Abstract
Liver fibrosis is an urgent clinical problem without effective therapies. Here we conducted a high-content screening on a natural Euphorbiaceae diterpenoid library to identify a potent anti-liver fibrosis lead, 12-deoxyphorbol 13-palmitate (DP). Leveraging a photo-affinity labeling approach, apolipoprotein L2 (APOL2), an endoplasmic reticulum (ER)-rich protein, was identified as the direct target of DP. Mechanistically, APOL2 is induced in activated hepatic stellate cells upon transforming growth factor-β1 (TGF-β1) stimulation, which then binds to sarcoplasmic/ER calcium ATPase 2 (SERCA2) to trigger ER stress and elevate its downstream protein kinase R-like ER kinase (PERK)–hairy and enhancer of split 1 (HES1) axis, ultimately promoting liver fibrosis. As a result, targeting APOL2 by DP or ablation of APOL2 significantly impairs APOL2–SERCA2–PERK–HES1 signaling and mitigates fibrosis progression. Our findings not only define APOL2 as a novel therapeutic target for liver fibrosis but also highlight DP as a promising lead for treatment of this symptom.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA sequencing data can be retrieved using accession number GSE250087. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with dataset identifiers PXD047924 and PXD047815. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Higashi, T., Friedman, S. L. & Hoshida, Y. Hepatic stellate cells as key target in liver fibrosis. Adv. Drug Deliv. Rev. 121, 27–42 (2017).
Kisseleva, T. & Brenner, D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 18, 151–166 (2021).
Zhao, M. Y. et al. Targeting fibrosis, mechanisms and cilinical trials. Signal Transduct. Target. Ther. 7, 206 (2022).
Schuppan, D., Ashfaq-Khan, M., Yang, A. T. & Kim, Y. O. Liver fibrosis: direct antifibrotic agents and targeted therapies. Matrix Biol. 68-69, 435–451 (2018).
Lee, Y. A., Wallace, M. C. & Friedman, S. L. Pathobiology of liver fibrosis: a translational success story. Gut 64, 830–841 (2015).
Henderson, N. C., Rieder, F. & Wynn, T. A. Fibrosis: from mechanisms to medicines. Nature 587, 555–566 (2020).
Benesic, A., Jalal, K. & Gerbes, A. L. Acute liver failure during pirfenidone treatment triggered by co-medication with esomeprazole. Hepatology 70, 1869–1871 (2019).
Verma, N. et al. Drug idiosyncrasy due to pirfenidone presenting as acute liver failure: case report and mini-review of the literature. Hepatol. Commun. 2, 142–147 (2018).
Atanasov, A. G., Zotchev, S. B., Dirsch, V. M., Supuran, C. T. & Taskforce, I. N. P. S. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 20, 200–216 (2021).
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803 (2020).
Li, Y. N. et al. Existing knowledge on Euphorbia fischeriana Steud. (Euphorbiaceae): traditional uses, clinical applications, phytochemistry, pharmacology and toxicology. J. Ethnopharmacol. 275, 114095 (2021).
Zhan, Z. J., Li, S., Chu, W. & Yin, S. Euphorbia diterpenoids: isolation, structure, bioactivity, biosynthesis, and synthesis (2013–2021). Nat. Prod. Rep. 39, 2132–2174 (2022).
Huang, J. L. et al. Discovery of highly potent daphnane diterpenoids uncovers importin-β1 as a druggable vulnerability in castration-resistant prostate cancer. J. Am. Chem. Soc. 144, 17522–17532 (2022).
Yan, X. L. et al. Discovery of the first Raptor (regulatory-associated protein of mTOR) inhibitor as a new type of antiadipogenic agent. J. Med. Chem. 66, 5839–5858 (2023).
Hu, X. R. et al. A natural small molecule mitigates kidney fibrosis by targeting Cdc42-mediated GSK-3β/β-catenin signaling. Adv. Sci. (Weinh.) 11, e23078501 (2024).
Altrock, E. et al. Inhibition of fibronectin deposition improves experimental liver fibrosis. J. Hepatol. 62, 625–633 (2015).
Nevzorova, Y. A., Boyer-Diaz, Z., Cubero, F. J. & Gracia-Sancho, J. Animal models for liver disease—a practical approach for translational research. J. Hepatol. 73, 423–440 (2020).
Tschaikowsky, M. et al. The articular cartilage surface is impaired by a loss of thick collagen fibers and formation of type I collagen in early osteoarthritis. Acta Biomater. 146, 274–283 (2022).
Xu, H. Y. et al. 12-Deoxyphorbol 13-palmitate inhibit VEGF-induced angiogenesis via suppression of VEGFR-2-signaling pathway. J. Ethnopharmacol. 146, 724–733 (2013).
Xu, H. Y. et al. 12-Deoxyphorbol 13-palmitate mediated cell growth inhibition, G2-M cell cycle arrest and apoptosis in BGC823 cells. Eur. J. Pharmacol. 700, 13–22 (2013).
Pan, S. J., Zhang, H. L., Wang, C. Y., Yao, S. C. L. & Yao, S. Q. Target identification of natural products and bioactive compounds using affinity-based probes. Nat. Prod. Rep. 33, 612–620 (2016).
Bunnage, M. E., Gilbert, A. M., Jones, L. H. & Hett, E. C. Know your target, know your molecule. Nat. Chem. Biol. 11, 368–372 (2015).
Seneviratne, U. et al. Photoaffinity labeling and quantitative chemical proteomics identify LXRβ as the functional target of enhancers of astrocytic apoE. Cell Chem. Biol. 28, 148–157 (2021).
Scales, S. J. et al. Apolipoprotein L1-specific antibodies detect endogenous APOL1 inside the endoplasmic reticulum and on the plasma membrane of podocytes. J. Am. Soc. Nephrol. 31, 2044–2064 (2020).
Molina, D. M. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Pant, J. et al. Apolipoproteins L1-6 share key cation channel-regulating residues but have different membrane insertion and ion conductance properties. J. Biol. Chem. 297, 100951 (2021).
Pays, E. The function of apolipoproteins L (APOLs): relevance for kidney disease, neurotransmission disorders, cancer and viral infection. FEBS J. 288, 360–381 (2021).
Meng, X. M., Nikolic-Paterson, D. J. & Lan, H. Y. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338 (2016).
Fu, S. N. et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531 (2011).
Kao, E., Shinohara, M., Feng, M., Lau, M. Y. & Ji, C. Human immunodeficiency virus protease inhibitors modulate Ca2+ homeostasis and potentiate alcoholic stress and injury in mice and primary mouse and human hepatocytes. Hepatology 56, 594–604 (2012).
Raffaello, A., Mammucari, C., Gherardi, G. & Rizzuto, R. Calcium at the center of cell signaling: interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem. Sci. 41, 1035–1049 (2016).
Liu, Z. K. et al. Transforming growth factor (TGF) cross-talk with the unfolded protein response is critical for hepatic stellate cell activation. J. Biol. Chem. 294, 3137–3151 (2019).
Ohyama, T. et al. Azelnidipine is a calcium blocker that attenuates liver fibrosis and may increase antioxidant defence. Br. J. Pharmacol. 165, 1173–1187 (2012).
Zhang, K. et al. Hes1, an important gene for activation of hepatic stellate cells, is regulated by Notch1 and TGF-β/BMP signaling. World J. Gastroenterol. 21, 878–887 (2015).
Szabat, M. et al. Musashi expression in β-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2, e232 (2011).
Lee, J. E., Morrison, W. & Hollien, J. Hairy and enhancer of split 1 (HES1) protects cells from endoplasmic reticulum stress-induced apoptosis through repression of GADD34. J. Biol. Chem. 293, 5947–5955 (2018).
Johnson, K. et al. A stem cell-based approach to cartilage repair. Science 336, 717–721 (2012).
Kakeya, H. Natural products-prompted chemical biology: phenotypic screening and a new platform for target identification. Nat. Prod. Rep. 33, 648–654 (2016).
Takahashi, S. et al. Association of SNPs and haplotypes in APOL1, 2 and 4 with schizophrenia. Schizophr. Res. 104, 153–164 (2008).
Liao, W. et al. A novel anti-apoptotic role for apolipoprotein L2 in IFN-γ-induced cytotoxicity in human bronchial epithelial cells. J. Cell. Physiol. 226, 397–406 (2011).
Amir, A. et al. Endoplasmic reticulum stress in liver diseases. Hepatology 77, 619–639 (2023).
Koo, J. H. et al. Endoplasmic reticulum stress in hepatic stellate cells promotes liver fibrosis via PERK-mediated degradation of HNRNPA1 and up-regulation of SMAD2. Gastroenterology 150, 181–193 (2016).
Loeuillard, E. et al. Endoplasmic reticulum stress induces inverse regulations of major functions in portal myofibroblasts during liver fibrosis progression. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3688–3696 (2018).
Akhurst, R. J. & Hata, A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 (2012).
Mederacke, I., Dapito, D. H., Affò, S., Uchinami, H. & Schwabe, R. F. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 (2015).
Ravi, M., Paramesh, V., Kaviya, S. R., Anuradha, E. & Solomon, F. D. P. 3D cell culture systems: advantages and applications. J. Cell. Physiol. 230, 16–26 (2015).
Seneviratne, U. et al. Photoaffinity labeling and quantitative chemical proteomics identify LXRβ as the functional target of enhancers of astrocytic apoE. Cell. Chem. Biol. 28, 148–157 (2021).
Yang, B. F. et al. Small molecule RL71 targets SERCA2 at a novel site in the treatment of human colorectal cancer. Oncotarget 6, 37622–37634 (2015).
Acknowledgements
This work was supported by the Open Program of Shenzhen Bay Laboratory (SZBL2021080601007 to S.Y.); the National Natural Science Foundation of China (82273804 to S.Y. and 32070767 to J.G.); the Guangdong Basic and Applied Basic Research Foundation, China (2021B1515140062 to S.Y.); the Science and Technology Program of Guangzhou, China (2024B03J1322 to S.Y.); and the Science and Technology Planning Project of Guangdong Province, China (2023A1111120025 to S.Y.). Cartoons in the graphical abstract were created with BioRender.
Author information
Authors and Affiliations
Contributions
S.Y., J.G., X.D.L. and Y.J. conceived and designed this study. L.G., Q.J. and J.H. developed the methods. L.G., Q.J., X.W. and X.Z. acquired the data (western blot, qRT–PCR and immunohistochemistry analysis, RNA-seq analysis and pulldown experiment analysis). D.H. and G.T. performed the chemical work (extraction and separation of compounds, structural modification and probe synthesis). L.G., Q.J., X.W., X.Z., L.W., W.X. and R.F. analyzed and interpreted the data (statistical analysis, biostatistics and computational analysis). S.Y., J.G. and L.G. wrote the paper. S.Y., J.G., X.D.L. and G.T. provided administrative, technical or material support (for example, reporting or organizing data). S.Y., J.G., X.D.L. and Y.J. were responsible for research supervision. All authors approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Peer review
Peer review information
Nature Chemical Biology thanks Sang Geon Kim, Zhengqiu Li and Jessica Maiers for their contributions to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
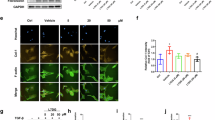
Extended Data Fig. 1 Discovery of DP as a potent inhibitor of HSCs activation.
a, 11 different skeletal types in Euphorbiaceae diterpenoid library and the sample size of each skeleton. b, Representative fluorescent photographs of immunofluorescence staining for fibronectin (FN) in TGF-β1-stimulated LX-2 cells treated with DMSO (CTR), compounds (10 μM) or PFD (500 μM) for 48 h (left). Quantification for fluorescence intensity of fibronectin in indicated groups (right). Scale bar, 100 μm. UND, untreated. Data presented are means ± s.d. c, Representative western blot for fibrotic markers in 10 ng/mL PDGF, 10 μM Angiotensin II (Ang II), 10 ng/mL CTGF or 1 μg/mL lipopolysaccharide (LPS) stimulated LX-2 cells treated with DP (10 μM) for 48 h. d, Representative western blot for FN, collagen I, and α-SMA in TGF-β1-stimulated NRK-49F and MRC-5 cells treated with indicated concentrations of DP or PFD for 48 h. e, IC50 curves of DP in different cell lines treated for 48 h. CI, confidence interval. For b−e, n = 3 independent experiments.
Extended Data Fig. 2 Toxicity evaluation on DP in mouse models.
a, b, Representative images of mouse kidney, lung, heart and spleen tissues stained with H&E in indicated groups. Scale bar, 250 μm. n = 5 independent mice. c, Representative images of mouse articulatio genus stained with H&E in indicated groups. Scale bar, 250 μm. n = 3 independent mice.
Extended Data Fig. 3 DP alleviates DDC-induced mouse liver fibrosis.
a, Representative images of mouse liver tissues stained with H&E, Sirus Red, Masson and α-SMA in indicated groups. Scale bar, 250 μm. n = 5 independent mice. b, Levels of ALP, TBIL, UREA and CREA in serum from mice in indicated groups. n = 7 independent mice. Data presented are means ± s.e.m. Statistical significance was determined by two-tailed unpaired Student’s t-test (ALP and TBIL) or one-way ANOVA followed by Dunnett’s multiple comparisons test (UREA and CREA). n.s., no significance. c, Representative western blot for fibrotic markers in indicated groups. n = 3 independent experiments.
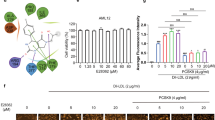
Extended Data Fig. 4 DP directly targets APOL2.
a, Representative western blot for fibrotic markers in TGF-β1-stimulated LX-2 cells treated with indicated concentrations of DP-PT (left) or DP-BOD (right) for 48 h. b, SDS-PAGE gels of fluorescence scanning imaging and coomassie brilliant blue (CBB) staining in vitro. c, Western blot analysis on endogenous APOL2 protein after DP-PT pull-down in LX-2 cell lysis. d, Western blot analysis on exogenous APOL2 protein after DP-PT pull-down in 293 T cells. 293 T cells were transfected with V5-tagged APOL2 for 6 h. After 24 h, the cells were incubated with indicated concentrations of DP-PT in the presence or absence of DP for 3 h. After photocross-linking, the probe-bound protein was precipitated by streptavidin beads and was immunoblotted with an antibody against V5. e, Western blot analysis on endogenous APOL1 protein after DP-PT pull-down in LX-2 cells. f, The influence of DP on APOL2 protein levels in LX-2 cells. g, Quantification for APOL2 protein levels in the cellular thermal shift assay. Data presented are means ± s.d. Statistical significance was determined by two-tailed unpaired Student’s t-test compared with untreated (UND) group. For a−g, n = 3 independent experiments.
Extended Data Fig. 5 Identification of modification sites by LC-MS/MS.
a, b, Recombinant APOL2 was incubated with DP-PT and was then successively subjected to photocross-linking. The labeled protein was digested with trypsin and analyzed by LC-MS/MS. The data was processed in Proteome Discoverer 2.2.0. c. Binding modes in the top ten scoring rankings between DP and APOL2 according to docking simulation using Schrodinger software. The different conformations of DP were labeled as blue, the amino acid residues involved in covalent interactions were labeled as green, the representative amino acid residues involved in interactions were labeled as yellow. d, Pull-down experiments of DP-PT on different APOL2 mutants in 293 T cells. V199/G200A: Val199 and Gly200 mutate into Ala; N212/V216A: Asn212 and Val216 mutate into Ala; L209A: Leu209 mutates into Ala; I227A: Ile227 mutates into Ala; I246A: Ile246 mutates into Ala. EV, empty vehicle. n = 3 independent experiments.
Extended Data Fig. 6 APOL2 overexpressed in activated HSCs is required for the progression of liver fibrosis and the antifibrotic effects of DP is APOL2 dependent.
a, APOL2 transcript expression analysis of GEO database. b, mRNA levels of Apol2 in mouse liver tissues. c, Representative western blot for APOL2 in CTGF-, Ang II-, LPS- or PDGF-stimulated LX-2 cells. d, Representative fluorescent photographs for co-staining of APOL2 with albumin, CD31, or F4/80 in mouse liver tissues. Scale bar, 50 μm. e,f, LX-2 cells were transfected with siAPOL2-1/2/3 (e) or APOL2 plasmid (f) for 6 h and subsequently treated with or without TGF-β1 or different concentrations of DP for 48 h. The indicated protein expressions were detected. g, Levels of mouse serum UREA and CREA in indicated groups. h, Representative images of mouse kidney, lung, heart, and spleen tissues stained with H&E in indicated groups. Scale bar, 250 μm. i−l, LX-2 cells were transfected with siAPOL2-1, APOL2 or APOL22A plasmid for 6 h and subsequently treated with or without TGF-β1 or different concentrations of DP for 48 h. The indicated protein expressions were detected. For c, d, e, f and i−l, n = 3 independent experiments. For b, g and h, n = 5 independent mice. For a, b and g, data presented are means ± s.e.m. Statistical significance was determined by two-tailed unpaired Student’s t-test (a,b) or one-way ANOVA followed by Dunnett’s multiple comparisons test (g).
Extended Data Fig. 7 DP alleviates liver fibrosis via the PERK-HES1 axis.
a, LX-2 cells were treated with indicated concentrations of DP (left) or transfected with 80 nM siAPOL2-1/2 or siControl (right) for 6 h and subsequently treated with or without TGF-β1 for 3 h. The protein expressions of total Smad2/3, phosphorylated Smad2/3 were determined by immunoblotting. b. The coomassie brilliant blue staining of interacting proteins of APOL2. c, Colocalization analysis of APOL2 and SERCA2. Plots of pixel intensity along the white line from left to the right of each plot, colors as in merged images. d, Immunofluorescence assay of APOL2 (red) and α-SMA (green) in indicated groups. Scale bar, 15 μm. e, Representative western blot for indicated proteins in LX-2 cells treated with DP (5 μM) at different time points. f, Representative western blot for indicated proteins in LX-2 cells transfected with siAPOL2-1/2 or siControl for 6 h and subsequently treated with TGF-β1 for 48 h. Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown in a plot graph below each blot. For a, b and d−f, n = 3 independent experiments.
Extended Data Fig. 8 The anti-ER stress efficacy of DP is APOL2 dependent.
a, LX-2 cells were transfected with siAPOL2-1 or siControl for 6 h and subsequently treated with TGF-β1 with or without different concentrations of DP for 48 h. The indicated protein expressions were detected. b, LX-2 cells were transfected with APOL2 or vector for 6 h and subsequently treated with or without DP (1 μM) for 48 h. The indicated protein expressions were detected. c, LX-2 cells were transfected with APOL22A or vector for 6 h and subsequently treated with or without DP (1 μM) for 48 h. The indicated protein expressions were detected. d, LX-2 cells were transfected with APOL2WT or APOL22A for 6 h and subsequently treated with or without DP (1 μM) for 48 h. The indicated protein expressions were detected. Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown in a plot graph below each blot. For a−d, n = 3 independent experiments.
Extended Data Fig. 9 The anti-fibrotic efficacy of DP or APOL2 loss was exerted via PERK-HES1 signaling.
a, Representative western blot for indicated proteins in TGF-β1-stimulated LX-2 cells treated with indicated concentrations of GSK2656157 (GSK) for 48 h. b, LX-2 cells were transfected with sieIF2α or siATF4 for 6 h and subsequently treated with or without indicated concentration of DP for 48 h. The indicated protein expressions were detected. c, LX-2 cells were transfected with siAPOL2-1 for 6 h and subsequently treated with or without sieIF2α or siATF4 for 6 h. After further cultivation for 48 h, the indicated protein expressions were detected. d, Representative western blot for indicated proteins in TGF-β1-stimulated LX-2 treated with indicated concentrations of DP with or without 10 μM GSK for 48 h. Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown in a plot graph below each blot. For a−d, n = 3 independent experiments.
Extended Data Fig. 10 DP treatment or Apol2 depletion inhibit the PERK-HES1 signaling in vivo.
a,b, Representative western blot for HES1, ATF4, PERK, p-PERK, eIF2α, p-eIF2α and APOL2 in (a) CCl4- or (b) DDC-induced mouse fibrotic liver tissues in indicated groups. c, Representative western blot for HES1, ATF4, PERK, p-PERK, eIF2α, p-eIF2α and APOL2 of mouse liver tissues in indicated groups. Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown in a plot graph below each blot. For a−c, n = 3 independent experiments.
Supplementary information
Supplementary Table 1
Antibodies for immunoblotting
Supplementary Table 2
Primers for qPCR
Supplementary Table 3
List of quantified proteins with abundance ratio from LX-2 cells
Supplementary Table 4
Quantification for α-SMA and APOL2 immunostaining in human liver tissues
Supplementary Table 5
List of proteins that interact with APOL2 in LX-2 cells
Supplementary Table 6
RNA-seq data in LX-2 cells treated with DP
Supplementary Table 7
Statistical source data for immunoblot quantitation
Supplementary Data 1
Raw figures for IHC or H&E photos
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots and gels.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots and gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots and gels.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gan, L., Jiang, Q., Huang, D. et al. A natural small molecule alleviates liver fibrosis by targeting apolipoprotein L2. Nat Chem Biol (2024). https://doi.org/10.1038/s41589-024-01704-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41589-024-01704-3