Abstract
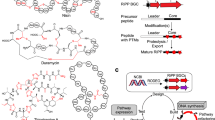
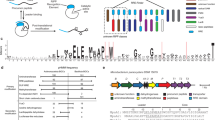
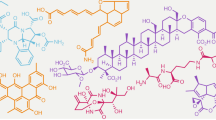
The combination of next-generation DNA sequencing technologies and bioinformatics has revitalized natural product discovery. Using a bioinformatic search strategy, we recently identified ∼600 gene clusters in otherwise overlooked streptococci that code for ribosomal peptide natural products synthesized by radical S-adenosylmethionine enzymes. These grouped into 16 subfamilies and pointed to an unexplored microbiome biosynthetic landscape. Here we report the structure, biosynthesis and function of one of these natural product groups, which we term enteropeptins, from the gut microbe Enterococcus cecorum. We show three reactions in the biosynthesis of enteropeptins that are each catalysed by a different family of metalloenzymes. Among these, we characterize the founding member of a widespread superfamily of Fe–S-containing methyltransferases, which, together with an Mn2+-dependent arginase, installs N-methylornithine in the peptide sequence. Biological assays with the mature product revealed bacteriostatic activity only against the producing strain, extending an emerging theme of fratricidal or self-inhibitory metabolites in microbiome firmicutes.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The Uniprot ID for KgrC is S1RLF6. Plasmids and strains used in this study are described in Supplementary Table 2. All oligonucleotides are shown in Supplementary Table 3. The sequences of codon-optimized gene fragments are provided in Supplementary Note 1. Other relevant data supporting the findings of this study are available within the paper and the supplementary material. NMR spectra are included in Supplementary Information. Raw NMR data used to elucidate natural product structures are available from the corresponding author upon reasonable request.
References
Fischbach, M. A. & Walsh, C. T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006).
Montalbán-López, M. et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 38, 130–239 (2021).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014).
Bushin, L. B., Clark, K. A., Pelczer, I. & Seyedsayamdost, M. R. Charting an unexplored streptococcal biosynthetic landscape reveals a unique peptide cyclization motif. J. Am. Chem. Soc. 140, 17674–17684 (2018).
Russell, A. H. & Truman, A. W. Genome mining strategies for ribosomally synthesised and post-translationally modified peptides. Comput. Struct. Biotechnol. J. 18, 1838–1851 (2020).
Hetrick, K. J. & van der Donk, W. A. Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr. Opin. Chem. Biol. 38, 36–44 (2017).
Kloosterman, A. M., Medema, M. H. & van Wezel, G. P. Omics-based strategies to discover novel classes of RiPP natural products. Curr. Opin. Biotechnol. 69, 60–67 (2021).
Tietz, J. I. & Mitchell, D. A. Using genomics for natural product structure elucidation. Curr. Top. Med. Chem. 16, 1645–1694 (2016).
Schramma, K. R., Bushin, L. B. & Seyedsayamdost, M. R. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nature Chem 7, 431–437 (2015).
Ibrahim, M. et al. Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J. Bacteriol. 189, 8844–8854 (2007).
Broderick, J. B., Duffus, B. R., Duschene, K. S. & Shepard, E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014).
Landgraf, B. J., McCarthy, E. L. & Booker, S. J. Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 85, 485–514 (2016).
Bassler, B. L. & Losick, R. Bacterially speaking. Cell 125, 237–246 (2006).
Whiteley, M., Diggle, P. S. & Greenberg, E. P. Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320 (2017).
Rued, B. E. et al. Quorum sensing in Streptococcus mutans regulates production of tryglysin, a novel RaS-RiPP antimicrobial compound. mBio 12, e02688–20 (2021).
Bushin, L. B., Covington, B. C., Rued, B. E., Federle, M. J. & Seyedsayamdost, M. R. Discovery and biosynthesis of streptosactin, a sactipeptide with an alternative topology encoded by commensal bacteria in the human microbiome. J. Am. Chem. Soc. 142, 16265–16275 (2020).
Caruso, A. & Seyedsayamdost, M. R. Radical SAM enzyme QmpB installs two 9-membered ring sactionine macrocycles during biogenesis of a ribosomal peptide natural product. J. Org. Chem. 86, 11284–11289 (2021).
Clark, K. A., Bushin, L. B. & Seyedsayamdost, M. R. Aliphatic ether bond formation expands the scope of radical SAM enzymes in natural product biosynthesis. J. Am. Chem. Soc. 141, 10610–10615 (2019).
Caruso, A., Bushin, L. B., Clark, K. A., Martinie, R. J. & Seyedsayamdost, M. R. Radical approach to enzymatic β-thioether bond formation. J. Am. Chem. Soc. 141, 990–997 (2019).
Caruso, A., Martinie, R. J., Bushin, L. B. & Seyedsayamdost, M. R. Macrocyclization via an arginine-tyrosine crosslink broadens the reaction scope of radical S-adenosylmethionine enzymes. J. Am. Chem. Soc. 141, 16610–16614 (2019).
Burkhart, B. J., Hudson, G. A., Dunbar, K. L. & Mitchell, D. A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 11, 564–570 (2015).
Kanyo, Z. F., Scolnick, L. R., Ash, D. E. & Christianson, D. W. Structure of a unique binuclear manganese cluster in arginase. Nature 383, 554–557 (1996).
Ash, D. E., Cox, J. D. & Christianson, D. W. Arginase: a binuclear manganese metalloenzyme. Met. Ions Biol. Syst. 37, 407–428 (2000).
Jung, A., Chen, L. R., Suyemoto, M. M., Barnes, H. J. & Borst, L. B. A review of Enterococcus cecorum infection in poultry. Avian Dis. 62, 261–271 (2018).
Do, T., Page, J. E. & Walker, S. Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes. J. Biol. Chem. 295, 3347–3361 (2020).
Agar, J. N. et al. IscU as a scaffold for iron–sulfur cluster biosynthesis: sequential assembly of [2Fe–2S] and [4Fe–4S] Clusters in IscU. Biochemistry 39, 7856–7862 (2000).
Davis, K. M. et al. Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition. Proc. Natl Acad. Sci. USA 114, 10420–10425 (2017).
Schramma, K. R. & Seyedsayamdost, M. R. Lysine–tryptophan-crosslinked peptides produced by radical SAM enzymes in pathogenic streptococci. ACS Chem. Biol. 12, 922–927 (2017).
Himes, P. M., Allen, S. E., Hwang, S. & Bowers, A. A. Production of sactipeptides in Escherichia coli: probing the substrate promiscuity of subtilosin A biosynthesis. ACS Chem. Biol. 11, 1737–1744 (2016).
Hudson, G. A. et al. Bioinformatic mapping of radical S-adenosylmethionine-dependent ribosomally synthesized and post-translationally modified peptides identifies new Cα, Cβ, and Cγ-linked thioether-containing peptides. J. Am. Chem. Soc. 141, 8228–8238 (2019).
Tronick, S. R. & Martinez, R. J. Methylation of the flagellin of Salmonella typhimurium. J. Bacteriol. 105, 211–219 (1971).
Stocker, B. A. D., McDonough, M. W. & Ambler, R. P. A gene determining presence or absence of e-N-methyllysine in Salmonella flagellar protein. Nature 189, 556–558 (1961).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Lankau, T., Kuo, T. N. & Yu, C. H. Computational study of the degradation of S-adenosyl methionine in water. J. Phys. Chem. A 121, 505–514 (2017).
Salyan, M. E. K. et al. A general liquid chromatography/mass spectroscopy-based assay for detection and quantitation of methyltransferase activity. Anal. Biochem. 349, 112–117 (2006).
Fleuchot, B. et al. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80, 1102–1119 (2011).
Chang, J. C., LaSarre, B., Jimenez, J. C., Aggarwal, C. & Federle, M. J. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 7, e1002190 (2011).
Flühe, L. & Marahiel, M. A. Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis. Curr. Opin. Chem. Biol. 17, 605–612 (2013).
Chiumento, S. et al. Ruminococcin C, a promising antibiotic produced by a human gut symbiont. Sci. Adv. 5, eaaw9969 (2019).
Balty, C. et al. Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus gnavus. J. Biol. Chem. 294, 14512–14525 (2019).
Rea, M. C. et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl Acad. Sci. USA 107, 9352–9357 (2010).
Lee, H., Churey, J. J. & Worobo, R. W. Biosynthesis and transcriptional analysis of thurincin H, a tandem repeated bacteriocin genetic locus, produced by Bacillus thuringiensis SF361. FEMS Microbiol. Lett. 299, 205–213 (2009).
Mo, T. et al. Thuricin Z: a narrow-spectrum sactibiotic that targets the cell membrane. Agnew. Chem. Int. Ed. 58, 18793–18797 (2019).
Kawulka, K. E. et al. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 125, 4726–4727 (2003).
Kawulka, K. E. et al. Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to α-carbon cross-links: formation and reduction of α-thio-α-amino acid derivatives. Biochemistry 43, 3385–3395 (2004).
Flühe, L. et al. Two [4Fe–4S] clusters containing radical SAM Enzyme SkfB catalyze thioether bond formation during the maturation of the sporulation killing factor. J. Am. Chem. Soc. 135, 959–962 (2013).
Lui, W.-T. et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl Acad. Sci. USA 107, 16286–16290 (2010).
Flühe, L. et al. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357 (2012).
Bösch, N. M. et al. Landornamides: antiviral ornithine-containing ribosomal peptides discovered through genome mining. Angew. Chem. Int. Ed. 59, 11763–11768 (2020).
Mordhorst, S., Morinaka, B. I., Vagstad, A. L. & Piel, J. Posttranslationally acting arginases provide a ribosomal route to non-proteinogenic ornithine residues in diverse peptide sequences. Angew. Chem. Int. Ed. 59, 21442–21447 (2020).
Walsh, C. T. Blurring the lines between ribosomal and nonribosomal peptide scaffolds. ACS Chem. Biol. 9, 1653–1661 (2014).
Agarwalla, S., Kealey, J. T., Santi, D. V. & Stroud, R. M. Characterization of the 23S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 277, 8835–8840 (2002).
Lee, T. T., Agarwalla, S. & Stroud, R. M. Crystal structure of RumA, an iron–sulfur cluster containing E. coli ribosomal RNA 5-methyluridine methyltransferase. Structure 12, 397–407 (2004).
Benjdia, A. et al. Post-translational modification of ribosomally synthesized peptides by a radical SAM epimerase in Bacillus subtilis. Nature Chem 9, 698–707 (2017).
Chen, Y., Wang, J., Li, G., Yang, Y. & Ding, W. Current advancements in sactipeptide natural products. Front. Chem. 9, 595991 (2021).
Claverys, J.-P., Martin, B. & Håvarstein, L. S. Competence-induced fratricide in streptococci. Mol. Microbiol. 64, 1423–1433 (2007).
Claverys, J.-P. & Håvarstein, L. S. Cannibalism and fratricide: mechanisms and raisons d’être. Nat. Rev. Microbiol. 5, 219–229 (2007).
Bushin, L. B. & Seyedsayamdost, M. R. Guidelines for determining the structures of radical SAM enzyme-catalyzed modifications in the biosynthesis of RiPP natural products. Methods Enzymol. 606, 439–460 (2018).
Gerlt, J. A. et al. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochimica et Biophysica 1854, 1019–1037 (2015).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Madeira, F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
van de Rijn, I. & Kessler, R. E. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun. 27, 444–448 (1980).
Acknowledgements
We thank the Edward C. Taylor-Eli Lilly Fellowship in Chemistry (to K.A.C.) and the National Science Foundation (NSF CAREER Award 1847932 to M.R.S.) for support of this work.
Author information
Authors and Affiliations
Contributions
K.A.C. and M.R.S. conceived of the study. K.A.C. carried out in vitro characterization and structural elucidation of the reactions of all enzymes. B.C.C. identified the mature natural products from the native organism. K.A.C. and B.C.C. carried out bioactivity assays. K.A.C. and M.R.S. wrote the manuscript, with contributions from B.C.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Qi Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note 1, Tables 1–18 and Figs. 1–15.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Clark, K.A., Covington, B.C. & Seyedsayamdost, M.R. Biosynthesis-guided discovery reveals enteropeptins as alternative sactipeptides containing N-methylornithine. Nat. Chem. 14, 1390–1398 (2022). https://doi.org/10.1038/s41557-022-01063-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-022-01063-3