scholar of the National Multiple Sclerosis Society. The TCW-1E44 iPSC line was generated with the support of NIH NIA K01AG062683 (J.T.) and the Druckenmiller Fellowship from the New York Stem Cell Foundation (J.T.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
K.L. and M. Kampmann conceptualized and led the overall project and wrote the manuscript, with input from all co-authors. K.L. performed the majority of experiments, with support from B.R., and performed all data analysis. I.V.L.R. performed immunostaining of mouse tissue provided by Y.A. and S.W., with guidance from M.V.S. H.K. performed immunostaining of AD tissues provided by M.S.S. and co-culture experiments, with guidance from E.S.L., and W.X. performed immunostaining of HIE tissue, with guidance from S.F. W.R.F. also performed immunostaining of AD tissues provided by M.S.S. E.L. generated the CRISPRi TCW-1E44 hiPSC line. J.T. supplied the TCW-1E44 hiPSC line, with guidance from A.G. M. Koontz generated hiPSC-derived astrocytes using the methods of Krencik et al. and Li et al., with guidance from E.M.U., who also provided the 162D iPSC line. M. Krawcyzk and Y.Z. supplied unpublished human astrocyte RNA-seq data for master regulator analysis.
Corresponding authors
Ethics declarations
Competing interests
M. Kampmann is an inventor on US Patent 11,254,933 related to CRISPRi and CRISPRa screening, serves on the scientific advisory boards of Engine Biosciences, Casma Therapeutics, Cajal Neuroscience and Alector and is an advisor to Modulo Bio and Recursion Therapeutics. J.T. co-founded Asmos Therapeutics, LLC, serves on the scientific advisory board of NeuCyte, Inc. and has consulted for FIND Genomics Inc., CareCureSystems Corporation, TheWell Biosciences Inc. and Aleta Neuroscience, LLC. A.G. serves on the scientific advisory board for Genentech and is a consultant to Muna Therapeutics. None of the other authors declare competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Valentina Fossati and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Additional characterization of iAstrocytes.
a, Empirical cumulative distribution functions of the mean expression of genes (averaged across experimental replicates, n = 3 wells) with astrocyte-specific expression (astrocyte fidelity > 40) or without astrocyte specific expression (astrocyte fidelity < 40) in iAstrocytes vs astrocytes generated using the TCW et al. protocol16 (TCW astrocytes). Genome-wide astrocyte fidelity scores were obtained from Kelley et al. TPM: transcripts per million. b, Relative expression (z-scored) of the top 50 genes with the highest astrocyte fidelity scores from Kelley et al.115 (organized by hierarchical clustering, see Methods) in iAstrocytes vs. TCW astrocytes (n = 3 experimental replicates corresponding to heatmap rows). Genes with statistically significant differential expression (adjusted P value < 0.1) between iAstrocytes and TCW astrocytes are marked with asterisks; P values were calculated and adjusted for multiple testing (false-discovery rate method) using DESeq2 (two-sided Wald test; see Methods). c, Heatmap of log-scaled transcripts per million (TPM) values of NFIA transcripts in human primary astrocytes from Zhang et al.93 d, Representative images from immunostaining of GFAP in iAstrocytes cultured alone or with iNeurons (n = 3 wells). In each case, an entire field of view is displayed (left) next to magnified sections (all at same scale) containing representative astrocyte morphologies (right). Scale bars correspond to 60 μm.
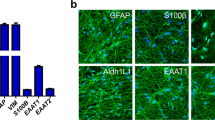
Extended Data Fig. 2 Validation of iAstrocyte differentiation from two additional hiPSC lines.
A, Representative images of immunofluorescence against GFAP, S100β, GLAST, Cx43, glutamine synthetase, or vimentin in iAstrocytes vs. TCW astrocytes derived from TCW-1E44 or 162D hiPSCs (scale bar: 60 μm). b,Quantification of GFAP, S100β, GLAST, Cx43, glutamine synthetase, or vimentin immunofluorescence intensity (n = 3 wells). c, Phagocytosis of pHrodo-labeled rat synaptosomes (median fluorescence intensity measured by flow cytometry) by iAstrocytes derived from TCW-1E44 or 162D hiPSCs in the absence (n = 5 wells) or presence (n = 1 well) of cytochalasin D (cytoD). d, Percent VCAM1+ cells in TCW-1E44 or 162D iAstrocytes treated with vehicle control vs. IL-1α+TNF+ C1q (n = 4 wells). e, Percentage of dead cells (measured by TO-PRO-3 permeability) in iNeurons incubated with conditioned media from TCW-1E44 or 162D iAstrocytes treated with vehicle control or IL-1α+TNF+ C1q (n = 12 wells). In panels b and c, P values were calculated using the two-sided Student’s t-test. In panels d and e, P values were calculated using the two-sided Mann-Whitney U test.
Extended Data Fig. 3 iAstrocytes respond to IL-1α+TNF+ C1q in a highly similar manner as hiPSC-derived astrocytes generated using different protocols and primary mouse astrocytes.
a, Principal component (PC) analysis plot of the gene expression profiles (top 5000 variable genes) of iAstrocytes vs. astrocytes derived using the protocols from TCW et al.16 (TCW astrocytes), Li et al.17 (Li astrocytes), or Krencik et al.89 (Krencik astrocytes), as well as iPSC-derived neurons (iNeurons) and neural progenitor cells (NPCs), treated with vehicle control or IL-1α+TNF+ C1q (n = 3 wells for astrocyte samples, n = 2 wells for iNeuron and NPC samples). b, Number of differential expressed genes (DEGs) induced by IL-1α+TNF+ C1q. c, Log2-fold-changes of pan-reactive, A1 reactive, and A2 reactive genes defined in Liddlelow et al.9 in hiPSC-derived astrocytes from this study. d, Overlap of upregulated and downregulated DEGs induced by IL-1α+TNF+ C1q among hiPSC-derived astrocytes from this study. e, Overlap of upregulated and downregulated DEGs induced by IL-1α+TNF+ C1q from hiPSC-derived astrocytes from this study compared to DEGs from inflammatory reactive astrocytes in other studies. f-g, Phagocytosis of pHrodo-labeled synaptosomes (f; n = 3 wells for −cytoD, n = 1 well for +cytoD) or induction of cell-surface VCAM1 (g; n = 6 wells) by iAstrocytes compared to Li and Krencik astrocytes. cytoD: cytochalasin D. h-i, Induction of VCAM1 expression by IL-1α+TNF+ C1q in primary mouse astrocytes measured by flow cytometry in this study (h; n = 4 wells) or primary mouse astrocytes measured by RNA-seq in Guttenplan et al.10 and Hasel et al.44 (i; n = 3 mice). In panel e, P values were calculated using the two-sided Fisher’s exact test and adjusted for multiple testing using the Benjamini-Hochberg method. In panel and i, P values were calculated using the two-sided Student’s t-test. In panels f-h, P values were calculated using beta regression (two-sided Wald test; see Methods).
Extended Data Fig. 4 sgRNA abundance distribution in CRISPRi screens, comparison of phenotypes from VCAM1 and phagocytosis CRISPRi screens, and validation of selected hits from CRISPRi screens.
a, sgRNA abundance distribution for the CRISPRi screens shown in Fig. 3. b, Gene scores (see Methods) from the phagocytosis vs. VCAM1 CRISPRi screen against transcription factors (left) or the druggable genome (right) in iAstrocytes treated with IL-1α+TNF+ C1q. c-d, Phagocytosis of pHrodo-labeled synaptosomes (c) or induction of cell-surface VCAM1 (d) by iAstrocytes transduced with non-targeting sgRNA (NTC) vs. sgRNAs targeting selected hits from the screens shown in Fig. 3, treated with vehicle control or IL-1α+TNF+ C1q (n = 6 wells for NTC, n = 3 wells for knockdowns). MFI: median fluorescence intensity measured by flow cytometry. In panels c and d, P values were calculated by linear regression (two-sided Wald test; see Methods) and adjusted for multiple testing (Padj; Holm’s method) per family of tests (all comparisons within a plot).
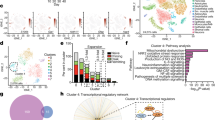
Extended Data Fig. 5 Additional analyses of CROP-seq data.
a, Expression levels of the top cluster markers of non-targeting control (NTC) sgRNA-transduced iAstrocytes shown in Fig. 4a. b-c, Cellular pathway (BioPlanet116) enrichment analysis of Cluster 3 and 4 markers (b) and cell type marker (Descartes117) enrichment analysis of Cluster 5 and 6 markers (c) of NTC sgRNA-transduced iAstrocytes shown in Fig. 4a. P values were calculated using the two-sided Fisher’s exact test and corrected for multiple testing using the Benjamini-Hochberg method. d, The degree of regulator knockdown (left) or the number of differentially expressed genes (DEGs) whose differential expression induced IL-1α+TNF+ C1q is significantly altered by regulator knockdown. e, Hierarchical clustering of the P-value-weighted log-fold-changes (gene score) of the union of knockdown-associated DEGs from panel d; DEGs associated with ABCE1 knockdown were excluded due to a significant number of DEGs also being caused by ABCE1 knockdown in vehicle control-treated iAstrocytes.
Extended Data Fig. 7 C3 and IFIT3 expression and cytokine production in iAstrocytes derived from multiple hiPSC lines.
a, Transcript levels of IFIT3 overlaid onto the UMAP embedding from Fig. 3a. b, Representative immunofluorescence images of C3 and IFIT3 staining (scale bar: 60 μm). c, Percent IFIT3−/C3+, IFIT3+ /C3−, or IFIT3+ /C3+ cells measured by immunofluorescence in iAstrocytes derived from multiple hiPSC lines (WTC11, TCW-1E44, 162D) treated with vehicle control vs. all possible combinations of IL-1α, TNF, and C1q, in the absence (n = 3 wells per condition) or presence of additional IL-6/IL6R chimera (25 ng/mL) or IFN-β (5 ng/mL) added concurrently (n = 4 wells per condition). D, Concentration of IFN-β, IL-6, CXCL10, or GM-CSF in conditioned media from iAstrocytes derived from multiple hiPSC lines (WTC11, 162D) treated with vehicle control vs. all possible combinations of IL-1α, TNF, and C1q (n = 4 wells). For panels c a, P values were calculated using beta regression (two-sided Wald test; see Methods). For panel d, P values were calculated using linear regression (two-sided Wald test; see Methods). P values were adjusted for multiple testing (Padj; Holm’s method) per family of tests (all comparisons within a plot).
Extended Data Fig. 8 Validation of STAT3, CEBPB, NFKB2, and IRF1 knockdown in iAstrocytes derived from multiple hiPSC lines.
a, Percent VCAM1−/C3+, VCAM1+ /C3−, or VCAM1+ /C3+ cells measured by flow cytometry)in iAstrocytes derived from multiple hiPSC lines (TCW-1E44 and 162D) transduced with non-targeting sgRNA (NTC) or sgRNAs targeting STAT3, CEBPB, NFKB2, or IRF1 (n = 6 wells). b, Combined statistical analysis of the effect of STAT3, CEBPB, NFKB2, or IRF1 knockdown compared to NTC in IL-1α+TNF+ C1q-treated iAstrocytes derived from multiple hiPSC lines (WTC11, TCW-1E44, 162D). For panels a and b, P values were calculated using beta regression (two-sided Wald test; see Methods) and adjusted for multiple testing (Padj; Holm’s method) per family of tests (all comparisons within a plot or table).
Extended Data Fig. 9 Effect of small molecule modulators of STAT3 or STAT1/2 activity.
a, Representative immunofluorescence images of phospho-STAT3 (Y705) staining in vehicle control vs. IL-1α+TNF+ C1q-treated iAstrocytes. Scale bar corresponds to 20 μm. b, Phospo-STAT3 (Y705) levels measured by flow cytometry in iAstrocytes treated with vehicle control vs. IL-1α+TNF+ C1q in the presence of increasing doses of napabucasin (n = 6 wells for 0 μM napabucasin, n = 3 for napabucasin > 0 μM). MFI: median fluorescence intensity. c, Percent VCAM1−/C3+, VCAM1+ /C3−, or VCAM1+ /C3+ cells measured by flow cytometry)in iAstrocytes treated with vehicle control vs. IL-1α+TNF+ C1q in the presence of increasing doses of napabucasin (n = 6 wells for 0 μM napabucasin, n = 3 for napabucasin > 0 μM). d, Percent VCAM1−/C3+, VCAM1+ /C3−, or VCAM1+ /C3+ cells measured by flow cytometry in iAstrocytes treated with vehicle control vs. IL-1α+TNF+ C1q, with or without concurrent RGFP966 treatment (n = 6 wells). In panels b-d, P values were calculated using linear regression for MFI values or beta regression (two-sided Wald test; see Methods) for percentages and adjusted for multiple testing (Padj; Holm’s method) per family of tests (all comparisons within a plot).
Extended Data Fig. 10 Overlap analysis of IRAS1 and IRAS2 markers with external datasets.
a-c, Overlap analysis (Fisher’s exact test; see Methods) of differentially expressed genes (DEGs) between IRAS1 vs. IRAS2 with DEGs between IRAS1- and IRAS2-co-clustering astrocytes from Barbar et al.81 (a), Wheeler et al.82 (b), or Hasel et al.44 (c). d, Overlap analysis of DEGs between IRAS1 vs. IRAS2 with DEGs between astrocytes from Stat3 astrocyte-specific conditional knockout (cKO) mice vs. wild-type (WT) mice subject to spinal cord injury (SCI) from Anderson et al.37. e-g, Module expression score (see Methods) of IRAS1 or IRAS2 markers overlaid onto the UMAP embedding of Barbar et al.81 (e), Wheeler et al.82 (f), or Hasel et al.44 (g) astrocytes from Fig. 7d, h, and l, respectively. h, Module expression score of upregulated vs. downregulated DEGs between astrocytes from Stat3 cKO SCI vs. WT SCI mice from Anderson et al.37 overlaid onto the UMAP embedding of iAstrocytes from Fig. 4a. i, Cellular pathway (MSigDB78) and upstream transcription factor (TRRUST79) enrichment analysis of upregulated vs. downregulated DEGs between astrocytes from Stat3 cKO SCI vs. WT SCI mice from Anderson et al.37. For panels a-d and i, P values were calculated using the two-sided Fisher’s exact test and corrected for multiple testing using the Benjamini-Hochberg method.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Supplementary Text
Supplementary Table 1
Bulk RNA-seq differential gene expression analysis results. From this study: iAstrocytes and hiPSC-derived astrocytes generated using the protocols from TCW et al.16, Li et al.17 and Krencik et al.88 treated with vehicle versus IL-1α+TNF+C1q. From external datasets: immunopanned primary mouse astrocytes treated with vehicle versus IL-1α+TNF+C1q from Guttenplan et al.10, hiPSC-derived astrocytes treated with vehicle versus IL-1β from Perriot et al.107 and human cerebral organoid-derived astrocytes treated with vehicle versus IL-1α+TNF+C1q from Barbar et al.18. For data from hiPSC-derived astrocytes in this study, expression was quantified using Salmon, and DEGs were called using DESeq2. For external datasets, the DEGs from Gutteplan et al. were downloaded from the supplementary table included in GSE143598, and raw data from Perriot et al. and Barbar et al. were analyzed with BioJupies106 to call DEGs. P values from DESeq2 or BioJupies were calculated using the two-sided Wald test and corrected for multiple testing using the Benjamini–Hochberg method.
Supplementary Table 2
Phenotype scores and P values from CRISPRi screens. See the first tab of the Excel file for a description of the contents of the remaining tabs and information regarding the columns in each tab. P values were calculated using the two-sided Mann–Whitney U-test.
Supplementary Table 3
Hits from MRA and associated scores and statistics. See Castro et al.26 for interpretation of the activity score. The mean LFC score of a regulon is calculated by averaging the LFC of all DEGs induced by IL-1α+TNF+C1q in that regulon. The P values reported were calculated from two-tailed gene set enrichment analysis as detailed in Castro et al.26 and Campbell et al.28.
Supplementary Table 4
Cluster markers of iAstrocytes in Fig. 4a and differential expression analysis between IRAS1 and IRAS2 iAstrocytes. The column ‘avg_diff’ contains the magnitude of the difference in the mean Pearson residual values of a given gene between cells in the cluster of interest versus all other cells or between IRAS2 (cluster 2) versus IRAS1 (cluster 1) iAstrocytes. The columns ‘pct.1’ and ‘pct.2’ contain the percent of cells expressing the gene of interest in the cluster of interest (‘pct.1’) or all other cells (‘pct.2’) or in IRAS2 (‘pct.1’) iAstrocytes or IRAS1 (‘pct.2’) iAstrocytes. P values were calculated using the two-sided Student’s t-test and adjusted for multiple testing using the Benjamini–Hochberg method.
Supplementary Table 5
Enrichment analysis of IRAS1 and IRAS2 markers. Enrichment results for the MSigDB79 and TRRUST80 libraries were downloaded from Enrichr81. See Chen et al.81 for how the ‘combined score’ is calculated. P values were calculated using the two-sided Fisher’s exact test and adjusted for multiple testing using the Benjamini–Hochberg method.
Supplementary Table 6
Differential gene expression analysis results from knockdown of regulators in CROP-seq experiment. The results shown correspond to the interaction term between cytokine treatment and regulator knockdown—that is, they reflect how regulator knockdown affects the change in gene expression induced by IL-1α+TNF+C1q. See the first tab of the Excel file for the number of cells recovered for each knockdown. P values were calculated using the two-sided Wald test and adjusted for multiple testing using the Benjamini–Hochberg method.
Supplementary Table 7
Gene modules and enrichment analysis of gene modules from Extended Data Fig. 4d. Enrichment results for the MSigDB79 and TRRUST80 libraries were downloaded from Enrichr81. See Chen et al.81 for how the ‘combined score’ is calculated.
Supplementary Table 8
Cluster markers and DEGs from integrated analysis of iAstrocytes with Barbar et al., Wheeler et al. or Hasel et al. astrocytes. Cluster markers and DEGs are derived using only astrocytes from Barbar et al.18, Wheeler et al.104 or Hasel et al.45. The column ‘avg_diff’ contains the magnitude of the difference in the mean Pearson residual values of a given gene between cells in the cluster of interest versus all other cells or between two clusters of interest (the first cluster minus the second cluster). The columns ‘pct.1’ and ‘pct.2’ contain the percent of cells expressing the gene of interest in the cluster of interest (‘pct.1’) or all other cells (‘pct.2’) or the two clusters of interest. P values were calculated using the two-sided Student’s t-test and adjusted for multiple testing using the Benjamini–Hochberg method.
Supplementary Table 9
Differential gene expression analysis between Stat3 cKO SCI versus WT SCI astrocyte transcriptomes from Anderson et al. and enrichment analysis of DEGs. The raw data from Anderson et al.38 were analyzed with BioJupies106 to call DEGs between Stat3 cKO SCI versus WT SCI astrocyte transcriptomes. Enrichment results for the MSigDB79 and TRRUST80 libraries were downloaded from Enrichr81. See Chen et al.81 for how the ‘combined score’ is calculated. P values were calculated using the two-sided Fisher’s exact test and adjusted for multiple testing using the Benjamini–Hochberg method.
Supplementary Table 10
Clinical metadata of human neuropathology samples. For the AD cohort, Braak stage is provided for each sample. Additional neuropathology scoring (for example, Thal and CERAD scores) are provided if available.
Supplementary Table 11
Metadata of astrocyte bulk RNA-seq samples used for co-expression network reconstruction in MRA. Samples from publicly available datasets can be cross-referenced with the GEO using their GSM accession numbers.
Supplementary Table 12
sgRNA information for CRISPRi libraries. sgRNA_ID, a unique ID for each sgRNA that includes the gene target, orientation, genomic coordinate and the TSS targeted. Gene_target, the gene targeted by the sgRNA. TSS, the transcriptional start site targeted by the sgRNA. sgRNA_sequence, the protospacer sequence of the sgRNA.
Supplementary Table 13
Information on mice used for immunostaining
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Leng, K., Rose, I.V.L., Kim, H. et al. CRISPRi screens in human iPSC-derived astrocytes elucidate regulators of distinct inflammatory reactive states. Nat Neurosci 25, 1528–1542 (2022). https://doi.org/10.1038/s41593-022-01180-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01180-9
This article is cited by
-
A phenotypic screening platform for identifying chemical modulators of astrocyte reactivity
Nature Neuroscience (2024)
-
Induced pluripotent stem cells (iPSCs): molecular mechanisms of induction and applications
Signal Transduction and Targeted Therapy (2024)
-
Functional roles of reactive astrocytes in neuroinflammation and neurodegeneration
Nature Reviews Neurology (2023)
-
Neural Differentiation and spinal cord organoid generation from induced pluripotent stem cells (iPSCs) for ALS modelling and inflammatory screening
Molecular Neurobiology (2023)
-
Towards elucidating disease-relevant states of neurons and glia by CRISPR-based functional genomics
Genome Medicine (2022)