Abstract
Cultivated peanut (Arachis hypogaea L.) is a widely grown oilseed crop worldwide; however, the events leading to its origin and diversification are not fully understood. Here by combining chloroplast and whole-genome sequence data from a large germplasm collection, we show that the two subspecies of A. hypogaea (hypogaea and fastigiata) likely arose from distinct allopolyploidization and domestication events. Peanut genetic clusters were then differentiated in relation to dissemination routes and breeding efforts. A combination of linkage mapping and genome-wide association studies allowed us to characterize genes and genomic regions related to main peanut morpho-agronomic traits, namely flowering pattern, inner tegument color, growth habit, pod/seed weight and oil content. Together, our findings shed light on the evolutionary history and phenotypic diversification of peanuts and might be of broad interest to plant breeders.
Similar content being viewed by others
Main
Cultivated peanut or groundnut (Arachis hypogaea L.) is a sustainable and affordable source of edible oil and proteins, which globally yields 54 million tons from a cultivated area of 32 million ha (http://www.fao.org/faostat, 2020). Its allotetraploid nature (genome AABB, size ~2.7 Gb) is thought to arise from the polyploidization of an interspecific hybrid between 2 of 81 wild species, currently described in the genus Arachis—Arachis duranensis Krapov. and W.C. Gregory (genome AA, size ~1.25 Gb, female parent) and Arachis ipaënsis Krapov. and W.C. Gregory (genome BB, size ~1.56 Gb, male parent)1,2.
A. hypogaea is commonly assumed to be domesticated from the wild tetraploid progenitor Arachis monticola, most probably in a region now encompassing part of southern Bolivia and northern Argentina1,3,4,5. The first archeological evidence of peanut cultivation traces back to 7,600 years ago6. In the 16th century, peanut cultivation diffused from South America through the Portuguese and the Spanish explorers7. Further migration routes from North America to Northern China and from South Asia to Southern China have been recently inferred from genetic data8. Nowadays, peanut is grown in more than 100 countries, with China being the first for production and India the first for the cultivated area.
A. hypogaea is a self-pollinating species characterized by low levels of genetic variation resulting from a series of domestication bottlenecks9,10; nonetheless, it displays large morphological variation. The absence or presence of flowers on the main axis and the flowering pattern, alternate or sequential, are at the basis of the classification of A. hypogaea in two subspecies, A. hypogaea subsp. hypogaea (Ahh) and A. hypogaea subsp. fastigiata (Ahf)11. Additional traits led to the distinction of two botanical varieties within Ahh (var. hypogaea and var. hirsuta) and four within Ahf (var. fastigiata, var. vulgaris, var. aequatoriana and var. peruviana)11. Breeding resulted in hybridization among these taxa and thus irregular morphologies. Today, a widely used peanut classification is in accordance with five main market types (Virginia, Runner, Peruvian Runner, Valencia and Spanish)12. Analysis of genetic structure resulted in clustering patterns approximately in accordance with both classifications13,14.
Recently, the International Peanut Genome Initiative and two research groups announced the release of cultivated peanut genome assemblies5,15,16, thus paving the way for in-depth exploration of peanut genetic diversity. Here aiming to define the genetic structure and evolutionary history of peanuts, we performed chloroplast and whole-genome sequencing of peanut accessions belonging to a global peanut collection, encompassing 18 diploid Arachis species, A. monticola and A. hypogaea. Mapping approaches, based on genome-wide association study (GWAS) and recombinant inbred line (RIL) population linkage analysis, were followed to identify candidate genes and genomic regions associated with peanut diversification, domestication and breeding.
Results
Sequencing and genotyping
Chloroplast de novo sequencing was performed on 36 wild Arachis accessions (34 from diploid wild species and 2 from the tetraploid species A. monticola) and a selection of 77 cultivated accessions that, based on the United States of Department of Agriculture (USDA) taxonomic descriptors17, could be unambiguously assigned to A. hypogaea subspecies and botanical varieties (Supplementary Tables 1 and 2). The length of the assembled chloroplast genomes ranged between 156,258 bp and 160,366 bp (Supplementary Table 3). In total, 1,884 polymorphisms (both SNPs and insertions/deletions (InDels)) were found between the 113 assembled chloroplast genomes. Most of the polymorphic sites occurred between wild and cultivated peanuts, whereas 14 polymorphisms were found within A. hypogaea (Supplementary Table 4). Eight additional polymorphic sites were found in a panel including, besides A. hypogaea, accessions representing six wild species of the AA genome section (Supplementary Table 4). Sanger sequencing and/or kompetitive allele-specific PCR (KASP) assays18 allowed the validation of five randomly chosen chloroplast polymorphisms detected by de novo sequencing (Supplementary Fig. 1 and Supplementary Table 5). As an independent approach to revealing chloroplast DNA polymorphisms, the 113 assembled chloroplast genomes were processed to identify mononucleotide repeat (MNR) loci, representing the most frequent class of microsatellite loci in chloroplast genomes19,20,21,22,23. On average, 10,515 MNR loci were detected across the analyzed genomes (Supplementary Table 6).
Whole-genome resequencing (WGR) was performed on 11 A. duranensis, 1 A. ipaensis, 2 A. monticola and 353 A. hypogaea accessions originating from different countries (Fig. 1a and Supplementary Tables 1 and 2), resulting in 160.46 billion reads and 14.54 terabase pairs of clean data. Following alignment against the peanut cv. Tifrunner genome assembly15, unique mapped reads of the 355 tetraploid A. hypogaea accessions were associated with 29.00× mean depth and 88.12% genome coverage (Supplementary Table 7). No significant difference was found between the two unique mapped read rates associated with accessions assigned to Ahh (88.087%) and Ahf (88.220%; Supplementary Table 8). In total, 864,179 SNPs and 71,052 InDels were obtained after quality control. About 40% of the variants were located on the first ten chromosomes (corresponding to the A subgenome), resulting in one variant every 3 kb on average, while 60% of the variants were located on the last ten chromosomes (the B subgenome), resulting in one variant every 2.6 kb on average. The application of KASP assays to a panel of 30 SNP loci and 10,650 data points resulted in the validation of 97.5% of the SNP calls (Supplementary Tables 9 and 10).
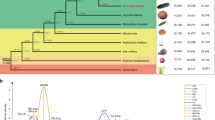
a, Geographic distribution of 355 Arachis accessions resequenced in this study. The color proportion of the circle is proportional to the number of accessions of different types. The map was generated using the mapPies function in the freely available rworldmap R package. b, Chloroplast phylogeny obtained by de novo sequencing of 36 wild Arachis species and 77 primitive landraces assigned to A. hypogaea subspecies and botanical varieties. c,d, Results of hierarchical clustering (c) and PCA (d) from WGR of the same tetraploid accessions in b. e–j, Polymorphic sites between (PB) and across (PA) two groups of individuals randomly sampled—one from Ahh and one from Ahf (e,f), both from Ahh (g,h) and both from Ahf (i,j). Results refer to 100 bootstrap replications and are presented separately for the A. hypogaea A genome (chromosomes 1–10) and B genome (chromosomes 11–20). k, Extent of LD decay in different A. hypogaea botanical varieties and types. l,m, PCA (l) and parametric clustering (m) of the 355 Arachis accessions resequenced in this study. mon, monticola; hyp, hypogaea; fas, fastigiata; per, peruviana; vul, vulgaris; hir, hirsuta.
The evolutionary history and genetic structure of peanuts
Chloroplast genomes are maternally inherited; therefore, chloroplast DNA sequences are widely used to infer maternal lineage(s), leading to the origin of allopolyploids24,25. Phylogenesis based on chloroplast genome SNPs and InDels indicated A. duranensis as the last wild species to diverge before A. hypogaea, in accordance with previous studies suggesting A. duranensis as the donor of the A. hypogaea maternal genome26. Remarkably, three A. duranensis accessions (PI219823, PI468201 and PI468202), together with one A. archeri accession (PI604844) previously shown to be most likely a misclassified A. duranensis27, were included with maximum bootstrap support in a phylogenetic clade-specific for Ahh, except for the Ahf accessions N524 and N530 (Fig. 1b). Pedigree notes indicated that N524, which was classified as Ahf based on morphologic traits, indeed inherited an Ahh chloroplast genome (Supplementary Fig. 2). Clustering based on MNR loci also confirmed the presence of two A. duranensis accessions (PI219823 and PI475883) in a clade mostly referable to Ahh (Supplementary Fig. 3). Both SNP/InDel and MNR-based phylogeneses also provided strong bootstrap support for the occurrence of a clade referable to Ahf germplasm, except for N496 (Fig. 1b and Supplementary Fig. 3). Overall, the clear-cut phylogenetic divergence between Ahh and Ahf, together with grouping of several A. duranensis accessions in the Ahh intraspecific clade, strongly indicate that different A. duranensis mother lineages, and thus allopolyploidization events, originated Ahh and Ahf.
Although A. monticola is thought to be the wild progenitor of A. hypogaea, the two A. monticola accessions genotyped in this study diverged after the split between Ahh and Ahf, as they clustered with Ahh in the chloroplast phylogenesis (Fig. 1b and Supplementary Fig. 3). This suggests that these two accessions are indeed feral forms originating from Ahh hybridization. Further studies, considering more accessions classified as A. monticola, might clarify the position of this species in the evolutionary history of peanuts.
Nuclear polymorphism data from the same tetraploid accessions used for chloroplast phylogenesis were also subjected to genetic structure analysis. Principal components analysis (PCA) and maximum likelihood hierarchical clustering provided further support for the clear-cut differentiation between the two A. hypogaea subspecies and, within Ahf, the botanical varieties fastigiata, vulgaris and peruviana (Fig. 1c,d).
Two additional nuclear trees were obtained for the A and B genomes (Supplementary Fig. 4). Inconsistencies between A genome hierarchical clustering (Supplementary Fig. 4a) and the chloroplast genome phylogenesis (Fig. 1b) can be explained by recombination between homeologous chromosomes, with this event being very common in angiosperm polyploids28,29. This would have caused the fixation of DNA segments from the B paternal genome in chromosomes 1–10 of cultivated peanuts. Significantly, homeologous chromosomal rearrangements were reported in A. hypogaea, which changed the genomic formula of specific chromosomal regions from the expected AABB to AAAA or BBBB15,30. In addition, misassemblies of homeologous regions in the reference genome might also affect nuclear phylogenesis. With this respect, the newly released Tifrunner v2 assembly reports several changes in correspondence of homeologous regions. Finally, inconsistencies between nuclear and chloroplast tree topologies have been commonly observed in plants31 and among nuclear peanut phylogenies32.
The analysis of genomic SNP distribution provided further evidence that different allopolyploids originated Ahh and Ahf. Indeed, bootstrap sampling of groups of individuals from Ahh and Ahf revealed a large excess of polymorphisms between groups (PB) compared with polymorphisms shared across groups (PA), in accordance with a scenario in which alleles that were polymorphic between different tetraploid progenitors were fixed in Ahh and Ahf (Fig. 1e,f). In contrast, sampling of group pairs within the same subspecies yielded opposite results (PB ≪ PA; Fig. 1g–j), in agreement with their descendance from a common tetraploid progenitor. With a few exceptions, we found a roughly even distribution of the PA and PB polymorphism classes in the genome (Supplementary Fig. 5).
Linkage disequilibrium (LD) decay significantly varied within A. hypogaea, as it was slower in var. hirsuta and hypogaea than in var. fastigiata and vulgaris (Fig. 1k), which is consistent with the lower level of genetic diversity found in var. hirsuta and var. hypogaea (Supplementary Fig. 6). The half-maximum decay distance was 99.4 kb within var. hypogaea, 174.5 kb within var. hirsuta, 5.6 kb within var. fastigiata and 15.8 kb within var. vulgaris.
To identify genomic regions that are highly divergent between the peanut subspecies Ahh and Ahf, thus contributing to their diversification, we estimated haplotypes and found specific haplotypes distinguishing the botanical varieties (Supplementary Fig. 7).
The effect of the recent breeding history on peanut genetic structure was investigated using the whole panel of 355 accessions sequenced in this study, also including cultivars derived from hybridization breeding programs. Parametric modeling, PCA and hierarchical clustering (Fig. 1l,m and Supplementary Table 11) defined additional levels of population stratification. In more detail, within var. hypogaea, one cluster was associated with several Chinese landraces (Cls8) and one (Cls1) with American varieties or derivatives. Within var. vulgaris, distinct clusters were found for African landraces (Cls6), Chinese landraces (Cls2) and cultivars from southern China (Cls7). Cls9 was found mainly for var. fastigiata. Finally, five clusters (Cls3, Cls5, Cls10, Cls11 and Cls12) were found for irregular-type peanuts, originating from hybridization between the two A. hypogaea subspecies, with Cls3 and Cls5 being morphologically more similar to Ahh and Ahf, respectively.
Genes associated with divergence between peanut subspecies
Different evolutionary histories of the peanut subspecies Ahh and Ahf were accompanied by the fixation of contrasting phenotypes for several traits, including the flowering pattern, the number of branches, the growth habit and the color of the inner seed tegument. The flowering pattern, sequential in Ahf and alternate in Ahh (Fig. 2a,b), is thought to have a major role in the adaptation to different ecosystems. Mapping this trait by two RIL populations originating from different parental lines identified, in one case, a major Quantitative Trait Locus (QTL) at the end of chromosome 12 and, in the other, two QTLs at the end of chromosomes 2 and 12 (Fig. 2c–e and Supplementary Table 12). Recombinant screening using a set of newly developed KASP markers allowed us to fine-map the QTL on chromosome 12 in a 514.83 kb region containing 52 genes (Supplementary Fig. 8 and Supplementary Table 13). Among them, a gene (arahy.BBG51B) encoding a phosphatidylethanolamine-binding protein was the only one associated with a frameshift mutation (Supplementary Table 13). Notably, based on phylogenetic reconstruction, this gene, named AhTFL1, was deemed as the putative orthologue of AtTFL1 (AT5G03840), involved in the control of inflorescence architecture in Arabidopsis33,34,35,36 (Fig. 2f,g and Supplementary Table 14), thus making AhTFL1 an obvious candidate to control the flowering pattern in peanut. GWAS confirmed the presence of strong signals for markers closely associated with AhTFL1 on the terminal regions of chromosome 2 (377.48 kb, −log10P = 34.05) and chromosome 12 (14.4 kb, −log10P = 27.31; Fig. 2h and Supplementary Table 15), suggesting that AhTFL1 homologs on the A and B genomes are both contributing to the flowering pattern phenotype. AhTFL1 sequencing in the GWAS population revealed the occurrence of three mutations (a MITE insertion, a 1,492 bp deletion and a 1 bp deletion; Fig. 2f and Supplementary Fig. 9) fully cosegregating with the sequential flowering pattern (Supplementary Tables 16 and 17). Notably, a recent work37 also reports full cosegregation between the MITE InDel described in our study and the peanut flowering pattern, as well as significantly lower expression of AhTFL1 in (1) Ahf compared with Ahh and (2) flowering compared with non-flowering branches. GWAS for the total number of branches (TNBs) resulted in the strongest signal colocalizing with AhTFL1, indicating that AhTFL1 may have a pleiotropic effect on this trait (Fig. 2i and Supplementary Table 18).
a, Alternate pattern. b, Sequential pattern. c–e, Likelihood of odd (LOD) score graphs obtained by QTL composite interval mapping using the YZ9102 x wt09-0023 RIL population (c) and the YH15 x W1202 RIL population (d,e). f, Structure of the AhTFL1 gene and features of the three mutations found in the GWAS population. g, Phylogenetic relationships among Arachis and Arabidopsis TFL homologs. AhTFL1 and AtTFL1 are highlighted in bold. h,i, GWAS Manhattan and quantile–quantile (Q–Q) plots for the flowering pattern (h) and TNBs (i). The MLM implemented in the R package GAPIT was used to test for marker–trait association. The horizontal line in each Manhattan plot indicates the −log10P threshold for significant association after the Bonferroni correction. The shaded area in the Q–Q plots indicates the 95% confidence interval under the null hypothesis of no association between the SNPs and the trait, under the assumption of a uniform [0, 1] distribution for the P values. Mu, mutations.
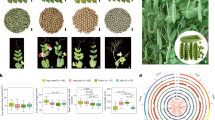
Another trait displaying divergent phenotypes between the two peanut subspecies is the color of the seed inner tegument, which is invariably yellow in Ahh (Fig. 3a) and white in Ahf (Fig. 3b). Both GWAS and RIL-based mapping highlighted the strong association between the tegument color and a genomic region on chromosome 5 (Fig. 3c–e and Supplementary Tables 12 and 19). Screening of recombinant RILs by KASP markers allowed to fine-map the QTL to an interval of 540.14 kb (Supplementary Fig. 10a), which was further refined to 107.88 kb by the screening of 7,900 segregant F2 individuals (Supplementary Fig. 10b). Within the interval, a gene (arahy.0C6ZNN) encoding a laccase-like protein, named AhLAC, was the only one associated with a frameshift mutation (Supplementary Table 20). Notably, this gene is the putative ortholog of the Arabidopsis gene AtLAC15 (also referred to as TRANSPARENT TESTA 10 or AtTT10, AT5G48100; Fig. 3f and Supplementary Table 21), which was shown to influence the color of the seed coat through its enzymatic role in the oxidative polymerization of flavonoids38. The strongest GWAS signal (−log10P = 22.17) was 68.96 kb from AhLAC (Fig. 3c and Supplementary Table 19). AhLAC sequencing in the GWAS population revealed the occurrence of two mutations (a MITE insertion and a 1 bp insertion, the latter only occurring in the two Ahf var. peruviana accessions; Fig. 3g and Supplementary Fig. 11). A KASP assay was designed on the MITE insertion (Supplementary Table 22) and verified to be fully cosegregating with the inner tegument color in both the GWAS population and the YZ9102 x wt09-0023 RIL population (Supplementary Tables 23–25). Heterologous overexpression of AhLAC partially complemented the Arabidopsis Attt10 loss-of-function mutant in four independent transgenic lines, with the level of seed lightness (expressed by the L* score) being inversely related to the transgene expression level (Fig. 3h and Supplementary Fig. 12a–c). Consistently, the yellow inner tegument accessions YH154 and YH37 displayed markedly higher AhLAC expression than the white inner tegument accessions YH76 and ZYH109 (Fig. 3i). Finally, the epicatechin content was significantly higher in YH76 and ZYH109 than in YH154 and YH37 (Supplementary Fig. 12d,e), consistent with the possibility that AhLAC causes pigmentation through epicatechin oxidative polymerization, similarly to Arabidopsis AtTT10 (ref. 38).
a, Yellow phenotype. b, White phenotype. c, GWAS Manhattan plot and Q–Q plot. The MLM implemented in the R package GAPIT was used to test for marker–trait association. The horizontal line in the Manhattan plot indicates the −log10P threshold for significant association after the Bonferroni correction. The shaded area in the Q–Q plot indicates the 95% confidence interval under the null hypothesis of no association between the SNPs and the trait, under the assumption of a uniform [0, 1] distribution for the P values. d,e, Chromosome 5 LOD score graphs obtained by QTL composite interval mapping using the YZ9102 x wt09-0023 RIL population (d) and the Zheng8903 x YH4 RIL population (e). f, Phylogenetic relationships among peanut and Arabidopsis LAC homologs. AhLAC and Arabidopsis TRANSPARENT TESTA 10 (AtTT10) are highlighted in bold. g, Structure of the AhLAC gene and features of the two mutations found in the GWAS population. h, Complementation of the Arabidopsis Atlac/Attt10 mutant with peanut AhLAC. The phenotype of the WT Col-0 accession is also shown. i, AhLAC expression levels in the seed coat of yellow (YH154 and YH37) and white tegument (YH76 and ZYH109) accessions at 45 and 85 days after flowering. Data in i are given as mean ± s.e.m.; n = 3 biologically independent samples; the two-tailed Student’s t test was carried out to compare means. WT, wild type.
Genetic dissection of main peanut economic traits
The peanut growth habit (erect or prostrate; Fig. 4a,b) strongly conditions cultivation practices39. Both GWAS and genetic mapping using two RIL populations resulted in signals for a genomic region on chromosome 15 (Fig. 4c–e and Supplementary Tables 12 and 26), in accordance with previous studies40,41,42. Recombinant screening using a set of newly developed KASP markers allowed to fine-map the QTL on a 299.11 kb region containing 20 genes (Supplementary Fig. 13 and Supplementary Table 27). Among them, a MADS-box gene (arahy.ATH5WE) was chosen as a candidate, as (1) it was the only one displaying a mutation within the coding sequence (a frameshift caused by a 1,870 bp deletion), and (2) the MADS-box family of transcription factors was previously associated with the plant growth habit43. The most significant GWAS signal on chromosome 15 (−log10P = 9.12) was only 2.07 kb apart from the same MADS- box homolog, which, based on phylogenetic analysis, was related to Arabidopsis AtPI (At5G20240) and AtAP3 (AT3G54340; Fig. 4g and Supplementary Table 28). Three gene mutations (a 2 bp insertion in the first exon, a 1,870 bp deletion in the first intron and a MITE insertion in the third intron) were characterized (Fig. 4f and Supplementary Fig. 14), and the polymorphisms were used to develop KASP and Integrative Genomics Viewer markers. With a few exceptions, at least one of the three mutations was found to cosegregate with the erect phenotype (Fig. 4h,i) in the GWAS population (Supplementary Tables 29 and 30). Considering that the growth habit might be influenced by environmental factors, further investigations are required to clarify the putative role of a MADS transcription factor as a determinant of the peanut growth habit.
a, Erect phenotype. b, Prostrate phenotype. c, GWAS Manhattan plot and Q–Q plot. The MLM implemented in the R package GAPIT was used to test for marker–trait association. The horizontal line in the Manhattan plot indicates the threshold for significant association after the Bonferroni correction. The shaded area in the Q–Q plot indicates the 95% confidence interval under the null hypothesis of no association between the SNPs and the trait, under the assumption of a uniform [0, 1] distribution for the P values. d,e, Chromosome 15 LOD score graphs obtained by QTL composite interval mapping using the YZ9102 x wt09-0023 RIL population (d) and the YH15 x W1202 RIL population (e). f, Structure of the AhMADS-box transcription factor 6 gene and features of the three mutations found in the GWAS population. g, Phylogenetic relationships among Arachis and Arabidopsis AGL homologs. AtPI, AtAP3 and AhMADS-box are highlighted in bold. h,i, The distribution of 353 accessions according to growth habit (h) and mutation types (i).
Pod and kernel dimensions, together with kernel oil content, are key peanut commercial traits. RIL-based mapping indicated that kernel weight, kernel length and pod weight are genetically correlated. The identification of QTLs on chromosomes 5 and 16 is in accordance with previous studies44,45. GWAS confirmed marker–trait associations on chromosomes 5 and 16 (significance peaks for −log10P = 13.05 and 15.91, respectively); however, a signal on chromosome 6 was also found (Fig. 5a,d,g and Supplementary Table 31). Finally, GWAS for oil content highlighted a main signal on chromosome 8 (−log10P = 8.94; Supplementary Fig. 15 and Supplementary Table 32), in correspondence with a previously mapped QTL46 and in accordance with the recent findings in ref. 8.
a,d,g, Manhattan plots for HPW (a), HKW (d) and SL (g). The MLM implemented in the R package GAPIT was used to test for marker–trait association. The horizontal line in each Manhattan plot indicates the −log10P threshold for significant association after the Bonferroni correction. b,e,h, Q–Q plots for HPW (b), HKW (e) and SL (h). The shaded area in each Q–Q plot indicates the 95% confidence interval under the null hypothesis of no association between the SNPs and the trait, under the assumption of a uniform [0, 1] distribution for the P values. c,f,i, Violin plots and boxplots describing the SL distribution in different accessions with genotypes AA (n = 65) and TT (n = 285) at the A05.85303877 locus (c), genotypes CC (n = 112) and TT (n = 233) at the A06.108072513 locus (f), and genotypes CC (n = 113) and TT (n = 229) at the A16.143683180 locus (i). In the boxplots, centerline indicates the median; box lower and upper edges indicate the 25% and 75% quartiles, respectively; whiskers indicate 1.5× interquartile range; the two-tailed Student’s t test was carried out to compare means. j,k,l, Pie-charts for the proportion of accessions falling in different A. hypogaea types and having alternative genotypes at the sites A05.85303877 (j), A06.108072513 (k) and A16.143683180 (l).
Discussion
This study reports the results of a massive DNA sequencing effort, allowing the fine-scale reconstruction of main events associated with the evolutionary history and phenotypic diversification of peanuts. Chloroplast genome sequencing and phylogenesis from a large germplasm panel, including several accessions of the chloroplast donor wild species A. duranensis, provided a solid indication that the peanut subspecies Ahh and Ahf result from distinct polyploidization and domestication events. This was confirmed by the characterization of genetic polymorphisms between and within the two taxonomic groups. Notably, multiple polyploidization events were reported to be at the basis of the evolution of several species25,47,48,49, in accordance with our findings. The independent origin of Ahh and Ahf explains the contradictory findings from ref. 5 and refs. 15,30, tracing back peanut polyploidization <10,000 years ago and 0.42–0.47 million years ago, respectively, which were previously debated50,51. Indeed, the two research groups based their evolutionary analyses on different reference genome sequences, from the Ahh cultivar Tifrunner and the Ahf cultivar Shitouqi. We predict that research methods used in this study might be transferred to other allopolyploid plant species whose origin is still elusive.
Two independent mapping approaches (GWAS and biparental linkage analysis) were used to investigate the genetic basis of phenotypic divergence between Ahh and Ahf and the genetic control of several economically important traits. This choice is justified by the need to increase the confidence of the results obtained by the GWAS approach that, while allowing a higher mapping resolution, might lead to false positive signals in the case of A and B genome homeologous regions showing high sequence similarity or in the case of homeologous recombination changing the genomic formula from the expected AABB to AAAA or BBBB15,30. The identification of AhTFL1 as the gene putatively controlling the peanut flowering pattern is in line with previous findings in Arabidopsis and other crop species, although the peanut raceme inflorescence bears distinctive features. Continuous flowering often coincides with early maturing, which is desirable in areas characterized by a shorter growing season. Notably, we also showed that the pigmentation of the inner tegument likely originates from a mutation of the laccase gene AhLAC, although further investigation is needed to clarify whether AhLAC promotes tegument pigmentation through the oxidative polymerization of flavonoids, as it was shown for its Arabidopsis homolog AtTT10 (ref. 38). The tegument color can affect several physiologic and economic traits in plants, including legumes, such as seed dormancy, response to pathogens and pests and seed nutritional traits52,53; thus, this finding can be of broad interest for plant scientists and breeders. It might be speculated that the occurrence of white inner tegument in Ahf contributes to the absence of seed dormancy in this subspecies, in contrast with Ahh. This trait makes Ahf more suitable for cultivation in warm environments and allows consecutive harvests.
Together, the data reported in this study provide an important genomic resource for further and faster peanut genetic improvement, and the results presented here might be of broad interest to the plant sciences community and plant breeding.
Methods
Plant material and DNA extraction
The germplasm panel used in this study included 34 accessions of wild diploid species, 2 accessions of wild tetraploid A. monticola, 353 accessions of cultivated tetraploid A. hypogaea and three previously described RIL populations of A. hypogaea (Supplementary Tables 1 and 2)54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75. The 353 A. hypogaea accessions were selected from more than 2,000 accessions collected from 27 countries and 18 China provinces based on tunable genotyping-by-sequencing (tGBS) sequencing and phenotype cluster analysis13. The diversity panel included five peanut botanical varieties (85 var. hypogaea, 12 var. hirsuta, 26 var. fastigiata, 84 var. vulgaris and 2 var. peruviana) and two kinds of irregular types (100 irregular hypogaea-type and 44 irregular fastigiata-type) associated with landraces, cultivars and breeding lines. Genomic DNA extraction was performed on the whole germplasm panel using the Plant Genomic DNA Kit (Tiangen Biotech).
Chloroplast de novo sequencing and variant identification
In total, 113 chloroplast genomes (77 from A. hypogea landraces representing var. hypogaea, var. hirsuta, var. fastigiata, var. vulgaris and var. peruviana; 2 from A. monticola accession; and 34 from wild diploid accessions) were de novo assembled using default pipeline settings (-R 15 -k 21,45,65,85,105) of the GetOrganelle toolkit version 1.7.3.5 (ref. 76). The chloroplast genomes and repeat_pattern1 that consist of two equimolar isomeric sequences and with the same direction of the small single-copy (SSC) regions were used for making alignments with the MAFFT program version 7 (ref. 77) for pairwise comparisons. The SNP and InDel variants between the chloroplast genomes were identified using MEGA X78 with the Chlorophycean Mitochondrial code set.
Genomic resequencing and variant identification
Paired-end DNA libraries with inserts of approximately 300 bp were constructed and sequenced using the Illumina HiSeq Xten (Illumina) platform with PE151. Raw data were cut with an average coverage of 20× per sample for further analysis. High-quality reads passing the quality check and filtering were aligned to the genome of cultivated peanut A. hypogaea cv. Tifrunner version 1 using minimap2 (v2.10)79 with the command ‘-ax sr -t 25 -K 5G’. BAM alignment files were then generated with sambamba (v0.6.8)80 by removing potential PCR duplications.
SNP and InDel calling were performed with the Genome Analysis Toolkit (v4.0.12.0)81 with the HaplotypeCaller method. Detected SNPs matching any of the following conditions were filtered out: QualByDepth <2.0, FisherStrand >60.0, RMSMappingQuality <40.0, MappingQualityRankSumTest <−12.5 and ReadPosRankSumTest <−8.0. The conditions used to filter out InDels were as follows: QualByDepth <2.0, FisherStrand >200.0 and ReadPosRankSumTest <−20.0. After applying the aforementioned filtering conditions, we obtained variationSet1. To further exclude variant calling errors, all variations with a missing rate >0.05 (alleles having less than five reads supporting them were marked as missing), minor allele frequency <0.01 and number of heterozygous genotypes >10 were filtered out using vcftools (v0.1.19)82 and bcftools (v1.10.2)83, which resulted in variationSet2.
Chloroplast phylogenesis
The 113 chloroplast genomes were configured, of which the SSC regions aligned in the same direction were used to construct the neighbor-joining tree with MEGA X78.
The multi-FASTA file containing the 113 assembled chloroplast genomes was analyzed with an in-house generated Python script to identify and count mononucleotide microsatellites. For each sequence entry in the FASTA file, the script identifies and counts occurrences of monorepetitions of the four nucleotide bases (A, T, G and C) that fall within the length range of 3–20 nucleotides. After counting these repeats, the script calculates the percentage abundance of each SSR type relative to the total sequence length. In the following step, the microsatellites that were not present in any of the samples were discarded, as well as those with the same abundance across all the samples. The obtained matrix of abundance was processed in R to generate an Euclidean distance matrix. Samples were clustered by hierarchical clustering based on the Pearson correlation of the distance values. Bootstrap values were obtained using the R package pvclust using 1,000 iterations.
Genomic distribution of SNPs between and across subspecies
Two groups of five individuals, one from Ahh and the other from Ahf, or both from the same subspecies, were extracted by performing 100 bootstrap replicates. SNP data were used to extract polymorphisms between groups (PB), occurring when alternative alleles are fixed in each group (that is, FST = 1), and polymorphisms shared across groups (PA), occurring when alternative alleles are present in both groups. Chromosomes 1–10 and 11–20 were analyzed separately. The density distribution of polymorphisms in 1 Mb genome windows was drawn using the R package Cmplot.
LD and haplotype block analyses
LD decay was calculated for all pairs of variations on var. hypogaea and irregular hypogaea-type (183 samples), var. hirsuta (12 samples), var. fastigiata (26 samples), var. vulgaris and irregular fastigiata-type (130 samples) from variationSet2 using PopLDdecay (v3.31) with default parameters84. Considering the influence of the different number of samples in LD decay calculation, we standardized the sample size of var. hypogaea and irregular hypogaea-type and var. vulgaris and irregular fastigiata-type to 12 and 26, respectively, using shuf (version 8.22) and repeated 100 times. Half-maximum decay distance was calculated based on averaging the r2 values of each 100-bp separation bin (that is, average r2 for SNPs separated by 1–100 bp, 101–200 bp, etc.). To calculate the half-maximum decay distance for var. hypogaea and var. vulgaris, all 100 standardized sample lists were used to calculate the half-maximum decay distance individually, and then the median value was taken (using the built-in quantile function in R with P = 0.5 and type = 1). To call haplotype blocks in 79 selected landraces, we used the R package HaploBlocker (v1.5.18)85 with adaptive mode and different subspecies as subgroups on variationSet2. All 79 samples were clustered with the binary matrix output from haplotype blocks using ade4 in R (v1.7-16)86 on the first ten chromosomes (subgenome A) and the second ten chromosomes (subgenome B) separately.
Population structure analysis
After clumping the remaining variants in variationSet2 using PLINK (v1.90b6.9)87 with ‘--clump-p1 1 --clump-p2 1 --clump-r2 0.5’, variations (variationSet3) were retained for population structure analysis. A maximum likelihood phylogenetic tree was constructed with IQ-TREE (v1.6.12)88 using the optimal model (GTR + F + ASC + R5) as determined by the Bayesian information criterion. Population structure was also studied using ADMIXTURE (v1.30)89 with k between 1 and 20. The program smartpca from the Eigenstrat package (v7.2.1)90 was used to calculate eigenvectors of variationSet2. Allelic differentiation between populations was measured by nucleotide diversity (π) of each subspecies group using vcftools (v0.1.19) with a 200 kb window and a step size of 100 kb for each subspecies on variationSet2.
QTL mapping and GWAS
The three RIL populations YZ9102 x wt09-0023, YH15 x W1202 and Zheng8903 x YH4, including 521, 318 and 212 lines, respectively, were used for QTL mapping. The YZ9102 x wt09-0023 RIL population was sequenced using the single digest restriction site-associated DNA sequencing protocol91, and the sequencing depths for the two parents and the RILs were approximately 25× and 5×, respectively75. SNP sites were used for genetic map construction and QTL mapping as previously reported75. The other two populations were sequenced using WGR, and the sequencing depths for the parents and the RILs were approximately 30× and 1×, respectively73,74. The sliding-window approach for genotype calling and recombination breakpoint determination92 was applied to convert SNPs into bin markers. The genetic maps of the YZ9102 x wt09-0023 and the Zheng8903 x YH4 populations were constructed using Joinmap (v5.0)93, whereas the genetic map of the YH15 x W1202 population was constructed using QTL Icimapping (v4.2)94. QTL analysis was performed using the multiple QTL mapping algorithm implemented in MapQTL (v6.0)95 by setting the mapping step size as 0.1 cM and the LOD threshold as 2.5. Fine mapping was performed by developing KASP markers in the QTL interval and screening recombinant RILs. As for the inner tegument color trait, further fine mapping was performed by screening recombinant individuals from an F2 population originating from the P573 x P602 cross.
GWAS was carried out on the 353 cultivated peanuts from variationSet2. Phenotypic data for flowering pattern, TNBs, color of the inner tegument, growth habit and oil content based on gas chromatography were collected in one environment (2019: Zhengzhou (2019ZZ)), whereas 100 kernel weight (HKW), 100 pod weight (HPW) and seed length (SL) were collected in seven environments (2017: Yuanyang (2017YY); 2018: Yuanyang (2018YY), Xinyang (2018XY), Weifang (2018WF); 2019: Zhengzhou (2019ZZ), Shangqiu (2019SQ), Weifang (2019WF)) using a randomized complete block design with two replicates. The mixed linear model (MLM)96 implemented in the R package GAPIT (v3.0)97 was used to identify significant associations (Supplementary Table 33), using population structure results from ADMIXTURE analysis (K), the first two principal components (PCs) and the flowering pattern as covariates. The genome-wide significance threshold for association was set as 0.05/n (where n is the number of markers). Significant SNPs in the candidate intervals were annotated using software snpEff (v4.5)98.
AhLAC functional characterization
A 35S overexpression vector (PBI121) was constructed by double digestion (XbaI and SacI) and ligation of the AhLAC gene into the vector. The Arabidopsis tt10 mutant (cs2105589) was transformed by Agrobacterium inflorescence immersion99, and mature seeds were collected. Transgenic-resistant plants were subsequently screened on Murashige and Skoog (MS) medium containing 50 mg l−1 kanamycin. The AhLAC gene expression level in transgenic plants was determined by real-time qPCR using the primer pair 5′-ATGAAATGTTGTTGCTTGG-3′ (F)/5′-TCAACAAGGAGGCAGATCTG-3′ (R) in combination with the primer pair 5′-TCCGGACCAGCGTCTCA-3′ (F)/5′-CCACCACGAAGACGCAGGA-3′ (R), the latter targeting the AtUBQ10 housekeeping gene100. The level of seed lightness (the L* score) was quantified on a 0–100 scale by the high-precision spectrophotometer NR110 (3nh).
To investigate the functional role of AhLAC in peanut, seed coat AhLAC expression levels were quantified at 45 and 85 days after flowering in peanut genotypes displaying yellow (YH154 and YH37) or white (YH76 and ZYH109) inner teguments. Real-time qPCR was performed using the primer pair 5′-CATGGAGTGAAGCAGCCAAGAA-3′ (F)/5′-AGTGGCTCTTGCCCAATCACT-3′ (R) in combination with the primer pair 5′-GACGCTTGGCGAGATCAACA-3′ (F)/5′-AACCGGACAACCACCACATG-3′ (R); the latter targeting the ADH3 housekeeping gene101. Epicatechin was extracted from the dry seed coat of YH154, YH37, YH76 and ZHY109 using the standard procedure102 and quantified by the high-performance liquid chromatograph series 6420A mass spectrometry AGILENT 1260 (Agilent Technologies).
Statistical testing
A two-tailed Student’s t test was conducted in basic R (v4.1.3) to compare means relative to unique mapped read rates, relative AhLAC expression, epicatechin content, SL and seed lightness.
Map generation
The map depicting the country of origin for the peanut accessions considered in this study was generated using the mapPies function in the freely available package rworldmap103 in R (v4.1.3).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets analyzed or generated by this study are available in Supplementary Information and the public repositories of the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov) and Zenodo (https://zenodo.org/). WGR data are available in the NCBI Sequence Read Archive database (Bioproject PRJNA605106). The assembled chloroplast genomes obtained in this study are available in the NCBI GenBank database (accessions from PP971404 to PP971516). Genomic SNPs and InDels identified in this study are available at the Zenodo repository (https://doi.org/10.5281/zenodo.12475904)104.
Code availability
The custom script used to extract polymorphisms between and across A. hypogaea subspecies Ahh and Ahf is available at the Zenodo repository (https://doi.org/10.5281/zenodo.12614808)105. The in-house generated Python scripts used to count SSRs, calculate their percentage and perform clustering based on SSR data are available at the Zenodo repository (https://doi.org/10.5281/zenodo.12191309)106.
References
Seijo, G. et al. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 94, 1963–1971 (2007).
Carvalho, P. A. S. V. et al. Presence of resveratrol in wild Arachis species adds new value to this overlooked genetic resource. Sci. Rep. 10, 12787 (2020).
Krapovickas, A. Origen, variabilidad y diffusion del Mani (Arachis hypogaea). Actas Y Memorias del Congreso Internacional de Americanistas 2517–2534 (1968).
Yin, D. et al. Genome of an allotetraploid wild peanut Arachis monticola: a de novo assembly. GigaScience 7, giy066 (2018).
Zhuang, W. et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 51, 865–876 (2019).
Dillehay, T. D., Rossen, J., Andres, T. C. & Williams, D. E. Preceramic adoption of peanut, squash, and cotton in northern Peru. Science 316, 1890–1893 (2007).
Hammons, R. O. et al. (eds) Peanuts Genetics, Processing, and Utilization 1–26 (Academic Press and AOCS Press, 2016).
Lu, Q. et al. A genomic variation map provides insights into peanut diversity in China and associations with 28 agronomic traits. Nat. Genet. 56, 530–540 (2024).
Varshney, R. K. et al. The first SSR-based genetic linkage map for cultivated groundnut (Arachis hypogaea L.). Theor. Appl. Genet. 118, 729–739 (2009).
Pandey, M. K. et al. (eds) Genetics, Genomics and Breeding of Peanuts 79–113 (CRC Press, 2014).
Krapovickas, A. & Gregory, W. C. Taxonomy of the genus Arachis (Leguminosae). Bonplandia 8, 1–186 (1994).
Archer, P. (ed.) Peanuts Genetics, Processing, and Utilization 253–266 (Academic Press and AOCS Press, 2016).
Zheng, Z. et al. Genetic diversity, population structure, and botanical variety of 320 global peanut accessions revealed through tunable genotyping-by-sequencing. Sci. Rep. 8, 14500 (2018).
Otyama, P. I. et al. Evaluation of linkage disequilibrium, population structure, and genetic diversity in the U.S. peanut mini core collection. BMC Genomics 20, 481 (2019).
Bertioli, D. J. et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884 (2019).
Chen, X. et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 12, 920–934 (2019).
Pittman, R. N. United States Peanut Descriptors (US Government Printing Office, 1995); https://archive.org/details/IND20479053
Semagn, K., Babu, R., Hearne, S. & Olsen, M. Single nucleotide polymorphism genotyping using kompetitive allele specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breed. 33, 1–14 (2014).
Wheeler, G. L., Dorman, H. E., Buchanan, A., Challagundla, L. & Wallace, L. E. A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Appl. Plant Sci. 2, apps.1400059 (2014).
Lian, C. et al. Comparative analysis of chloroplast genomes reveals phylogenetic relationships and intraspecific variation in the medicinal plant Isodon rubescens. PLoS ONE 17, e0266546 (2022).
Mao, L., Zou, Q., Sun, Z., Dong, Q. & Cao, X. Insights into chloroplast genome structure, intraspecific variation, and phylogeny of Cyclamen species (Myrsinoideae). Sci. Rep. 13, 87 (2023).
Mu, Z. et al. Intraspecific chloroplast genome variation and domestication origins of major cultivars of Styphnolobium japonicum. Genes 14, 1156 (2023).
Zhang, W. et al. Comparative analysis of 17 complete chloroplast genomes reveals intraspecific variation and relationships among Pseudostellaria heterophylla (Miq.) Pax populations. Front. Plant Sci. 14, 1163325 (2023).
Chen, N. et al. Evolutionary patterns of plastome uncover diploid–polyploid maternal relationships in Triticeae. Mol. Phylogenet. Evol. 149, 106838 (2020).
Brock, J. R., Mandáková, T., McKain, M., Lysak, M. A. & Olsen, K. M. Chloroplast phylogenomics in Camelina (Brassicaceae) reveals multiple origins of polyploid species and the maternal lineage of C. sativa. Hortic. Res. 9, uhab050 (2022).
Grabiele, M., Chalup, L., Robledo, R. & Seijo, G. Genetic and geographic origin of domesticated peanut as evidenced by 5S rDNA and chloroplast DNA sequences. Plant Syst. Evol. 298, 1151–1165 (2012).
Du, P. et al. Development of an oligonucleotide dye solution facilitates high throughput and cost-efficient chromosome identification in peanut. Plant Methods 15, 69 (2019).
Deb, S. K., Edger, P. P., Pires, J. C. & McKain, M. R. Patterns, mechanisms, and consequences of homoeologous exchange in allopolyploid angiosperms: a genomic and epigenomic perspective. New Phytol. 238, 2284–2304 (2023).
Mason, A. S. & Wendel, J. F. Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 11, 1014 (2020).
Bertioli, D. J. et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446 (2016).
Hodel, R. G. J., Zimmer, E. A., Liu, B. B. & Wen, J. Synthesis of nuclear and chloroplast data combined with network analyses supports the polyploid origin of the apple tribe and the hybrid origin of the maleae-gillenieae clade. Front. Plant Sci. 12, 820997 (2022).
Tian, X. et al. Chloroplast pylogenomic analyses reveal a maternal hybridization event leading to the formation of cultivated peanuts. Front. Plant Sci. 12, 804568 (2021).
Shannon, S. & Meeks-Wagner, D. R. A. Mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3, 877–892 (1991).
Severin, A. J. et al. RNA-seq atlas of glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10, 160 (2010).
Dhanasekar, P. & Reddy, K. S. A novel mutation in TFL1 homolog affecting determinacy in cowpea (Vigna unguiculata). Mol. Genet. Genomics 290, 55–65 (2015).
Krylova, E. A., Khlestkina, E. K., Burlyaeva, M. O. & Vishnyakova, M. A. Determinate growth habit of grain legumes: role in domestication and selection, genetic control. Ecol. Genet. 18, 43–58 (2020).
Kunta, S. et al. Identification of a major locus for flowering pattern sheds light on plant architecture diversification in cultivated peanut. Theor. Appl. Genet. 135, 1767–1777 (2022).
Pourcel, L. et al. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17, 2966–2980 (2005).
Butzler, T. M., Bailey, J. & Beute, M. K. Integrated management of sclerotinia blight in peanut: utilizing canopy morphology, mechanical pruning, and fungicide timing. Plant Dis. 82, 1312–1318 (1998).
Kayam, G. et al. Fine-mapping the branching habit trait in cultivated peanut by combining bulked segregant analysis and high-throughput sequencing. Front. Plant Sci. 8, 467 (2017).
Pan, J. et al. BSA-seq and genetic mapping identified candidate genes for branching habit in peanut. Theor. Appl. Genet. 135, 4457–4468 (2022).
Fang, Y. et al. Identification of quantitative trait loci and development of diagnostic markers for growth habit traits in peanut (Arachis hypogaea L.). Theor. Appl. Genet. 136, 105 (2023).
Rosin, F. M., Hart, J. K., Onckelen, H. V. & Hannapel, D. J. Suppression of a vegetative MADS box gene of potato activates axillary meristem development. Plant Physiol. 131, 1613–1622 (2003).
Luo, H. et al. Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.). Theor. Appl. Genet. 131, 267–282 (2018).
Gangurde, S. S. et al. Nested-association mapping (NAM)-based genetic dissection uncovers candidate genes for seed and pod weights in peanut (Arachis hypogaea). Plant Biotechnol. J. 18, 1457–1471 (2020).
Liu, N. et al. High-resolution mapping of a major and consensus quantitative trait locus for oil content to a ~0.8-Mb region on chromosome A08 in peanut (Arachis hypogaea L.). Theor. Appl. Genet. 133, 37–49 (2020).
Wolfe, T. M. et al. Recurrent allopolyploidizations diversify ecophysiological traits in marsh orchids (Dactylorhiza majalis s.l.). Mol. Ecol. 32, 4777–4790 (2023).
Mavrodiev, E. V. et al. Multiple origins and chromosomal novelty in the allotetraploid Tragopogon castellanus (Asteraceae). N. Phytol. 206, 1172–1183 (2015).
Soltis, D. E. & Soltis, P. S. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 14, 348–352 (1999).
Bertioli, D. J. et al. Evaluating two different models of peanut’s origin. Nat. Genet. 52, 557–559 (2020).
Zhuang, W. et al. Reply to: evaluating two different models of peanut’s origin. Nat. Genet. 52, 560–563 (2020).
Hradilová, I. P. et al. Variation in wild pea (Pisum sativum subsp. elatius) seed dormancy and its relationship to the environment and seed coat traits. PeerJ 7, e6263 (2019).
Smýkal, P., Vernoud, V., Blair, M. W., Soukup, A. & Thompson, R. D. The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 5, 351 (2014).
Robledo, G. & Seijo, G. Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theor. Appl. Genet. 121, 1033–1046 (2010).
Du, P. et al. Chromosome painting of telomeric repeats reveals new evidence for genome evolution in peanut. J. Integr. Agric. 15, 2488–2496 (2016).
Stalker, H. T. Utilizing wild species for peanut improvement. Crop Sci. 57, 1102–1120 (2017).
Robledo, G., Lavia, G. I. & Seijo, G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theor. Appl. Genet. 118, 1295–1307 (2009).
Li, C. et al. Development and application of whole-chromosome painting of chromosomes 7A and 8A of Arachis duranensis based on chromosome-specific single-copy oligonucleotides. Genome 67, 178–188 (2024).
Stalker, H. T. A new species in section Arachis of peanuts with A D genome. Am. J. Bot. 78, 630–637 (1991).
Valls, J. F. M. & Simpson, C. E. (eds) Biology and Agronomy of Forage Arachis 1–18 (CIAT, 1994).
Shandong Peanut Research Institute. Peanut Varieties of China (Agriculture Press, 1987).
GRIN-Global. U.S. National Plant Germplasm System. npgsweb.ars-grin.gov/gringlobal/search (2024).
The First Seed Industry. www.a-seed.cn/ (2024).
National Peanut Center. Peanut varieties database. China Peanut Data Center http://peanut.cropdb.cn/variety/index.htm (2024).
Yu, S. L. Chinese Peanut Varieties and Their Pedigree (Shanghai Science and Technology Press, 2008).
Banks, D. J. & Kirby, J. S. Registration of Pronto peanut (reg no. 28). Crop Sci. 23, 184 (1983).
Oil Crops Research Institute, Chinese Academy of Agricultural Sciences. Directory of Peanut Variety Resources in China (Agriculture Press, 1993).
Shandong Peanut Research Institute. Directory of Peanut Variety Resources in China (Penglai County Printing Factory, 1978).
Bailey, W. K. & Hammons, R. O. Registration of Chico peanut germplasm (reg. no. GP 2). Crop Sci. 15, 105 (1975).
Belamkar, V. et al. A first insight into population structure and linkage disequilibrium in the US peanut minicore collection. Genetica 139, 411–429 (2011).
Alyr, M. H. et al. Fine-mapping of a wild genomic region involved in pod and seed size reduction on chromosome A07 in peanut (Arachis hypogaea L.). Genes (Basel) 11, 1402 (2020).
Shrestha, A., Srinivasan, R., Sundaraj, S., Culbreath, A. K. & Riley, D. G. Second generation peanut genotypes resistant to thrips-transmitted tomato spotted wilt virus exhibit tolerance rather than true resistance and differentially affect thrips fitness. J. Econ. Entomol. 106, 587–596 (2013).
Liu, H. et al. QTL mapping of web blotch resistance in peanut by high-throughput genome-wide sequencing. BMC Plant Biol. 20, 249 (2020).
Sun, Z. et al. QTL mapping of quality traits in peanut using whole-genome resequencing. Crop J. 10, 177–184 (2022).
Qi, F. et al. QTL identification, fine mapping, and marker development for breeding peanut (Arachis hypogaea L.) resistant to bacterial wilt. Theor. Appl. Genet. 135, 1319–1330 (2022).
Jin, J. et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241 (2020).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. Preprint at bioRxiv https://doi.org/10.1101/201178 (2018).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
Zhang, C., Dong, S. S., Xu, J. Y., He, W. M. & Yang, T. L. PopLD decay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788 (2019).
Pook, T. et al. HaploBlocker: creation of subgroup-specific haplotype blocks and libraries. Genetics 212, 1045–1061 (2019).
Bougeard, S. & Dray, S. Supervised multiblock analysis in R with the ade4 package. J. Stat. Softw. 86, 1–17 (2018).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Alexander, D. H. & Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12, 246 (2011).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Ruperao, P. et al. Apilot-scale comparison between single and double-digest RAD markers generated using GBS strategy in sesame (Sesamum indicum L.). PLoS ONE 18, e0286599 (2023).
Huang, X. et al. High-throughput genotyping by whole genome resequencing. Genomes Res. 19, 1068–1076 (2009).
Van Ooijen, J. W. JoinMap 5: software for the calculation of genetic linkage maps in experimental populations of diploid species. https://www.kyazma.nl/index.php/JoinMap/ (2018).
Meng, L., Li, L. L., Zhang, L. Y. & Wang, J. K. QTL IciMapping, integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283 (2015).
Van Ooijen, J. W. et al. MapQTL 6: software for the mapping of quantitative trait loci in experimental populations of diploid species (2009); https://www.scienceopen.com/book?vid=9e9eabc7-f089-43be-831d-2d086fa52646
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208 (2006).
Lipka, A. E. et al. GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399 (2012).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Martinez-Trujillo, M. et al. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Rep. 22, 63–70 (2004).
Souček, P. et al. Stability of housekeeping gene expression in Arabidopsis thaliana seedlings under differing macronutrient and hormonal conditions. J. Plant Biochem. Biotechnol. 26, 415–424 (2017).
Brand, Y. & Hovav, R. Identification of suitable internal control genes for quantitative real-time PCR expression analyses in peanut (Arachis hypogaea). Peanut Sci. 37, 12–19 (2010).
Aguirre-Hernández, E. et al. HPLC/MS analysis and anxiolytic-like effect of quercetin and kaempferol flavonoids from Tilia americana var. Mexicana. J. Ethnopharmacol. 127, 91–97 (2010).
South, A. rworldmap: a new R package for mapping global data. R J. 3, 35–43 (2011).
Zheng, Z. SNPs and InDels identified in 353 peanut accessions. Zenodo https://doi.org/10.5281/zenodo.12475904 (2024).
Zheng, Z. Custom scripts for calculating the polymorphisms between groups (PB) and across groups (PA) from Ahh and Ahf. Zenodo https://doi.org/10.5281/zenodo.12614808 (2024).
Zheng, Z. In-house generated python script to identify and count mononucleotide microsatellites. Zenodo https://doi.org/10.5281/zenodo.12191309 (2024).
Acknowledgements
We thank the financial support from the National Key Research and Development Program (2023YFD1202800 to X.Z.), Special Project for National Supercomputing Zhengzhou Center Innovation Ecosystem Construction (201400210600 to Z. Zheng), Henan Provincial R&D Projects of Inter-regional Cooperation for Local Scientific and Technological Development Guided by Central Government (YDZX20214100004191 to B.H.), Major Science and Technology Projects of Henan Province (201300111000 and 221100110300 to X.Z.), China Agriculture Research System of Ministry of Finance People's Republic of China (MOF) and Ministry of Agriculture and Rural Affairs (MARA) (CARS-13 to X.Z.), Henan Provincial Agriculture Research System, China (S2012-5 to W.D.), the Thousand Top Talent Youth in Zhongyuan (ZYQR201912171 to Z. Zheng) and Key Research Project of the Shennong Laboratory (SN01-2022-03 to X.Z.). The work from S. Pavan was partially carried out within the framework of the Agritech National Research Center, receiving funding from the European Union Next-Generation EU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)—MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4—D.D. 1032 (17 June 2022), CN00000022 to S.P.). We would also like to express our gratitude to the Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Shandong Peanut Research Institute, and other research institutions for providing peanut germplasms for this study.
Author information
Authors and Affiliations
Contributions
Z. Zheng designed the experiments and wrote the paper. Z.S. and F.Q. prepared the DNA, performed field experiments and analyzed the candidate genes. Y.F. and K.L. analyzed genetic variation and performed GWAS. S.P. contributed to experimental design, data analysis and interpretation, and paper preparation and revision. B.H. and W.D. provided help to design the experiments. P.D. provided the wild accessions. M.T., L.S., J.X., S.H., H.L., L.Q., Z. Zhang, X.D., L.M., R.Z. and J.W. provided help in laboratory and field experiments. A.L., J.R., Y.L., C.M., C.D. and R.A.C. contributed to the analysis of genetic polymorphisms between and within subspecies. Y.B. and R.G.F.V. revised the paper and offered suggestions. X.Z. conceived and facilitated the project, developed the RIL populations and revised the paper. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Hon-Ming Lam and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15.
Supplementary Tables
Supplementary Tables 1–33.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zheng, Z., Sun, Z., Qi, F. et al. Chloroplast and whole-genome sequencing shed light on the evolutionary history and phenotypic diversification of peanuts. Nat Genet (2024). https://doi.org/10.1038/s41588-024-01876-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41588-024-01876-7