Abstract
Gene expression is tightly regulated, with many genes exhibiting cell-specific silencing when their protein product would disrupt normal cellular function1. This silencing is largely controlled by non-coding elements, and their disruption might cause human disease2. We performed gene-agnostic screening of the non-coding regions to discover new molecular causes of congenital hyperinsulinism. This identified 14 non-coding de novo variants affecting a 42-bp conserved region encompassed by a regulatory element in intron 2 of the hexokinase 1 gene (HK1). HK1 is widely expressed across all tissues except in the liver and pancreatic beta cells and is thus termed a ‘disallowed gene’ in these specific tissues. We demonstrated that the variants result in a loss of repression of HK1 in pancreatic beta cells, thereby causing insulin secretion and congenital hyperinsulinism. Using epigenomic data accessed from public repositories, we demonstrated that these variants reside within a regulatory region that we determine to be critical for cell-specific silencing. Importantly, this has revealed a disease mechanism for non-coding variants that cause inappropriate expression of a disallowed gene.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All non-clinical data analyzed during this study are included in this published article (and its Supplementary Information). Clinical and genotype data can be used to identify individuals and are therefore available only through collaboration to experienced teams working on approved studies examining the mechanisms, cause, diagnosis and treatment of diabetes and other beta cell disorders. Requests for collaboration will be considered by a steering committee following an application to the Genetic Beta Cell Research Bank (https://www.diabetesgenes.org/current-research/genetic-beta-cell-research-bank/). Contact by email should be directed to S. Flanagan (s.flanagan@exeter.ac.uk). All requests for access to data will be responded to within 14 d. Accession codes and DOI numbers for all ChIP–seq, ATAC-seq, RNA-seq and scRNA-seq datasets are provided in Supplementary Table 5. We used the Genome Reference Consortium Human Build 37 (GRCh37) to annotate genetic data (accession number GCF_000001405.13). Details of this assembly are provided at https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/. For Fig. 3 and Extended Data Figs. 3 and 5–8, further data can be found at https://github.com/owensnick/HK1FigureNotebook.jl and https://doi.org/10.5281/zenodo.6815326. Source data are provided with this paper.
Code availability
The code used in this study can be freely downloaded from https://github.com/rdemolgen/SavvySuite, https://github.com/owensnick/HK1FigureNotebook.jl, https://github.com/owensnick/GenomeFragments.jl, https://github.com/kjgaulton/pipelines/tree/master/islet_snATAC_pipeline, https://github.com/4dn-dcic/hic2cool, https://github.com/STOR-i/GaussianProcesses.jl, https://github.com/exeter-tfs/MotifScanner.jl and https://qupath.github.io.
References
Pullen, T. J. et al. Identification of genes selectively disallowed in the pancreatic islet. Islets 2, 89–95 (2010).
Spielmann, M. & Mundlos, S. Looking beyond the genes: the role of non-coding variants in human disease. Hum. Mol. Genet. 25, R157–R165 (2016).
Ferreira, C. R. The burden of rare diseases. Am. J. Med. Genet. A 179, 885–892 (2019).
McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University. Online Mendelian Inheritance in Man, OMIM https://omim.org/ (2021).
Turro, E. et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 583, 96–102 (2020).
Shashi, V. et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet. Med. 16, 176–182 (2014).
Sawyer, S. L. et al. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin. Genet. 89, 275–284 (2016).
Kapoor, R. R. et al. Clinical and molecular characterisation of 300 patients with congenital hyperinsulinism. Eur. J. Endocrinol. 168, 557–564 (2013).
Snider, K. E. et al. Genotype and phenotype correlations in 417 children with congenital hyperinsulinism. J. Clin. Endocrinol. Metab. 98, E355–E363 (2013).
Halldorsson, B. V. et al. The sequences of 150,119 genomes in the UK Biobank. Nature 607, 732–740 (2022).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Robey, R. B. & Hay, N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene 25, 4683–4696 (2006).
Becker, T. C., BeltrandelRio, H., Noel, R. J., Johnson, J. H. & Newgard, C. B. Overexpression of hexokinase I in isolated islets of Langerhans via recombinant adenovirus. Enhancement of glucose metabolism and insulin secretion at basal but not stimulatory glucose levels. J. Biol. Chem. 269, 21234–21238 (1994).
Bianchi, M. & Magnani, M. Hexokinase mutations that produce nonspherocytic hemolytic anemia. Blood Cells Mol. Dis. 21, 2–8 (1995).
Pasquali, L. et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46, 136–143 (2014).
Chiou, J. et al. Single-cell chromatin accessibility identifies pancreatic islet cell type- and state-specific regulatory programs of diabetes risk. Nat. Genet. 53, 455–466 (2021).
Greenwald, W. W. et al. Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nat. Commun. 10, 2078 (2019).
Weng, C. et al. Single-cell lineage analysis reveals extensive multimodal transcriptional control during directed beta-cell differentiation. Nat. Metab. 2, 1443–1458 (2020).
Balboa, D. et al. Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat. Biotechnol. 40, 1042–1055 (2022).
Geusz, R. J. et al. Pancreatic progenitor epigenome maps prioritize type 2 diabetes risk genes with roles in development. eLife 10, e59067 (2021).
Cebola, I. et al. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 17, 615–626 (2015).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Bramswig, N. C. et al. Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. J. Clin. Invest. 123, 1275–1284 (2013).
Lawlor, N. et al. Multiomic profiling identifies cis-regulatory networks underlying human pancreatic β cell identity and function. Cell Rep. 26, 788–801.e6 (2019).
Giri, D. et al. Novel FOXA2 mutation causes hyperinsulinism, hypopituitarism with craniofacial and endoderm-derived organ abnormalities. Hum. Mol. Genet. 26, 4315–4326 (2017).
Papizan, J. B. et al. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 25, 2291–2305 (2011).
Dhawan, S. et al. DNA methylation directs functional maturation of pancreatic beta cells. J. Clin. Invest. 125, 2851–2860 (2015).
Keller, M. P. et al. The transcription factor Nfatc2 regulates beta-cell proliferation and genes associated with type 2 diabetes in mouse and human islets. PLoS Genet. 12, e1006466 (2016).
Fang, Z. et al. Single-cell heterogeneity analysis and CRISPR screen identify key beta-cell-specific disease genes. Cell Rep. 26, 3132–3144 (2019).
Xin, Y. et al. Pseudotime ordering of single human β-cells reveals states of insulin production and unfolded protein response. Diabetes 67, 1783–1794 (2018).
Lemaire, K., Thorrez, L. & Schuit, F. Disallowed and allowed gene expression: two faces of mature islet beta cells. Annu. Rev. Nutr. 36, 45–71 (2016).
Pinney, S. E. et al. Dominant form of congenital hyperinsulinism maps to HK1 region on 10q. Horm. Res. Paediatr. 80, 18–27 (2013).
Henquin, J. C. et al. Congenital hyperinsulinism caused by hexokinase I expression or glucokinase-activating mutation in a subset of beta-cells. Diabetes 62, 1689–1696 (2013).
Otonkoski, T. et al. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic beta cells. Am. J. Hum. Genet. 81, 467–474 (2007).
Almeida, A. M. et al. Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat. Med. 12, 846–851 (2006).
Thornton, P. S. et al. Recommendations from the Pediatric Endocrine Society for evaluation and management of persistent hypoglycemia in neonates, infants, and children. J. Pediatr. 167, 238–245 (2015).
Ellard, S. et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia 56, 1958–1963 (2013).
Campbell-Thompson, M. et al. Network for Pancreatic Organ Donors with Diabetes (nPOD): developing a tissue biobank for type 1 diabetes. Diabetes Metab. Res. Rev. 28, 608–617 (2012).
Laver, T. W. et al. SavvyCNV: genome-wide CNV calling from off-target reads. PLoS Comput. Biol. 18, e1009940 (2022).
Han, B. et al. Enhanced islet cell nucleomegaly defines diffuse congenital hyperinsulinism in infancy but not other forms of the disease. Am. J. Clin. Pathol. 145, 757–768 (2016).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Abdennur, N. & Mirny, L. A. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316 (2020).
Owens, N. D. L. et al. Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep. 14, 632–647 (2016).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020).
Song, J. Y. et al. HIC2, a new transcription activator of SIRT1. FEBS Lett. 593, 1763–1776 (2019).
Luu, L. et al. The loss of Sirt1 in mouse pancreatic beta cells impairs insulin secretion by disrupting glucose sensing. Diabetologia 56, 2010–2020 (2013).
Lee, J. H., Lee, J. H. & Rane, S. G. TGF-β signaling in pancreatic islet beta cell development and function. Endocrinology 162, bqaa233 (2021).
Acknowledgements
We thank P. Leete (IBEx, University of Exeter) for technical assistance with confocal microscopy. S.E.F. has a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (105636/Z/14/Z). M.N. Wakeling and M.B.J. are the recipients of an Independent Fellowship and T.W.L. and N.D.L.O. have a lectureship from the Exeter Diabetes Centre of Excellence funded by Research England’s Expanding Excellence in England (E3) fund. E.D.F. is the recipient of a Diabetes UK RD Lawrence fellowship (19/0005971). K.A.P. has a Career Development fellowship funded by the Wellcome Trust (219606/Z/19/Z). I.B. and M.J.D. were supported by the Northern Congenital Hyperinsulinism charitable fund, by the Manchester Academic Health Sciences Centre and by the University of Manchester MRC Confidence in Concept (CiC) Award (MC_PC_18056). S.J.R. and N.G.M. are grateful to the JDRF for funding (2-SRA-2018-474-S-B). I. Barić is member of the European Reference Network for Rare Hereditary Metabolic Disorders (MetabERN) (project 739543). This research was performed with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD; RRID SCR_014641), a collaborative type 1 diabetes research project supported by the JDRF (nPOD, 5-SRA-2018-557-Q-R) and the Leona M. & Harry B. Helmsley Charitable Trust (grants 2018PG-T1D053, G-2108-04793). The content and views expressed are the responsibility of the authors and do not necessarily reflect the official view of the nPOD. Organ Procurement Organizations partnering with the nPOD to provide research resources are listed at http://www.jdrfnpod.org/for-partners/npod-partners/. This research has been conducted using the UK Biobank resource. This work was carried out under UK Biobank project numbers 9055 and 9072. Targeted next-generation sequencing to exclude known disease-causing variants was funded by Congenital Hyperinsulinism International (a 501(c)3 organization) for 20 individuals within the discovery cohort. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. This paper is dedicated to the memory of our colleague and friend, Professor Mark Dunne, who will be greatly missed. Mark was an inspirational scientist whose pioneering research served to improve the lives of children and families affected by CHI. For the purpose of open access, the author has applied a CC BY public copyright licence to any author accepted manuscript version arising from this submission.
Author information
Authors and Affiliations
Consortia
Contributions
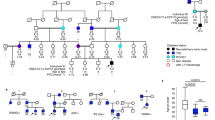
S.E.F. designed the study. A.D., S.E., I. Banerjee, S.E.F. and the International Congenital Hyperinsulinism Consortium recruited patients to the study. M.N. Wakeling, M.B.J., J.A.L.H., R.v.H., S.E. and S.E.F. performed molecular genetic analysis and interpretation of the resulting data. M.N. Wakeling and M.N. Weedon performed the bioinformatic analysis; T.I.H., J.J.H., I. Banerjee, A.T.H. and S.E.F. analyzed the clinical data. E.C. and M.J.D. performed pathological analysis of the pancreatic tissue; J.R.H., C.S.F., R.C.W., N.G.M. and S.J.R. designed and performed the immunohistochemistry studies; N.D.L.O. designed and performed epigenomic data analysis; M.N. Wakeling, N.D.L.O., J.R.H., M.B.J., T.W.L., E.D.F., K.A.P., A.T.H., M.J.D., S.J.R. and S.E.F. prepared the draft manuscript. All authors contributed to the discussion of the results and to manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks David van Heel and Christopher Wright for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
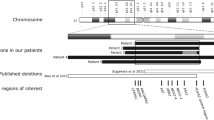
Extended Data Fig. 1 Integrative Genomics Viewer (IGV) screenshot of the ~4.5Kb deleted regions encompassing the regulatory element in intron 2 of HK1 in patients 1 and 2.1.
Both deletions are heterozygous, so reads are present in the deleted regions from the other haplotype, although the read depth is reduced. Reads highlighted in red have a long insert size. Both features are consistent with a deletion. The deleted region in patient 2.1 has detectable break points, as shown by the discontinuity in read depth and the reads clipped at the deletion boundary. In patient 1, the breakpoints do not result in read depth discontinuity or clipped reads as the deletion occurs between two matching repeating sequences.
Extended Data Fig. 2 Histological appearance of islets from HK1-CHI and ABCC8-HI tissue.
Panels a and b illustrate islets from two HK1 CHI patients (patients 1 and 6 Supplementary Table 2) in comparison to age-matched tissue from ABCC8 CHI patients with diffuse pancreatic disease (c, d). The yellow dashed line delineates a single islet in each sample. Nuclear enlargement (B, arrowhead) and nucleomegaly (C, D arrows) was virtually undetected in control islets (n = 12/72,145 islet cells) but found in 0.8% of HK1 islets (n = 374/46,885 cells) and 4.9% of ABCC8 islet cells (n = 70,929). Inserts illustrate the corresponding haematoxylin and eosin images. Scale bar 50 µm.
Extended Data Fig. 3 HK1 expression in GTEx.
a) Boxplots describing gene level HK1 expression in Genotype-Tissue Expression GTEx project ordered by expression with least expression in liver and pancreas. Data shows sum of GTEx v8 isoform level data calculated with RSEM. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 08/26/21. Boxplot central line denotes median, box limit the interquartile range, and whiskers extend to furthest point within 1.5 interquartile range, data points are outliers exceeding whiskers, median sample size n = 291, see source data/GTex for tissue sample sizes. b) Isoform level HK1 expression in GTEX tissues as a), top 3 most expressed isoforms are given. Long HK1 isoform only expressed in testis, short isoform ubiquitously expressed across all tissues. c) Transcript models of top three HK1 isoforms given in b). d) Expression of HK1 and GCK for comparison in scRNA-seq across alpha and beta-cell maturation, all data quantified by (GSE167880)20, which quantified their own data over maturation and adult endocrine cells (GSE114297)31. e) HK1 expression in human islet cell types by scRNA-seq (GSE101207)30. Data points from independent human donors, the central lines in the boxplots correspond to the median, boxes span from interquartile range, and whiskers extend to the furthest data point within 1.5xIQR from the boxes. HK1 expression only in duct and pancreatic stellate cells, expression absent in endocrine cell types.
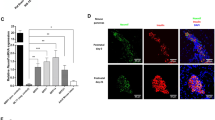
Extended Data Fig. 4 HK1 expression is present in the beta-cells of donors with HK1-CHI, but not ABCC8-Hyperinsulinism or Non-HI controls.
a) Confocal imaging of HK1 (red); glucagon (yellow); insulin (cyan) and DAPI (dark blue) in pancreatic tissue resected from patient 6 (Supplementary Table 2) with a HK1 variant. HK1 co-localises with insulin, but not with glucagon. Staining in tissue resected from an individual with ABCC8-Hyperinsulinism (HI-control) and a non-HI control, showed no expression of HK1 in either alpha (glucagon-positive) or beta (insulin-positive cells). Scale bar – 100 µm. b) Enlarged image of a portion of the islet shown in (a) from an individual with HK1-HI (Patient 6). This image confirms the expression of HK1 specifically in beta cells (white arrows) and absence of HK1 in alpha cells (orange arrows). Scale bar – 20 µm. c) HK1 expression (Median fluorescence intensity (MFI)) is higher in the HK1-HI donor beta-cells within HK1 positive islets, when compared to alpha cells (Patient 1 - beta-cells (n = 1580); alpha-cells (n = 161); and Patient 6 - beta-cells (n = 2284); alpha-cells (n = 9321)). Two-tailed Mann-Whitney (****p < 0.0001), p < 3.2 × 10−9 and p < 6.4 × 10−172 for Patient 1 and Patient 6 respectively. This suggests the impact of the HK1 variant is beta-cell specific. Violin plots shown with the median (dashed line) and IQR (dotted lines) presented. Source data provided in Supplementary Information.
Extended Data Fig. 5 Human Islet chromatin conformation at loci spanning HK1 and secreted endocrine genes.
a) Histogram of contact strengths of loops in human islet Hi-C data18, data shown for all loops called in that study. Vertical line marks the strength of strongest and smallest loop spanning HK1 gene (marked with white arrow in c)). b) Cumulative frequency plot of contacts shown in a), vertical line marks HK1 loop (white arrow in c)), the HK1 loop is in the 86th percentile of all islet HiC loop strengths. c) Heatmap of HiC data shown in a) for region ± 850 kb of HK1 as Fig. 3d, now with additional loops within region marked. White arrow gives principal loop spanning HK1 gene marked in a, b). d–f) HiC data and loops for endocrine cell secreted genes d) INS, e) SST, and f) GCG, shown at same resolution and scale as HK1 locus c) for context.
Extended Data Fig. 6 Chromatin accessibility, Transcription Factor binding and Epigenetic modifications at the HK1 locus over pancreatic differentiation.
a) snATAC-seq data in human islets showing mean chromatin accessibility over nuclei assigned to alpha, beta and delta-cell clusters in hormone high and hormone low conditions (GSE160472)17. Critical region containing variants encompassed within grey box. Alpha, beta and delta-cells each show accessibility in hormone high state, whereas only beta-cells retain accessibility in hormone low cells. Hormone high and low states characterised by level of accessibility over endocrine cell secreted hormone promoter, alpha – GCG, beta – INS, delta – SST. Data show in reads per million (RPM). b) Chromatin accessibility of pancreatic differentiation assessed by ATAC-seq (GSE149148)21, wider locus view of same data shown in Fig. 3c. Critical region containing variants encompassed within grey box. Stages: ES–embryonic stem cell, DE–definitive endoderm, Early/Late PP–pancreatic progenitor. c) Transcription factor binding in pancreatic progenitors (GSE149148)21 and in Carnegie Stage 16–18 liver bud for FOXA2 (E-MTAB-3061)22, data shown as a) and b). Liver bud data reveals that whilst FOXA2 binds critical region in pancreatic progenitors and human islets (Fig. 3b), it does not bind in liver bud, suggesting that critical region encodes pancreas specific regulation of HK1. d) Epigenetic modifications over HK1 locus, shown are active marks H3K27ac and H3K4me1 and repressive mark H3K27me3 over in vitro pancreatic cell differentiation (GSE149148)21 stages as in b) in vitro beta-cell line EndoC-BH1 (GSE118588)25 and human islets H3K27ac and H3K4me1 (E-MTAB-1919)16 and H3K27me3 (E087 – roadmap epienomics23); cell-sorted pancreatic alpha, beta and exocrine cells (GSE50386)24. Grey box encompasses critical region containing variants. Data reveals that H3K27ac broadly marks short isoform promoter (Extended Data Fig. 3) at ES cells stage is reduced by DE stage, whilst focal mark over regulatory region is present in EndoC-BH1 and human islets. Similarly, H3K4me1 shows string focal enrichment over regulatory region in EndoC-BH1 and islets. Finally, repressive mark H3K27me3 is absent over the course of pancreatic cell differentiation and is present in a broad domain in adult islets, EndoC-BH1’s, beta-cells and to reduced extent in exocrine cells.
Extended Data Fig. 7 Expression of family members of disrupted transcription factor binding motifs in critical region.
NFAT family, NKX2-2 and FOXA2: a) In single-cell RNA-seq in human islets (GSE101207)30, boxplots of mean normalised counts from independent islet donors in cells assigned to islet cell type clusters (data shown as in Extended Data Fig. 3e). Data points from independent human donors, the central lines in the boxplots correspond to the median, boxes span from interquartile range, and whiskers extend to the furthest data point within 1.5x IQR from the boxes. b) Over the course of pancreatic differentiation from embryonic stem cells to maturing beta-cells, expression data over a beta-cell differentiation psuedotime19, Gaussian process regression median and 95% CI shown. ES – embryonic stem cell, DE – definitive endoderm, GT – gut tube, FG – foregut, PE – pancreatic endoderm, EP – endocrine precursors, IB – immature beta-cells, MB – maturing beta-cells. c) scRNA-seq across alpha and beta-cell maturation, all data quantified by (GSE167880)20, which quantified their own data over maturation and adult endocrine cells (GSE114297)31.
Extended Data Fig. 8 Additional transcription factor motifs disrupted by variants.
a, b) Sanger sequencing identified 2 in cis de novo variants 12 base pairs apart (g.71,108,648 C > G and g.71,108,660 G > A) in patient 11. g.71,108,648 is affected by 3 different de novo substitutions in 6 individuals and resides within a predicted NFATC binding motif. Two further de novo variants identified in 3 patients are also predicted to impact on NFATC binding (Fig. 3). In contrast, further variants affecting g.71,108,660 have not been identified, this suggests that the g.71,108,648 C > G is causative of the disease in this patient. Nevertheless, it is of note that g.71,108,660 G > A is predicted to affect a HIC family motif a) and human islet scRNA-seq (GSE101207)30 determines HIC2 is expressed in beta-cells b) Interestingly, HIC2 is a transactivator of SIRT147 and the loss of SIRT1 impairs glucose sensing in beta-cells in mice48. c-e) Tier 2, secondary candidate motifs for disruption have lower normalised motif scores than Tier 1 matches. c) Two additional motif families are implicated TEAD and SMAD. TEAD shares partial consensus with NFAT (TTCCA) and is alternative candidate to NFAT. TEAD1 but not TEAD2 is expressed in beta-cells e) and TEAD1 plays a critical role in pancreatic progenitors22, however it should be noted that TEAD1 does not bind the critical region in pancreatic progenitors when the region is bound by FOXA2 (Fig. 3b). The SMAD family are signal transducers of TGF-beta signalling and play an important role in beta-cell development, function, and proliferation49. For all boxplots, in panels (b, d, e), central line denotes median, box limit the interquartile range, and whiskers extend to furthest point within 1.5 interquartile range.
Supplementary information
Supplementary Information
Supplementary Tables 1–5
Supplementary Table 6
Table describing the primary and secondary candidate disrupted transcription factor motifs by each variant.
Source data
Source Data Fig. 2
Mean fluorescence intensity values for HK1 expression within individual beta cells.
Source Data Extended Data Fig. 4
Mean fluorescence intensity values for HK1 expression within individual alpha and beta cells.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wakeling, M.N., Owens, N.D.L., Hopkinson, J.R. et al. Non-coding variants disrupting a tissue-specific regulatory element in HK1 cause congenital hyperinsulinism. Nat Genet 54, 1615–1620 (2022). https://doi.org/10.1038/s41588-022-01204-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01204-x
This article is cited by
-
Chromosome 20p11.2 deletions cause congenital hyperinsulinism via the loss of FOXA2 or its regulatory elements
European Journal of Human Genetics (2024)
-
Zooming into process-specific risk
Nature Metabolism (2023)
-
Noncoding variants alter GATA2 expression in rhombomere 4 motor neurons and cause dominant hereditary congenital facial paresis
Nature Genetics (2023)