Abstract
Methylation of cytosine residues in DNA, the best studied epigenetic modification, is associated with gene transcription and nuclear organization, and ultimately the function of a cell. DNA methylation can be influenced by various factors, including changes in neighbouring genomic sites such as those induced by transcription factor binding. The DNA methylation profiles in relevant cell types are altered in most human diseases compared with the healthy state. Given the physical stability of DNA and methylated DNA compared with other epigenetic modifications, DNA methylation is an ideal marker for clinical purposes. However, few DNA methylation-based markers have made it into clinical practice, with the notable exception of some markers used in the field of oncology. Autoimmune rheumatic diseases are genetically complex entities that can vary widely in terms of prognosis, subtypes, progression and treatment responses. Increasing reports showing strong links between DNA methylation profiles and different clinical outcomes and other clinical aspects in autoimmune rheumatic diseases reinforce the usefulness of DNA methylation profiles as novel clinical markers. In this Review, we provide an updated discussion on DNA methylation alterations in autoimmune rheumatic diseases and the advantages and disadvantages of using these markers in clinical practice.
Key points
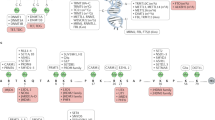
DNA methylation patterns are cell type specific, and particular patterns are associated with gene transcription and cellular function; aberrant DNA methylation profiles are associated with pathogenic cell phenotypes.
Genetic susceptibility variants and environmental cues, in conjunction with nuclear factors, can influence the DNA methylation profiles of immune cells.
Particular DNA methylation alterations are associated with the subtype, activity, progression and/or response to therapy of various autoimmune rheumatic diseases.
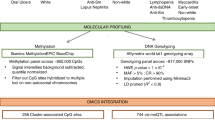
The stability of methylated cytosines, and their physical association with the DNA, make these markers ideally suited for clinical purposes.
For the future development of practical and useful DNA methylation-based markers, further studies that include large cohorts of patients and relevant cell types are needed.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Rakyan, V. K., Down, T. A., Balding, D. J. & Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12, 529–541 (2011).
Javierre, B. M. et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20, 170–179 (2010).
Zhao, M. et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J. Autoimmun. 54, 127–136 (2014).
Mok, A. et al. Hypomethylation of CYP2E1 and DUSP22 promoters associated with disease activity and erosive disease among rheumatoid arthritis patients. Arthritis Rheumatol. 70, 528–536 (2018).
Rodríguez-Ubreva, J. et al. Inflammatory cytokines shape a changing DNA methylome in monocytes mirroring disease activity in rheumatoid arthritis. Ann. Rheum. Dis. 78, 1505–1516 (2019).
Lian, X. et al. DNA demethylation of CD40L in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum. 64, 2338–2345 (2012).
Imgenberg-Kreuz, J. et al. Genome-wide DNA methylation analysis in multiple tissues in primary Sjögren’s syndrome reveals regulatory effects at interferon-induced genes. Ann. Rheum. Dis. 75, 2029–2036 (2016).
Zhao, M. et al. IL-6/STAT3 pathway induced deficiency of RFX1 contributes to Th17-dependent autoimmune diseases via epigenetic regulation. Nat. Commun. 9, 583 (2018).
Ai, R. et al. Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes. Nat. Commun. 9, 1921 (2018).
Tsou, P. S. et al. Inhibition of EZH2 prevents fibrosis and restores normal angiogenesis in scleroderma. Proc. Natl Acad. Sci. USA 116, 3695–3702 (2019).
Heyn, H., Méndez-González, J. & Esteller, M. Epigenetic profiling joins personalized cancer medicine. Expert. Rev. Mol. Diagnostics 13, 473–479 (2013).
Jones, P. A., Ohtani, H., Chakravarthy, A. & De Carvalho, D. D. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 19, 151–161 (2019).
McClay, J. L. et al. High density methylation QTL analysis in human blood via next-generation sequencing of the methylated genomic DNA fraction. Genome Biol. 16, 291 (2015).
Van Dongen, J. et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat. Commun. 7, 11115 (2016).
Imgenberg-Kreuz, J. et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 77, 736–743 (2018).
Hammaker, D. et al. LBH gene transcription regulation by the interplay of an enhancer risk allele and DNA methylation in rheumatoid arthritis. Arthritis Rheumatol. 68, 2637–2645 (2016).
Banovich, N. E. et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 10, e1004663 (2014).
Olsson, A. H. et al. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 10, e1004735 (2014).
Hannon, E. et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 19, 48–54 (2015).
Wagner, J. R. et al. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 15, R37 (2014).
Heyn, H. et al. DNA methylation contributes to natural human variation. Genome Res. 23, 1363–1372 (2013).
Michels, K. B. et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat. Methods 10, 949–955 (2013).
Gopalan, S. et al. Trends in DNA methylation with age replicate across diverse human populations. Genetics 206, 1659–1674 (2017).
Husquin, L. T. et al. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol. 19, 222 (2018).
Galanter, J. M. et al. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. eLife 6, e20532 (2017).
Rui, J. et al. Methylation of insulin DNA in response to proinflammatory cytokines during the progression of autoimmune diabetes in NOD mice. Diabetologia 59, 1021–1029 (2016).
Sanderson, S. M., Gao, X., Dai, Z. & Locasale, J. W. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat. Rev. Cancer 19, 625–637 (2019).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
McCarthy, N. S. et al. Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns. BMC Genomics 15, 981 (2014).
Golden, L. C. et al. Parent-of-origin differences in DNA methylation of X chromosome genes in T lymphocytes. Proc. Natl Acad. Sci. USA 116, 26779–26787 (2019).
Generali, E., Ceribelli, A., Stazi, M. A. & Selmi, C. Lessons learned from twins in autoimmune and chronic inflammatory diseases. J. Autoimmunity 83, 51–61 (2017).
Ulff-Møller, C. J. et al. Twin DNA methylation profiling reveals flare-dependent interferon signature and B cell promoter hypermethylation in systemic lupus erythematosus. Arthritis Rheumatol. 70, 878–890 (2018).
Gomez-Cabrero, D. et al. High-specificity bioinformatics framework for epigenomic profiling of discordant twins reveals specific and shared markers for ACPA and ACPA-positive rheumatoid arthritis. Genome Med. 8, 124 (2016).
Webster, A. P. et al. Increased DNA methylation variability in rheumatoid arthritis-discordant monozygotic twins. Genome Med. 10, 64 (2018).
Ramos, P. S. et al. Integrative analysis of DNA methylation in discordant twins unveils distinct architectures of systemic sclerosis subsets. Clin. Epigenetics 11, 58 (2019).
Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019).
Charras, A. et al. Cell-specific epigenome-wide DNA methylation profile in long-term cultured minor salivary gland epithelial cells from patients with Sjögren’s syndrome. Ann. Rheum. Dis. 76, 625–628 (2017).
Lu, Q. et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 46, 1282–1291 (2002).
Oelke, K. et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 50, 1850–1860 (2004).
Lu, Q. et al. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 179, 6352–6358 (2007).
Tseng, W. Y. et al. TNF receptor 2 signaling prevents DNA methylation at the Foxp3 promoter and prevents pathogenic conversion of regulatory T cells. Proc. Natl. Acad. Sci. USA 116, 21666–21672 (2019).
Sun, X. et al. All-trans retinoic acid induces CD4+CD25+Foxp3+ regulatory T cells by increasing Foxp3 demethylation in systemic sclerosis CD4+ T cells. J. Immunol. Res. 2018, 8658156 (2018).
Richardson, B. Effect of an inhibitor of DNA methylation on T cells. II. 5-azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum. Immunol. 17, 456–470 (1986).
Patel, D. R. & Richardson, B. C. Epigenetic mechanisms in lupus. Curr. Opin. Rheumatol. 22, 478–482 (2010).
Li, H. et al. Precision DNA demethylation ameliorates disease in lupus-prone mice. JCI Insight 3, e120880 (2018).
Zhao, M. et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J. Autoimmun. 35, 58–69 (2010).
Zhang, Q. et al. Increased Set1 binding at the promoter induces aberrant epigenetic alterations and up-regulates cyclic adenosine 5′-monophosphate response element modulator alpha in systemic lupus erythematosus. Clin. Epigenetics 8, 126 (2016).
Miller, S. et al. Hypomethylation of STAT1 and HLA-DRB1 is associated with type-I interferon-dependent HLA-DRB1 expression in lupus CD8+ T cells. Ann. Rheum. Dis. 78, 519–528 (2019).
Hedrich, C. M. et al. CAMP responsive element modulator (CREM) α mediates chromatin remodeling of CD8 during the generation of CD3+CD4−CD8− T cells. J. Biol. Chem. 289, 2361–2370 (2014).
Franks, S. E., Getahun, A., Hogarth, P. M. & Cambier, J. C. Targeting B cells in treatment of autoimmunity. Curr. Opin. Immunol. 43, 39–45 (2016).
Julià, A. et al. Epigenome-wide association study of rheumatoid arthritis identifies differentially methylated loci in B cells. Hum. Mol. Genet. 26, 2803–2811 (2017).
Absher, D. M. et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 9, e1003678 (2013).
Klein, K. & Gay, S. Epigenetics in rheumatoid arthritis. Curr. Opin. Rheumatol. 27, 76–82 (2015).
Vento-Tormo, R. et al. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 17, 4 (2016).
Garcia-Gomez, A. et al. TET2- and TDG-mediated changes are required for the acquisition of distinct histone modifications in divergent terminal differentiation of myeloid cells. Nucleic Acids Res. 45, 10002–10017 (2017).
Frank-Bertoncelj, M. et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun. 8, 14852 (2017).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. https://doi.org/10.1038/s41590-019-0378-1 (2019).
Lettre, G. & Rioux, J. D. Autoimmune diseases: insights from genome-wide association studies. Hum. Mol. Genet. 17, R116–R121 (2008).
Acosta-Herrera, M. et al. Genome-wide meta-analysis reveals shared new loci in systemic seropositive rheumatic diseases. Ann. Rheum. Dis. 78, 311–319 (2018).
Márquez, A. et al. A combined large-scale meta-analysis identifies COG6 as a novel shared risk locus for rheumatoid arthritis and systemic lupus erythematosus. Ann. Rheum. Dis. 76, 286–294 (2017).
Imgenberg-Kreuz, J. et al. Shared and unique patterns of DNA methylation in systemic lupus erythematosus and primary Sjögren’s syndrome. Front. Immunol. 10, 1686 (2019).
Carnero-Montoro, E. et al. Epigenome-wide comparative study reveals key differences between mixed connective tissue disease and related systemic autoimmune diseases. Front. Immunol. 10, 1880 (2019).
de la Rica, L. et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 14, R99 (2013).
Coit, P. et al. Epigenetic reprogramming in naive CD4+ T cells favoring T cell activation and non-Th1 effector T cell immune response as an early event in lupus flares. Arthritis Rheumatol. 68, 2200–2209 (2016).
Joseph, S. et al. Epigenome-wide association study of peripheral blood mononuclear cells in systemic lupus erythematosus: identifying DNA methylation signatures associated with interferon-related genes based on ethnicity and SLEDAI. J. Autoimmun. 96, 147–157 (2019).
Zhao, M. et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 75, 1998–2006 (2016).
Saxena, R., Mahajan, T. & Mohan, C. Lupus nephritis: current update. Arthritis Res. Ther. 13, 240 (2011).
Van Der Helm-Van Mil, A. H. M. et al. A prediction rule for disease outcome in patients with recent-onset undifferentiated arthritis: how to guide individual treatment decisions. Arthritis Rheum. 56, 433–440 (2007).
Kocijan, R., Harre, U. & Schett, G. ACPA and bone loss in rheumatoid arthritis. Curr. Rheumatol. Rep. 15, 366 (2013).
Daha, N. A. & Toes, R. E. M. Rheumatoid arthritis: are ACPA-positive and ACPA-negative RA the same disease? Nat. Rev. Rheumatol. 7, 202–203 (2011).
Renauer, P. et al. DNA methylation patterns in naïve CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus Sci. Med. 2, e000101 (2015).
Coit, P. et al. Renal involvement in lupus is characterized by unique DNA methylation changes in naïve CD4+ T cells. J. Autoimmun. 61, 29–35 (2015).
Breitbach, M., Ramaker, R. C., Roberts, K., Kimberly, R. P. & Absher, D. Population-Specific Patterns of Epigenetic Defects in the B Cell Lineage in Patients With Systemic Lupus Erythematosus. Arthritis Rheumatol. 72, 282–291 (2019).
Coit, P. et al. Ethnicity-specific epigenetic variation in naïve CD4+ T cells and the susceptibility to autoimmunity. Epigenetics Chromatin 8, 49 (2015).
Altorok, N., Tsou, P. S., Coit, P., Khanna, D. & Sawalha, A. H. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheum. Dis. 74, 1612–1620 (2015).
Nair, N. et al. Differential DNA methylation correlates with response to methotrexate in rheumatoid arthritis. Rheumatology 59, 1364–1371 (2019).
Gosselt, H. R. et al. Higher baseline global leukocyte DNA methylation is associated with MTX non-response in early RA patients. Arthritis Res. Ther. 21, 157 (2019).
Glossop, J. R. et al. DNA methylation at diagnosis is associated with response to disease-modifying drugs in early rheumatoid arthritis. Epigenomics 9, 419–428 (2017).
Plant, D. et al. Differential methylation as a biomarker of response to etanercept in patients with rheumatoid arthritis. Arthritis Rheumatol. 68, 1353–1360 (2016).
Chappell, L., Russell, A. J. C. & Voet, T. Single-cell (multi)omics technologies. Annu. Rev. Genomics Hum. Genet. 19, 15–41 (2018).
Der, E. et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. 20, 915–927 (2019).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Rahmani, E. et al. Cell-type-specific resolution epigenetics without the need for cell sorting or single-cell biology. Nat. Commun. 10, 3417 (2019).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
Lähnemann, D. et al. Eleven grand challenges in single-cell data science. Genome Biol. 21, 31 (2020).
Bock, C. et al. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat. Biotechnol. 34, 726–737 (2016).
Wang, Q. et al. Tagmentation-based whole-genome bisulfite sequencing. Nat. Protoc. 8, 2022–2032 (2013).
Gu, H. et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6, 468–481 (2011).
Acknowledgements
The authors thank O. Morante-Palacios and C. de la Calle-Fabregat for their help with the figures. The authors also thank CERCA Programme/Generalitat de Catalunya and the Josep Carreras Foundation for institutional support. E.B. is funded by the Ministry of Science, Innovation and Universities (MCIU) (grant numbers SAF2017-88086-R; AEI/FEDER, UE). Q.L. is funded by the National Natural Science Foundation of China (grant numbers 81830097 and 81861138016) and the Research and Development Plan of key areas in Hunan Province (grant number 2019WK2081). A.H.S is funded by the National Institutes of Health (grants numbers R01AI097134 and R01AR070148) and the Lupus Research Alliance.
Author information
Authors and Affiliations
Contributions
E.B. wrote the article. All authors researched data for the article, provided substantial contributions to discussions of content and reviewed and/or edited the article before submission.
Corresponding author
Additional information
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Rheumatology thanks C. Hedrich, M. Alarcon-Riquelme and P. Ramos for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- DNA methylation
-
The covalent attachment of a methyl group to a DNA residue (most commonly to a cytosine nucleotide followed by a guanine nucleotide, known as CpG sites).
- Methylation quantitative trait loci
-
Genetic variants that are associated with DNA methylation levels at particular CpG sites.
- Synovial fibroblasts
-
Also known as fibroblast-like synoviocytes, synovial fibroblasts are the main stromal cells of the joint synovium. These cells produce the extracellular matrix components of the synovial fluid and are critical for cartilage integrity and lubrication of the joint.
- Regulatory T cells
-
(Treg cells). A subpopulation of T cells that are immunosuppressive and generally suppress proliferation of effector T cells; these cells are essential for preventing autoimmune disease.
- Genome-wide association studies
-
Observational studies of genome-wide sets of genetic variants in different individuals to identify variants associated with a trait; these studies typically focus on associations between single nucleotide polymorphisms and the occurrence of a disease.
- Differentially methylated region
-
Genomic regions with a differential DNA methylation status across different biological samples. Differentially methylated regions usually involve adjacent sites or a group of sites close together.
- Whole-genome bisulfite sequencing
-
A next-generation sequencing-based method for assessing the DNA methylation level of all cytosines in a genome, using sodium bisulfite treatment and DNA sequencing.
- Reduced representation bisulfite sequencing
-
A method for analysing the DNA methylation profiles of genome regions that have a high CpG content using a combination of restriction enzyme digestion and bisulfite next-generation sequencing.
- DNA methylation bead arrays
-
A high-throughput microarray-based method for measuring DNA methylation levels at single CpG site resolution.
- Bisulfite pyrosequencing
-
A method designed to quantitatively determine the methylation status of individual cytosines in CpG sites from short PCR amplicons of DNA pre-treated with sodium bisulfite.
- Epigenome-wide association study
-
An observational study examining a genome-wide set of epigenetic marks, including DNA methylation, and their association with a particular phenotype or trait (such as the occurrence of a disease).
Rights and permissions
About this article
Cite this article
Ballestar, E., Sawalha, A.H. & Lu, Q. Clinical value of DNA methylation markers in autoimmune rheumatic diseases. Nat Rev Rheumatol 16, 514–524 (2020). https://doi.org/10.1038/s41584-020-0470-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41584-020-0470-9
This article is cited by
-
A comparison of feature selection methodologies and learning algorithms in the development of a DNA methylation-based telomere length estimator
BMC Bioinformatics (2023)
-
Cross-tissue correlations of genome-wide DNA methylation in Japanese live human brain and blood, saliva, and buccal epithelial tissues
Translational Psychiatry (2023)
-
Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential
Experimental & Molecular Medicine (2023)
-
TET (Ten-eleven translocation) family proteins: structure, biological functions and applications
Signal Transduction and Targeted Therapy (2023)
-
Demethylation of CDKN2A in systemic lupus erythematosus and rheumatoid arthritis: a blood biomarker for diagnosis and assessment of disease activity
Clinical Rheumatology (2023)