Abstract
During pregnancy the maternal–fetal interface plays vital roles in fetal development. Its disruption is frequently found in pregnancy complications. Recent studies show increased incidences of adverse pregnancy outcomes in patients with COVID-19; however, the mechanism remains unclear. Here we analysed the molecular impacts of SARS-CoV-2 infection on the maternal–fetal interface. Generating bulk and single-nucleus transcriptomic and epigenomic profiles from patients with COVID-19 and control samples, we discovered aberrant immune activation and angiogenesis patterns in distinct cells from patients. Surprisingly, retrotransposons were also dysregulated in specific cell types. Notably, reduced enhancer activities of LTR8B elements were functionally linked to the downregulation of pregnancy-specific glycoprotein genes in syncytiotrophoblasts. Our findings revealed that SARS-CoV-2 infection induced substantial changes to the epigenome and transcriptome at the maternal–fetal interface, which may be associated with pregnancy complications.
Similar content being viewed by others
Main
The coronavirus disease 2019 (COVID-19) pandemic, brought on by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, has afflicted more than 760 million people (WHO (https://covid19.who.int), May 2023). Infection during pregnancy is associated with adverse outcomes including preeclampsia, pre-term birth and stillbirth1,2. Although subpopulations of trophoblasts co-express the angiotensin-converting enzyme 2 (ACE2) receptor and transmembrane serine protease 2 (TMPRSS2) in early pregnancy3,4, placental infection and vertical viral transmission are rare5. Therefore, pregnancy complications are likely to result from maternal immune responses.
During pregnancy a dynamic balance is maintained between pathogen defence and preserving the fetus. This primarily occurs at the maternal–fetal interface (MFI), where fetal cells extensively invade and interact with maternal decidua6. About 40% of the maternal decidual cells during early pregnancy are immune cell types7. Altered composition and functionality of these cells underlie various pathologies8,9.
In the placenta retrotransposons provide an abundant source of functional sequences for host genomes10. Retrovirus-derived genes, including the SYNCYTINs, are necessary for normal placenta development and function11,12,13. Moreover, retrotransposons pervasively shape placental cis-regulatory element (CRE) landscapes across species14,15,16. Strikingly, viral infections, including SARS-CoV-2, are associated with aberrant derepression of retrotransposons17, which may cause widespread transcriptional dysregulation.
The molecular mechanism of COVID-19-associated pregnancy complications is unclear. Given the complex makeup of the MFI, cell type-specific analysis is necessary. Although there have been single-nucleus transcriptomic studies on placental samples from patients infected with COVID-19 (refs. 18,19,20), the associated epigenomic alterations remain unknown. Here we investigated the cell type-specific molecular dysregulation at the MFI in patients infected with SARS-CoV-2 by mapping the transcriptomes and epigenomes at both bulk and single-nucleus resolutions. We detected global transcriptomic and epigenomic changes in patients, which included misregulation of immune-response and angiogenesis genes. Interestingly, we discovered dysregulated retrotransposons, specifically LTR8B-derived enhancers, which were linked to the reduced expression of pregnancy-specific glycoprotein (PSG) genes. Collectively, we generated extensive profiles from the MFI of patient and control samples, and highlighted the involvement of epigenetic regulation of CREs and retrotransposons in COVID-19-related pregnancy complications.
Results
Multi-omic profiling of the MFI in patients with COVID-19
To elucidate molecular changes at the MFI following SARS-CoV-2 infection, we assessed the transcriptome and chromatin states of patient and control samples at both bulk and single-nucleus levels (Fig. 1a). We enrolled seven patients who were infected with SARS-CoV-2 during pregnancy and seven healthy pregnant donors (Extended Data Fig. 1 and Supplementary Tables 1 and 2). The patients tested positive for SARS-CoV-2 by quantitative PCR with reverse transcription (RT–qPCR; Ct ≤ 35) during late pregnancy (31.6–39.6 weeks). Among the patients, six displayed mild symptoms and one had severe symptoms, requiring admission to the intensive care unit and invasive ventilation. All MFI samples tested negative for SARS-CoV-2 by RT–qPCR (Supplementary Table 3) and as well as N protein staining (Extended Data Fig. 2a). We performed sensitivity analyses to confirm that factors including modes of delivery and gestational ages at delivery did not significantly impact the transcriptomic profiles (Extended Data Fig. 2b,c).
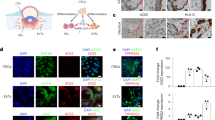
a, Schematic of the experiment design. Frozen tissue samples were pulverized and subjected to RNA extraction for stranded total RNA-seq. The remaining tissue powder underwent nuclei preparation for the following assays: bulk ATAC-seq, snRNA-seq, snATAC-seq and CUT&Tag for H3K27ac and H3K27me3 histone modifications. We then carried out integrative analysis on the multi-omic datasets derived from patient and control MFI samples. b, UMAP of the snRNA-seq showing 21 identified cell types. c, Bubble plot of marker genes for each of the 21 cell types identified in the snRNA-seq. The size of the bubbles represents the percentage of cells within a cluster that expresses the marker gene and the colour of the bubble represents the average expression level of the marker gene calculated by Seurat. d, UMAP of the snATAC-seq showing nine identified cell types. b,d, Each dot represents a nucleus and the colours correspond to its annotated cell type. e, Heatmap of the marker gene scores calculated by ArchR for all nine cell types in the snATAC-seq datasets. Each column represents the gene score of a marker gene and each row represents a cell type. Ctrl, control and Cov, COVID-19; the cell-type abbreviations in c–e are defined in b.
We conducted single-nucleus RNA-sequencing (snRNA-seq) on all patient and control samples (Supplementary Table 4). After filtering we analysed 62,132 nuclei (patients, n = 27,480 and controls, n = 34,652). Sequencing reads from each nucleus were mapped to both the human GRCh38/hg38 reference and the SARS-CoV-2 genomes21. To delineate cell-type identities, we performed unsupervised clustering with uniform manifold approximation and projection (UMAP). We identified 21 distinct cell types in the patient and control samples, which aligned with known cell categories at the MFI: trophoblasts, T cells, B cells, NK cells, macrophages, dendritic cells, endothelial cells, perivascular cells, fibroblasts and stroma cells (Fig. 1b,c, Extended Data Fig. 2d,e and Supplementary Fig. 1a). Concordant with the RT–qPCR and staining results, we found no detectable enrichment of viral transcripts in any samples (Supplementary Fig. 1b). Consistent with recent findings18, ACE2 and TMPRSS2 were expressed at low-to-undetectable levels in all term samples (Extended Data Fig. 2f).
To investigate gene regulation, we mapped the chromatin accessibility at the MFI by single-nucleus assay for transposase-accessible chromatin with sequencing (snATAC-seq; Supplementary Table 4). We profiled 68,786 individual nuclei from six patients and seven control samples (patients, n = 22,926 and controls, n = 45,860; Fig. 1a). Based on these maps, we defined nine cell types, including trophoblasts, immune cells, endothelial cells and fibroblasts (Fig. 1d). Each cell type showed distinct open chromatin patterns at the transcriptional start sites of marker genes (Fig. 1e and Extended Data Fig. 2g). Similar to snRNA-seq, syncytiotrophoblasts (STs) were the most abundantly profiled (Extended Data Fig. 2h).
To increase the depth of our epigenomic analysis, we also conducted RNA-seq, ATAC-seq and CUT&Tag targeting acetylation and tri-methylation of histone H3 lysine 27 (H3K27ac and H3K27me3) modifications on bulk patient and control samples (Fig. 1a and Supplementary Table 4). H3K27ac is a key histone modification found at active CREs and H3K27me3 is an important repressive mark regulating development genes22. We investigated whether epigenomic pathways are impacted by COVID-19. We observed a high correlation between the pseudo-bulk single-nucleus and bulk assays, confirming reproducibility between different data modalities (Supplementary Fig. 1c,d and Supplementary Data). Together, we generated extensive single-nucleus and bulk multi-omic profiles of the MFI from control and SARS-CoV-2-infected study participants, which we utilized for subsequent integrative analyses.
COVID-19 impacts the transcriptome and epigenome of the MFI
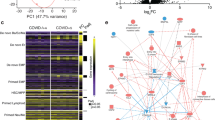
Next, we investigated the cell type-specific molecular changes at the MFI following SARS-CoV-2 infection (Supplementary Tables 5 and 6). We identified hundreds of differentially expressed genes (DEGs) from the bulk RNA-seq of patients (Fig. 2a; upregulated, n = 211 and downregulated, n = 605), suggesting extensive transcriptomic dysregulation. We discovered downregulation of placenta developmental and pregnancy-related genes, including PLAC1 and several PSG genes, whereas immune-related and angiogenesis genes such as IFI6, IFI27, CSF3 and VEGFA were upregulated (Fig. 2a). From snRNA-seq, we further found cell type-specific DEGs (Fig. 2b and Extended Data Fig. 3a). Selected DEGs were validated by immunohistochemical staining (Extended Data Fig. 3b). We noted a high consistency of DEG expression across individual patients (Extended Data Fig. 3c). As expected, some DEGs identified in the bulk data were ubiquitously dysregulated across different cell types—for example, IFI6—whereas others were contributed by specific cell types—for example, the upregulation of VEGFA was mainly observed in stroma cells and fibroblasts (Extended Data Fig. 3d and Supplementary Fig. 2a).
a, Dysregulated genes from the bulk RNA-seq. Each dot represents a gene. The negative log10-transformed two-tailed Wald test P values adjusted for multiple testing using the Benjamini and Hochberg method (Padj) and log2-transformed fold change (patient/control) were calculated by DESeq2. Upregulated (Padj < 0.05 and log2(fold change) > 1; n = 211) and downregulated (Padj < 0.05 and log2(fold change) < −1; n = 605) genes are labelled in red and blue, respectively. Dashed lines represent the indicated thresholds. b, Heatmap showing dysregulated genes from the snRNA-seq. For each cell type, upregulated and downregulated genes (one-tailed MAST model Padj < 0.05 and log2(fold change) > 0.25) were called by comparing the patient and control samples using Seurat. c, MA plot showing differentially accessible regions from the bulk ATAC-seq. Each dot represents a peak. The negative log2-transformed fold change (patient/control) and log2(average signal, baseMean) were calculated by DESeq2. Increased (two-tailed Wald test Padj < 0.01 and log2(fold change) > 1; n = 8,223) and decreased (two-tailed Wald test Padj < 0.001 and log2(fold change) < −1; n = 7,142) peaks are labelled in red and blue, respectively. d, Differentially accessible regions from the snATAC-seq. For each cell type, differential peaks (one-tailed Poisson test Padj < 0.01, fold change > 1.5 and reads per kilobase per million mapped reads (RPKM) > 1 in the tested group) were called by comparing the patient and control samples. b,d, Each row represents a cell type from a patient or control and each column represents a gene (b) or differentially accessible region (d). The colour scale indicates the calculated average gene expression (b) or RPKM (d) column z-score. e, Epigenetic modifiers that were differentially expressed between patient and control samples for each cell type from the snRNA-seq. The Padj was calculated using the one-tailed MAST model from Seurat. Ctrl, control and Cov, COVID-19; cell-type abbreviations as per Fig. 1b.
Epigenomic analysis revealed thousands of regions with altered chromatin accessibility or aberrant H3K27ac/H3K27me3 enrichment (Fig. 2c, Extended Data Fig. 4a and Supplementary Fig. 2b), indicating that chromatin states were impacted by SARS-CoV-2 infection. Cell type-specific differentially accessible regions were defined by snATAC-seq (Fig. 2d, Extended Data Fig. 4b and Supplementary Fig. 2c). Intriguingly, regions with increased chromatin accessibility were enriched with CTCF motifs, suggesting involvement with cis-regulatory higher-order chromatin interactions (Extended Data Fig. 4c). Concordant with the downregulation of pregnancy-related genes, loci with reduced chromatin accessibility or H3K27ac enrichment harboured placental transcription factor motifs, including TEAD4 and GRHL2 (Extended Data Fig. 4d). In line with the upregulation of interferon genes, regions with increased H3K27ac were enriched with motifs for interferon regulatory factors (Extended Data Fig. 4d).
To investigate the non-cell-autonomous epigenomic disruptions by COVID-19 (ref. 23), we analysed the expression of epigenetic modifying enzymes and identified 17 factors that were differentially expressed in at least one cell type (Fig. 2e). Concordant with H3K27ac and H3K27me3 enrichment changes in patients, we observed significant dysregulation of histone deacetylase (HDACs; HDAC4, HDAC5 and HDAC8) and H3K27 histone methyltransferase (enhancer of zeste homolog 2, EZH2) genes in different cell types (Fig. 2e). Notably, STs exhibited altered expression of additional chromatin modifiers, including the upregulation of nuclear receptor coactivator 1 (NCOA1; Fig. 2e and Extended Data Fig. 4e), which can be transcriptionally induced by interferon treatment24. Our results suggest that SARS-CoV-2 infection potentially disrupts host epigenetic pathways via non-cell-autonomous manners like activation of immune responses. Such transcriptomic and epigenomic effects at the MFI can influence angiogenesis and other important processes following SARS-CoV-2 infection during pregnancy.
Aberrant immune activation at the MFI following infection
Cytokine storms and adverse immune-related outcomes were previously reported in patients with COVID-19 (ref. 25). We investigated whether an analogous phenomenon was present at the MFI of patients. We detected upregulation of interferon-related genes in the patient samples (Fig. 2a). Correspondingly, upregulated genes were enriched with gene ontology (GO) terms relating to interferon signalling, inflammatory response and defence against viruses (Fig. 3a–c and Extended Data Fig. 5a). De-repressed loci that lost H3K27me3 in patients were associated with GO terms for inflammatory responses and interferon pathways (Extended Data Fig. 5b). Similarly, regions with increased H3K27ac were enriched with interferon regulatory factor binding motifs (Extended Data Fig. 4d). Conversely, loci with reduced chromatin accessibility were associated with GO terms related to negative regulation of immune cells (Extended Data Fig. 5c), in line with aberrant immune activation in patients.
a–c, GO analysis of upregulated genes from snRNA-seq in dendritic (a), Hofbauer (b) and M2 macrophage (c) cells. The size and colour of the bubble represent the number of upregulated genes and the ratio to total genes under each GO term, respectively. P values were calculated using a one-tailed hypergeometric test. d, Hierarchically clustered heatmap showing the log2(fold change) of selected interferon-induced gene expression in immune cell types from the snRNA-seq. e, Genome browser screenshot of the genomic region containing IFI27. The pseudo-bulk tracks for patient and control samples of immune and trophoblast cell types from the snRNA-seq are displayed as reads per million (RPM). The y axes for all tracks range from 0 to 75. f, Change in CellPhoneDB receptor–ligand interactions. Each row and column correspond to a cell type from the snRNA-seq and the colour indicates the log2(fold change) of the number of interactions between the patient and control samples. Ctrl, control and Cov, COVID-19; cell-type abbreviations as per Fig. 1b.
From the snRNA-seq analysis, most interferon-inducible (IFI) and interferon-induced transmembrane (IFITM) genes were upregulated in at least one immune cell type (Fig. 3d), which was confirmed in the bulk RNA-seq and by RT–qPCR (Extended Data Fig. 5d,e). For instance, IFI27, which encodes for an interferon-α-inducible protein, was upregulated in most immune cell types and trophoblasts in patient samples (Figs. 2a and 3d,e). Interestingly, overexpression of IFI27 in peripheral blood cells was reported as a potential biomarker for SARS-CoV-2 infection26. Our results suggest that IFI27 upregulation could be a non-cell type-restrictive response following SARS-CoV-2 infection and is associated with epigenomic reprogramming in patients.
To further characterize the dysregulation of immune-related pathways, we employed CellPhoneDB to find alterations in receptor–ligand interactions within our snRNA-seq datasets27. CellPhoneDB predicts cell population interactions by measuring the expression of ligands and receptors of all known pairs across distinct cell types27. We detected altered interactions relative to controls in multiple cell types of patients, including interactions between different immune cell types as well as interactions between immune and other cell types, which is indicative of immune dysregulation (Fig. 3f). We then filtered for patient-specific receptor–ligand interactions in which the ligand expression is upregulated in particular cell types. Interestingly, we found those COVID-19-induced interactions to be associated with GO terms including cytokine production (Extended Data Fig. 5f,g). Together, we report a significant increase in immune response, characterized by the upregulation of interferon-induced and cytokine-signalling genes in a cell type-specific manner, at the MFI following SARS-CoV-2 infection.
Angiogenesis dysregulation due to COVID-19 infection during pregnancy
Placental angiogenesis and remodelling of maternal spiral arteries in the MFI by invasive trophoblasts is vital for successful pregnancy28. Abnormal angiogenesis is frequently observed in placental disorders and infections29,30. We investigated whether SARS-CoV-2 infection would affect the angiogenesis pathway on a molecular level. We discovered the upregulation of blood vessel development and VEGF signalling genes in vascular endothelial cells and fibroblasts (Fig. 4a,b). Concordantly, increased ATAC-seq peaks in these two cell types were associated with GO terms for vasculogenesis and epithelial cell migration, suggesting epigenetic dysregulation of relevant pathways (Fig. 4c,d). This was also observed by bulk ATAC-seq (Extended Data Fig. 5c). Importantly, sensitivity analyses confirmed that our finding was not confounded by other clinical conditions (Extended Data Fig. 6a). Furthermore, from CellPhoneDB analysis, we found that COVID-19-induced receptor–ligand interactions are significantly associated with the angiogenesis GO term (Extended Data Fig. 5f). These receptor–ligand pairs between endothelial cells, fibroblasts and trophoblasts were further analysed (Fig. 4e). Cell type-specific receptor–ligand pairs relating to angiogenesis—including VEGFA–FLT1 and FGF–FGFR (Fig. 4e), which are known to be critical in angiogenesis during placental development— were significantly upregulated in patients. VEGFA was upregulated in fibroblasts and stroma cells (Fig. 4f).
a,b, GO analysis of upregulated genes from snRNA-seq in villous fibroblast cluster 1 (V.FB1; a) and villous vascular endothelial (V.Endo1; b) cells. The size and colour of the bubble represent the number of upregulated genes and the ratio to total genes under each GO term, respectively. P values were calculated using a one-tailed hypergeometric test. c,d, GREAT analysis of increased peaks in the V.FB1 (c) and V.Endo1 (d) cells from the snATAC-seq. The top relevant GO terms are shown. e, Significant patient-specific receptor–ligand (R–L) pairs under the GO term of angiogenesis. The colour of the bubbles indicates the log10(mean expression) and the size of the bubbles represents the significance in negative log10(one-tailed Wilcoxon test P value + 0.001) from CellPhoneDB. f, Genome browser screenshot of the genomic region upstream of VEGFA. The pseudo-bulk tracks for patient and control samples of V.FB1 from the snATAC-seq and the snRNA-seq are displayed as RPM. The grey shading highlights the promoter region of VEGFA and the yellow shading highlights a potential cis-regulatory enhancer called by the peak-to-gene linkage analysis (orange arc). This potential enhancer region is defined with significantly increased ATAC-seq signal in V.FB1 cells (fold change (COVID-19/control)) = 1.8 and one-tailed Poisson test Padj = 1.26 × 10−5). The y axes for all cell type tracks range from zero to two. Ctrl, control and Cov, COVID-19; cell-type abbreviations as per Fig. 1b.
Next, we conducted peak-to-gene linkage analysis using ArchR, which integrates snATAC-seq and snRNA-seq data to predict potential enhancer-gene pairs31. We identified a candidate enhancer upstream of the VEGFA promoter, coinciding with an ENCODE candidate CRE (cCRE) definition (Fig. 4f). Together with VEGFA upregulation, the candidate element gained chromatin accessibility in fibroblasts (Fig. 4f), suggesting that transcriptional dysregulation of angiogenesis pathways may involve epigenetic alterations of associated CREs. We also discovered the upregulation of the ENG gene, which encodes the endoglin glycoprotein that is vital for blood vessel development, in villous endothelial cells (Extended Data Fig. 6b). Elevated plasma concentration of soluble ENG is a marker for preeclampsia, which is frequently accompanied by increased circulating VEGF and soluble fms-like tyrosine kinase-1 (sFLT1)32. Concomitantly, we detected increased chromatin accessibility at its candidate enhancer in the corresponding cell types (Extended Data Fig. 6b). Furthermore, hypoxia response genes, such as NOS2 and EGLN3, were also upregulated in the patient samples (Extended Data Fig. 6c)33,34. Collectively, we found that SARS-CoV-2 induced the dysregulation of angiogenesis genes in vascular endothelial cells and fibroblasts at the MFI, which involved differential activities of putative CREs.
SARS-CoV-2 induces retrotransposon dysregulation at the MFI
Viral infections, including SARS-CoV-2, are associated with retrotransposon dysregulation17,35. However, the exact consequence during pregnancy remains elusive. Therefore, we analysed the transcriptional and epigenomic states of these sequences in patient and control samples. Due to their repetitive natures, next-generation sequencing reads from retrotransposons suffer from poor mappability and are routinely discarded. To circumvent this issue, we utilized our iterative alignment approach termed Subfamily Assignment for Multiple Alignment (SAMA)36, which rescues multiple-aligned reads and uniquely anchors them to retrotransposon subfamilies, thereby enabling us to measure transcriptomic and epigenomic changes with higher precision.
From bulk RNA-seq, we discovered four upregulated retrotransposon subfamilies and 38 downregulated subfamilies among patients, including the downregulation of the HERV17-int subfamily from which the SYNCYTIN-1 gene is derived (Fig. 5a). Interestingly, most dysregulated subfamilies are endogenous retroviruses (ERVs; Fig. 5a). Altered expression of many subfamilies was similarly detected by snRNA-seq (Extended Data Fig. 7a). We also identified cell type-specific dysregulated subfamilies (Fig. 5b). For example, the LTR16A1 subfamily was specifically downregulated in STs, suggesting a cell type-specific regulatory mechanism for these elements (Extended Data Fig. 7b). We then analysed the activities of individual retrotransposons and defined 1,324 upregulated and 7,117 downregulated elements. A Genomic Regions Enrichment of Annotation Tool (GREAT) analysis demonstrated that upregulated elements were associated with the angiogenesis GO term, whereas downregulated elements were related to pregnancy genes (Fig. 5c and Extended Data Fig. 7c). We found several downregulated HERV3-int elements in patients, including an ERV-derived gene (ERV3-1), which were specifically downregulated in STs (Extended Data Fig. 7d and Fig. 5d). Intriguingly, peak-to-gene analysis identified two ERV-derived putative enhancers for ERV3-1. Concomitantly, the ERV3-1 gene promoter and the candidate enhancers all lost chromatin accessibility in STs. Moreover, CUT&Tag revealed H3K27ac reduction at these enhancers in patients, which is indicative of reduced cis-regulatory activity. Notably, ERV3 class elements are expressed at high levels in the placenta and their decreased expression is linked to pregnancy disorders such as intra-uterine growth restriction37. Our findings suggest that pregnancy-related ERVs are dysregulated in the MFI in patients with COVID-19, which is associated with epigenetic reprogramming of ERV-derived CREs.
a, Dysregulated retrotransposon subfamilies from the bulk RNA-seq. Subfamilies that were significantly upregulated (Padj < 0.05 and log2(fold change) > 0.25; n = 4) or downregulated (Padj < 0.05 and log2(fold change) < −0.25; n = 38) are labelled in red and blue, respectively. b, Expression of retrotransposon subfamilies in the 21 cell types from the snRNA-seq. c, GREAT analysis of significantly downregulated individual retrotransposons from the bulk RNA-seq analysis. d, Genome browser screenshot of ERV3-1. The pseudo-bulk tracks from the snRNA-seq and snATAC are displayed as RPM with y axes ranging from zero to eight. The CUT&Tag H3K27ac tracks are displayed as aggregated RPM values with y axes ranging from zero to four. Blue shadings highlight the candidate enhancers defined by peak-to-gene analysis. e, Retrotransposon subfamilies with differential chromatin accessibility from the bulk ATAC-seq. Retrotransposon subfamilies with significantly increased (Padj < 0.05 and log2(fold change) > 0.25; n = 44) or decreased (Padj < 0.05 and log2(fold change) < −0.25; n = 44) accessibility are labelled in red and blue, respectively. a,e, The negative log10(two-tailed Wald test Padj) and log2(fold change, patient/control) values for each subfamily were calculated by DESeq2. Dashed lines represent the indicated thresholds. f, Heatmap showing chromatin accessibility of retrotransposon subfamilies across the nine cell types from the snATAC-seq. b,f, Each row is a retrotransposon subfamily and the colour scale represents the RPM-calculated row z-score. g, GREAT analysis of individual retrotransposons with decreased chromatin accessibility from the bulk ATAC-seq. c,g, The top relevant GO terms are shown. Ctrl, control and Cov, COVID-19; cell-type abbreviations as per Fig. 1b.
We further investigated retrotransposon-derived CREs in patients by assessing their chromatin accessibility. Utilizing SAMA, we identified retrotransposon subfamilies with varied bulk ATAC-seq signals in patients (Fig. 5e). Furthermore, snATAC-seq uncovered cell type-specific dysregulation, which was obfuscated in the bulk data (Fig. 5f). We found subfamilies, including HERV-K/LTR5, that significantly gained chromatin accessibility specifically in STs (Extended Data Fig. 7e). In addition, downregulated subfamilies identified in snRNA-seq showed decreased chromatin accessibility in corresponding cell types. For instance, HERV17-int showed both transcriptional downregulation and chromatin accessibility loss in STs (Extended Data Fig. 7f). Focusing on elements with altered snATAC-seq signal (Extended Data Fig. 7g), we discovered association with the inflammatory response GO term (Extended Data Fig. 7h). Interestingly, retrotransposons with decreased chromatin accessibility were also associated with the female pregnancy GO term (Fig. 5g). From snATAC-seq, elements with decreased chromatin accessibility in STs are enriched with important placental transcription factor binding motifs, including GATA2 and GRHL2 (Extended Data Fig. 7i). Notably, the expression levels of GATA2 and GRHL2 were also reduced in STs, consistent with loss of chromatin accessibility at their promoters (Extended Data Fig. 7j). Together, these results demonstrate changes in retrotransposon activities at the MFI following SARS-CoV-2 infection, which may influence important processes in pregnancy.
PSG downregulation coincides with reduced LTR8B activity
We observed a downregulation of pregnancy-related genes, including PSG, at the MFI of the pregnant patients with SARS-CoV-2 infection (Fig. 2a). The human genome contains ten bona fide PSG genes, PSG1–9 and PSG11, which are clustered on chromosome 19q13 (ref. 38). They are expressed at high levels during pregnancy and are involved in the processes of immune modulation and angiogenesis38. Reduced PSG protein levels in serum are associated with adverse pregnancy outcomes, including pregnancy loss and preeclampsia39,40. However, their precise regulatory mechanism remains poorly understood. We found that seven PSG genes were downregulated in the STs of patients infected with COVID-19 (Fig. 6a,b and Extended Data Fig. 8a,b). We validated their transcriptional and protein level changes using RT–qPCR and immunohistochemical staining, respectively (Fig. 6c,d). Intriguingly, all PSG genes harbour intronic LTR8B elements, which have high open chromatin signals and H3K27ac enrichment in control samples (Fig. 6a,e). We found the downregulation of PSG genes occurred concomitantly with decreased chromatin accessibility and H3K27ac levels at intronic LTR8B elements (Fig. 6e and Extended Data Fig. 8c). Moreover, these retrotransposons showed trophoblast-specific open chromatin states (Extended Data Fig. 8d). We postulate that these elements serve as trophoblast-specific CREs for the PSG genes.
a, Genome browser screenshot of the PSG gene cluster in the STs from the snRNA-seq and bulk RNA-seq. The purple shading highlights LTR8B elements within each PSG gene. The pseudo-bulk tracks from the snRNA-seq and bulk RNA-seq tracks are displayed as RPM with y axes ranging from 0 to 300 and 0 to 100, respectively. b, Average expression levels, calculated by Seurat, of the PSG1 (top), PSG4 (middle) and PSG9 genes (bottom) in trophoblast cell types from the snRNA-seq in patient and control samples. c, Expression levels, determined using RT–qPCR, of eight PSG genes in patient and control samples. Expression was normalized to TBP. Each dot represents a sample (n = 7 for both patient and control); P values were calculated using a one-tailed Wilcoxon test. The centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively. d, Immunohistochemistry staining (left) and H scores (right) of PSG9 protein in patient and control samples. Red arrowheads indicate STs and green arrowheads indicate cytotrophoblasts. Scale bars, 50 µm. Each dot in the bar chart represents a sample (n = 7 for patient and n = 6 for control). Data are presented as the mean values with error bars showing the s.e.m.; P values were calculated using a two-tailed Student’s t-test. e, Genome browser screenshot showing the PSG4 gene and its intronic LTR8B element, highlighted by the blue shading. The pseudo-bulk tracks from the snRNA-seq and snATAC-seq are displayed as RPM. The H3K27ac CUT&Tag tracks are displayed as aggregated RPM. The y axes range from 0 to 60 for the snRNA-seq, 0 to 4 for the snATAC-seq and 0 to 2 for the CUT&Tag. Ctrl, control and Cov, COVID-19; cell-type abbreviations as per Fig. 1b.
LTR8B elements function as enhancers in placental cells
We next investigated the enhancer potentials of the intronic LTR8B elements in regulating the PSG genes. Publicly available RNA-seq and chromatin immunoprecipitation-sequencing (ChIP–seq) datasets from primary placenta cell types were analysed41,42. We found that high expression of PSG genes in STs was accompanied by enrichment of active enhancer histone modification signatures (high H3K27ac and H3K4me1, and low H3K4me3) at the intronic LTR8B elements (Fig. 7a). As expected, repressive modifications—such as H3K9me3, H3K27me3 and DNA methylation—were depleted at these loci (Fig. 7a). Notably, another ERV1 cluster in the same intron of PSG8 was marked by H3K9me3, suggesting that the activation of these LTR8Bs did not result from positional effects (Fig. 7a). We validated the enhancer functionality of LTR8Bs using a luciferase assay in which most elements demonstrated strong enhancer activities (Fig. 7b and Extended Data Fig. 8e).
a, Genome browser screenshot showing the transcriptional and epigenetic states of the PSG8 gene in primary STs. The displayed datasets were obtained from the International Human Epigenome Consortium data repository41,42. The intronic ERV1 element is shaded in grey and the LTR8B elements within PSG8 are shaded in blue. All tracks are displayed as RPM, with y axes ranging from 0 to 200 for RNA-seq, 0 to 100 for H3K27ac ChIP–seq, 0 to 150 for H3K4me1 ChIP–seq, 0 to 250 for H3K4me3 ChIP–seq, 0 to 20 for H3K9me3 ChIP–seq, 0 to 40 for H3K27me3 ChIP–seq and 0 to 1 for whole-genome bisulphite sequencing (WGBS). b, Luciferase assay enhancer activity of individual intronic LTR8B elements within different PSG genes normalized to the background control. Each dot represents an independent experiment (n = 3) and data are presented as mean values with error bars showing the s.d. c, Peak-to-gene linkage analysis indicating a potential interaction between the PSG1 promoter and its intronic LTR8B (highlighted by the red box). Tracks are displayed as normalized pseudo-bulk coverage signals of each cell type from the snATAC-seq ranging from 0 to 0.25. Ctrl, control and Cov, COVID-19; cell-type abbreviations as per Fig. 1b.
We found that the degree of chromatin accessibility and H3K27ac enrichment at the LTR8Bs are highly correlated with the expression of corresponding PSG genes (Extended Data Fig. 8f,g), providing support for their CRE identity. Moreover, peak-to-gene linkage analysis from snATAC-seq identified strong linkages between PSG promoters and their intronic LTR8B elements (Fig. 7c and Extended Data Fig. 9a). To elucidate the precise targets of LTR8B enhancers, we performed Hi-C analysis of the human expanded potential stem cell-derived trophoblast stem cells (TSCs)43, which had high expression of most PSG genes and enrichment of active enhancer marks at the intronic LTR8Bs (Extended Data Fig. 9b). Strong three-dimensional interactions between the intronic LTR8B elements and multiple PSG gene promoters were defined (Fig. 8a), suggesting that these retrotransposons formed a regulatory network with PSG genes.
a, Screenshot of the virtual 4C results (top) and Hi-C interaction frequency (bottom) from TSCs at the PSG cluster showing the interaction of PSG gene promoters and intronic LTR8B elements. The virtual 4C shows interaction frequencies using each intronic LTR8B element as an anchor. The intronic LTR8B elements and the promoters of PSG genes are highlighted with blue and yellow shading, respectively. The tracks are displayed as RPKM, with y axes ranging from 0 to 20,000. The white arrowheads in the Hi-C heatmap point to interactions within the PSG cluster, indicating a high interaction frequency between intronic LTR8B and the promoters of PSG genes. b, Pseudo-bulk RPKM signal of all active LTR8B elements defined by co-accessibility analysis (n = 83) in the nine cell types from the snATAC-seq. Each dot represents one active LTR8B element; P values were calculated using a one-tailed paired Student’s t-test. The centre and bounds of the boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively. c, HOMER motif analysis of active LTR8B elements defined by the co-accessibility analysis (n = 83). P values were calculated using a one-tailed hypergeometric test. Ctrl, control and Cov, COVID-19; cell-type abbreviations as per Fig. 1b.
To determine whether LTR8B elements broadly functioned as placental enhancers and whether they were commonly affected by SARS-CoV-2, we interrogated other elements within the subfamily. The LTR8B subfamily possessed high chromatin accessibility in trophoblasts compared with other cell types, which was significantly reduced in the COVID-19 samples (Extended Data Fig. 9c,d). We defined 83 LTR8B elements as putative genic enhancers from peak-to-gene linkage analysis, which had high chromatin accessibility in trophoblasts (Fig. 8b). Following SARS-CoV-2 infection, a substantial loss of ATAC-seq signal was detected in all cell types, with the most significant change in STs (Fig. 8b). Moreover, these active elements were enriched with placental developmental transcription factor motifs, including GATA, JUNB and FOSL2 (Fig. 8c). For example, an LTR8B element was predicted to be a potential enhancer for the downregulated STS gene in STs (Extended Data Fig. 9e). This element was enriched with H3K27ac and H3K4me1 in primary ST cells. Concordant with STS downregulation, the LTR8B element lost chromatin accessibility in the COVID-19 samples. Furthermore, Hi-C showed three-dimensional chromatin interactions between the LTR8B element and the STS promoter in TSCs. Together, our results indicate that a subset of LTR8B retrotransposons function as enhancers in regulating placental genes and SARS-CoV-2 infection is linked to decreased activity of both these CREs and their targets.
COVID-19 infection during pregnancy induces epigenetic reprogramming of LTR8Bs
Given the transcriptional and chromatin state changes at the LTR8Bs and PSG genes, we further aimed to delineate the underlying molecular mechanism. We analysed the H3K27me3 and H3K27ac CUT&Tag data from control and patient samples. Although no significant change in H3K27me3 was detected, we found reduced H3K27ac at LTR8Bs in the patients with COVID-19 infection (Extended Data Fig. 10a), which correlated with PSG expression (Extended Data Figs. 8g and 10a). These results revealed that the cell type-specific downregulation of LTR8B elements was coupled with H3K27ac loss and was probably unrelated to H3K27me3 enrichment.
Reduced levels of transcription factor binding to CREs may also explain the downregulation of PSG genes. The motif for GATA was the most significantly enriched transcription factor motif in active LTR8B elements (Fig. 8c). GATA proteins are master regulators in placenta development44. Interestingly, we found that GATA2 expression and promoter chromatin accessibility were significantly reduced in the STs of the patients with COVID-19 (Extended Data Fig. 10b,c). Furthermore, known target genes of GATA2 also exhibited loss of ATAC-seq signals (Extended Data Fig. 10d)45. Together, we surmised that LTR8B-derived enhancers, which are normally accessible and enriched with H3K27ac in STs, regulate the PSG genes via GATA2 binding. However, these retrotransposons lose activity following SARS-CoV-2 infection, which could be due to aberrant epigenetic regulation and/or reduction of GATA2 recruitment.
Discussion
Throughout the COVID-19 pandemic, hospitals have seen increasing numbers of pregnant patients who are infected with SARS-CoV-2. Although mounting evidence suggests that patients face increased risks of adverse pregnancy outcomes, the underlying molecular mechanism remains unclear. In this study the participating patients were diagnosed with COVID-19 during late pregnancy. To discriminate the effect of maternal immune response from a direct viral infection, we confirmed that the patient MFI samples were free from the virus at the time of analysis. Nonetheless, immune and angiogenesis dysregulation was observed at the MFI of patients. This could expose the fetus to inflammation, hypoxia and oxidative stress, which can substantially impact the fetal epigenome and developmental process. Our findings suggest that patients with COVID-19 have a higher potential for placenta-related complications, and specific care and management should be instigated.
The complex immunomodulation at the MFI is important for successful pregnancies. Aberrant interferon expression is known to be a common cause of pregnancy disorders46. Here we detected upregulation of interferon-related genes across different cell types. IFITM proteins were demonstrated to inhibit syncytin-mediated ST formation in mouse models, which results in placental abnormalities47. The process of ST formation occurs from implantation to full term48. Therefore, the overexpression of IFITM genes in late pregnancy can still potentially be detrimental. Furthermore, type I interferon response towards SARS-CoV-2 can cause lung damage by cGAS–STING activation49, which may similarly apply to placental endothelial cells.
Abnormal angiogenesis is found in a variety of pregnancy complications; however, their regulatory pathways in individuals with COVID-19 have not been fully explored. We found that angiogenesis genes are upregulated in endothelial cells and fibroblasts, concomitant with altered chromatin accessibility at CREs. We discovered putative enhancers upstream of VEGFA and ENG. Both genes and their CREs showed higher expression and chromatin accessibility in patients with COVID-19, respectively. VEGFA and ENG are known to be vital to early placental vascular development and are transcriptionally dysregulated in patients with preeclampsia32. The dysregulation of these genes may be associated with endothelial dysfunction in patients infected with COVID-19.
We also discovered retrotransposon dysregulation in patient samples. Strikingly, hundreds of elements were differentially expressed and/or had altered chromatin states. Downregulated elements were associated with pregnancy genes and enriched with motifs for important transcription factors, indicating involvement in normal placental functions. For instance, a subset of LTR8B elements serve as enhancers for the PSG genes. PSG genes encode glycoproteins that have immunoregulatory, pro-angiogenic and anti-platelet functions. Their disruption is associated with pregnancy complications including preeclampsia38,40. We found that the deregulation of LTR8B-derived enhancers is potentially responsible for PSG downregulation in the STs of patients with COVID-19. Hi-C data confirmed the strong higher-order chromatin interactions between the retrotransposons and multiple PSG promoters, indicating potential PSG regulatory modules. The H3K27ac enrichment at LTR8B elements is correlated with PSG expression levels. These downregulated elements are enriched with binding motifs for GATA2, which is also transcriptionally downregulated in patients. We propose that reduced H3K27ac at these retrotransposon-derived enhancers and downregulation of GATA2 point to reduced enhancer activities that results in decreased PSG expression in SARS-CoV-2 infection. Such changes potentially cause impaired immunoregulation and angiogenesis at the MFI.
Collectively, we generated extensive multi-omic datasets of the MFI from patients infected with SARS-CoV-2 and control individuals. Our findings uncovered the critical role of epigenetic regulation and defined retrotransposon-derived enhancers associated with the altered expression of important pregnancy genes. Further studies investigating whether new SARS-CoV-2 variants could induce similar immune activation and angiogenesis dysregulation should be carried out.
Methods
Patient consent and sample collection
This study complies with all relevant ethical regulations. Approval was obtained from the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee (CREC ref. no. 2020.210) and Hong Kong University of Science and Technology (ref. no. HREP2021-0100 and HREC548).
Our case-control study included seven consecutively sampled pregnant patients who tested positive for SARS-CoV-2 by RT–qPCR (Ct ≤ 35) of deep throat saliva or nasopharyngeal swabs beyond 24 weeks of gestation and seven uninfected women. All participants gave written informed consent to participate in the study and for the results to be published. No compensation was provided to participants. Blood and cord blood were collected from the patients immediately after delivery and serum antibodies to SARS-CoV-2 were analysed. Qualitative detection of the anti-SARS-CoV-2 immunoglobulin G (IgG) to the SARS-CoV-2 nucleocapsid protein (N-protein) was performed using an Elecsys Anti-SARS-CoV-2 assay (Roche) on a Cobas e411 analyser.
Tissues from the MFI were collected within 2 h after delivery as previously described50. All procedures were performed in a certified Class II Biosafety cabinet. For diagnostic RT–qPCR, RNA was extracted from tissues taken midway between the umbilical cord insertion site and the edge of the placental disk. The tissue was placed with the basal plate on the uppermost orientation and several random sampling sites without frank pathology were identified. Visible blood vessels and blood clots were removed. The collected tissues were gently rinsed in sterile cold PBS to minimize maternal-blood carryover. All tissues were collected by the same person to avoid interpersonal variation in the collection practice. The samples were immersed in 10 ml RNAlater (Ambion) for 24 h at ambient temperature and stored in a freezer at −80 °C until RNA extraction.
For immunohistochemical assays, the washed samples were fixed with 4% paraformaldehyde solution for 24–48 h and dehydrated to enable embedding with paraffin51.
For other assays, the washed samples were snap-frozen in liquid nitrogen and stored in a −80 °C freezer. The frozen specimens were pulverized using a mortar and pestle with added liquid nitrogen. The resulting powdered samples were stored in a freezer at −80 °C until further experiments.
Detection of SARS-CoV-2 in patient samples
Three pieces of tissue were used from each case and amplified in duplicate. Total RNA was extracted using an RNeasy mini kit (Qiagen). The detection of SARS-CoV-2 RNA was performed with the FDA-authorized CDC 2019-novel coronavirus (2019 nCoV) real-time RT-PCR diagnostic panel (EUA 200001). The nucleocapsid genes (both N1 and N2) were assayed with human RNase P as an endogenous reference control. The DNA sequence of the SARS-CoV-2 N gene (10006625, IDT) was used as the positive control. A set of in vitro-synthesized RNA transcripts including three quantification positive controls (1,000, 100 and 10 copies genome equivalent) were also assayed.
Immunohistochemistry
Formalin‐fixed paraffin-embedded tissue blocks were sectioned to a thickness of 5 μm for standard haematoxylin and eosin as well as specific immunohistochemical staining. The slides were deparaffinized with xylene and rehydrated with ethanol. For the immunohistochemical staining, the slides were treated with 3% hydrogen peroxide, followed by heating in antigen retrieval buffer (pH 6.0; Abcam), incubation with Protein Block (Abcam) and overnight incubation at 4 °C with primary antibodies to FLNB (ab282106, Abcam), PAPPA (ab174314, Abcam) and PSG9 (AP53483PU-N, Origene). After washing with 0.1% TBS in Tween‑20, the slides were incubated with the secondary antibody horseradish peroxidase (HRP)-conjugated mouse anti-rabbit (Sigma-Aldrich), stained with the substrate 3,3′-diaminobenzidine (EnVision) and counterstained with haematoxylin (Sigma-Aldrich).
Human TSC culture
The TSCs were a gift from P. Liu (School of Biomedical Sciences, The University of Hong Kong)43. The TSCs were cultured as previously described52 with minor modifications. Briefly, the cells were maintained in TSC medium (DMEM/F12 supplemented with 0.1 mM 2-mercaptoethanol, 0.2% fetal bovine serum, 0.5% penicillin–streptomycin, 0.3% BSA, 1% ITS-X supplement, 150 µM l-ascorbic acid, 50 ng ml−1 EGF, 2 mM CHIR99021, 0.5 mM A83-01, 1 mM SB431542, 0.8 mM VPA and 5 mM Y27632). Tissue culture plates were coated with 5 µg ml−1 collagen IV (Corning) at 37 °C for 1 h. The cells were cultured at 37 °C with 5% CO2. TSCs at 20–30 passages were harvested for analyses.
Bulk RNA-seq and RT–qPCR
Total RNA from 10–20 mg of pulverized tissue or 1 – 2 × 106 pelleted TSCs was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s manual. The extracted total RNA was used for generating bulk RNA-seq libraries and RT–qPCR. For RNA-seq, 1 µg total RNA underwent ribosomal RNA depletion using a Ribo-off rRNA depletion kit (Vazyme), followed by RNA-seq library preparation using a QIAseq stranded total RNA library kit (Qiagen), as described in the manufacturer’s manual, and sequenced on an Illumina NextSeq 500 platform.
For RT–qPCR analyses of DEGs, which were performed separately from the SARS-CoV-2 diagnostic RT–qPCR, 1 µg total RNA was treated with DNase I (NEB) and purified with RNAClean XP beads (Beckman Coulter). First-strand synthesis was performed using a Superscript III reverse transcription system (Thermo Fisher Scientific) according to the manufacturer’s manual. The complementary DNA was analysed by qPCR on a LightCycler 480 Instrument II. The RT–qPCR primers are listed in Supplementary Table 7.
Bulk ATAC-seq library preparation
The ATAC-seq protocol was adopted from Buenrostro et al.53, with minor modifications. Briefly, 10–20 mg pulverized tissue was resuspended in 1 ml nuclei permeabilization buffer (5% BSA, 0.2% NP-40, 1 mM dithiothreitol and 1×protease inhibitors in PBS), incubated for 10 min at 4 °C with rotation and filtered through a 40-µm Cell Strainer (Corning). The nuclei were pelleted at 500g and 4 °C for 5 min and resuspended in 50 μl of chilled tagmentation buffer (Vazyme). Nuclei in suspension were counted using a haemocytometer and the concentration was adjusted to 2,000–5,000 nuclei µl−1. Vazyme V50 Tn5 transposase (0.5 µl) was added to 9.5 µl of the nuclei suspension, followed by incubation in a thermomixer at 37 °C with mixing at 500 r.p.m. for 30 min. The tagged DNA was amplified using KAPA HiFi hotstart ready mix (Roche) for 5–10 PCR cycles, followed by size selection with Ampure XP beads (Beckman Coulter) and sequenced on an Illumina NextSeq 500 platform.
CUT&Tag library preparation
CUT&Tag libraries were built based on a published protocol54. Briefly, the pulverized tissues were washed once with wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine and 1×protease inhibitor cocktail). Activated ConA beads (30 µl) were added to about 150,000 nuclei, followed by resuspension in 150 µl antibody buffer and division into three tubes. Primary antibody (1 µl; anti-H3K27me3 Active motif 39155, anti-H3K27ac Active motif 39133 or rabbit IgG antibody Sigma-Aldrich, I5006) was added to each tube and the samples were incubated overnight at 4 °C. The next day, the buffer was changed to Dig-Wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 1×protease inhibitor cocktail and 0.05% digitonin) containing 1:100 secondary antibody (donkey anti-rabbit IgG; Abcam, ab6701) and incubated at room temperature for 1 h. The beads were washed with Dig-Wash buffer and resuspended in 100 µl Dig-300 buffer (0.05% digitonin, 20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine and 1×protease inhibitor cocktail) containing 1:40 pA-Tn5 adaptor complex. These tubes were incubated at room temperature for 1 h, washed with Dig-300 buffer, resuspended in 300 µl Tagmentation buffer and incubated at 37 °C for 1 h. The tagged DNA was purified using a Qiagen MinElute PCR purification kit. The libraries were PCR amplified for 13 cycles using NEBNext HiFi 2× PCR master mix, purified with Ampure XP beads and sequenced on an Illumina NextSeq 500 platform.
Preparation of snRNA-seq and snATAC-seq libraries
Pulverized tissue (10–20 mg) was resuspended in 1 ml nuclei permeabilization buffer, incubated with rotation for 10 min at 4 °C and filtered through a 40-µm Cell Strainer (Corning). The nuclei were pelleted at 500g and 4 °C for 5 min and used for snRNA-seq library preparation using a Chromium Next GEM Single Cell 3ʹ V3.1 kit and snATAC library preparation using a Chromium next GEM single cell ATAC v1.1 kit. The libraries were converted for sequencing on the MGI sequencing platform using an MGIEasy universal library conversion kit. The converted libraries were sequenced on the MGISEQ-2000RS platform.
Hi-C library preparation
The TSC Hi-C library was generated using an Arima-Hi-C kit according to the manufacturer’s protocol. The library was converted for sequencing on the MGI sequencing platform with an MGIEasy universal library conversion kit and sequenced on the MGISEQ-2000RS platform.
Micro-ChIP–seq library preparation
TSC micro-ChIP–seq libraries were prepared as described previously55, with minor modifications. Briefly, 5 × 105 crosslinked cells were resuspended in lysis buffer and sonicated using a Covaris S220 sonicator continuously for 400 s at 175 W. Fragmented chromatin was added to Protein A Dynabeads (Thermo Fisher Scientific) bound to antibodies to H3K27ac (AM39133, Active Motif), H3K4me3 (AM39915, Active Motif) or H3K4me1 (AM91289, Active Motif). The mixtures were incubated at 4 °C for 40 h with rotation. The captured chromatin was washed four times with RIPA buffer. Reverse crosslinking was performed by incubation at 68 °C for 4 h with proteinase K. The eluted DNA was purified using a Qiagen MinElute PCR purification kit. Libraries were prepared using a KAPA HyperPrep Kit (Roche) and sequenced on an Illumina NextSeq 500 platform.
Luciferase assays
The PSG intronic LTR8B elements or GFP sequences (negative control) were cloned into the pGL3-promoter and pGL3-enhancer vectors. The primers used for amplification of target sequences are listed in Supplementary Table 7. Two days before transfection, TSCs were seeded at 4 × 104 cells per well in 24-well plates. The cells were co-transfected with the Renilla luciferase vector and pGL3 vector containing an LTR8B element or GFP sequence with Lipofectamine 3000 transfection reagent (Thermo Fisher). Luciferase activity was measured using Dual-luciferase reporter assay system reagents (Promega, E1960) according to the manufacturer’s protocol with a Spectronic Genesys 5 UV/Visible Spectrophotometer (ALT) 48 h after transfection. The luciferase activity was calculated using the firefly-to-Renilla signal ratio and each LTR8B activity was normalized to the background negative control.
Bioinformatic analyses
Bulk RNA-seq data analysis
The RNA-seq reads were aligned to the GRCh38/hg38 genome assembly and the GENCODE V39 transcriptome assembly separately using STAR v2.5.3a56 with the parameters --outFilterMultimapNmax 1 to only keep uniquely mapped reads in both alignments. Transcriptome alignments were quantified using RSEM v1.3.3 (ref. 57). Genomic alignments were combined to generate RPM signals for visualization. DEGs were defined by DESeq2 v1.38.2 (ref. 58) with default settings. For the quantification of individual retrotransposons, aligned reads were overlapped with retrotransposon annotations (RepeatMasker). Differentially expressed retrotransposons were defined by DESeq2 v1.38.2 using size factor estimated from gene count matrix.
Bulk ATAC-seq data analysis
Low-quality ATAC-seq reads (Phred score < 20) were removed and adaptors were trimmed using trim_galore v0.4.3 under paired-end mode. The reads were aligned to the GRCh38/hg38 genome assembly using Bowtie2 v2.3.3.1 (ref. 59) with the parameters -N 1 -L 25 -X 2000 --no-discordant --no-mixed. PCR duplicates were removed using Picard MarkDuplicates v2.9.0. To make the start site of each read represent the centre of the transposase binding60, the alignments were adjusted using MACS2 v2.1.0 (ref. 61) with the parameters --shift -75 --extsize 150.
S3norm62 was applied for signal normalization using the default setting (bin size, 20 bp). Peaks were called by MACS2 with normalized signal. Peaks called from all samples were merged and differential peaks were defined by DESeq2 v1.22.1 with the S3norm normalized read counts. Individual retrotransposons with differential chromatin accessibility were assigned by overlapping the retrotransposon coordinate with the differential peaks.
CUT&Tag data analysis
CUT&Tag reads were trimmed using trim-galore with the default parameters, followed by mapping to GRCh38/hg38 and the SARS-CoV-2 genome21 using bowtie2 v2.4.2 (--end-to-end --very-sensitive --no-mixed --no-discordant --no-overlap --no-dovetail --phred33 -I 10 -X 2000). SEACR v1.3 (ref. 63) was used to call H3K27ac peaks using ‘norm’ and ‘stringent’ modes for normalization and thresholding, respectively. SICER v1.0.2 was used to call H3K27me3 peaks. The corresponding IgG libraries were used as a control for peak calling. Peaks for the same histone modifications were merged for differential analysis with DEseq2 v1.28.0.
Analysis of snRNA-seq data
The snRNA-seq raw FASTQ files generated from the MGISEQ-2000RS platform were converted to 10x Cell Ranger-compatible format. The converted FASTQ files were mapped to GRCh38/hg38 pre-mRNA genome and the SARS-CoV-2 genome (SARS-CoV-2 isolate Wuhan-Hu-1, GenBank NC_045512.2)21 using Cell Ranger v6.0.1 with the default setting. The output was processed using Seurat v4.0.1 (ref. 64). Doublets (doublet score > 0.75) were removed using the computeDoubletDensity (d = 15) function from scDblFinder v.1.2.0 (ref. 65). We then applied standard filtering (nUMI ≥ 1,000, nGene ≥ 1,000, nUMI < 25,000, nGene < 4,000 and mitochondria ratio < 0.05) and removed clusters with no marker gene expression. The quality control statistics are listed in Supplementary Table 9. Nuclei that passed the filtering (n = 62,132) were normalized and transformed using the SCTransform function of Seurat with the glmGamPoi method. The nuclei were then subclustered and annotated based on manual curation of markers and reference to placenta marker databases66,67,68.
Genes that were differentially expressed between patients and controls were defined for each cluster using the FindMarker function of Seurat. CellPhoneDB v2.1.7 (ref. 27) was used to analyse receptor–ligand interactions using standard processing and subsampling of 8,000 cells was done for both the patient and control samples. Patient-specific receptor–ligand pairs were defined by P < 0.05, as calculated by CellPhoneDB, and significant upregulation of the ligand in patient cells from the snRNA-seq differential analysis.
For pseudo-bulk signal, raw reads of individual nuclei were split into separate files based on the cell barcode and aligned to the GRCh38/hg38 genome assembly using STAR v2.7.10b, as described in bulk RNA-seq data analysis. Aligned reads with the same UMI were removed using the UMI_Tools v1.1.2 dedup function69. Genomic alignments of all nuclei from the same cell type from patient or control samples were merged to generate the pseudo-bulk RPM signals for visualization. Individual retrotransposon analysis was performed as described in bulk RNA-seq data analysis.
Analysis of snATAC-seq data
The snATAC-seq raw FASTQ files generated from the MGISEQ-2000RS platform were converted to 10x Cell Ranger ATAC-compatible format. The converted FASTQ files were mapped to the GRCh38/hg38 assembly by Cell Ranger ATAC v2.1.0 with default settings. Mapped snATAC-seq was processed using ArchR v1.0.1 (ref. 31). Briefly, nuclei were filtered (nFrags ≥ 1,000, TSSEnrichment ≥ 4 and PromoterRatio ≥ 0.075) and clusters with low overall transcriptional-start-site signals were removed24. Nuclei that passed filter (n = 68,786) were clustered and annotated by label transfer from snRNA-seq and manual curation of marker genes. The quality control statistics are listed in Supplementary Table 9.
For peak analysis, pseudo-bulk signals were generated. Raw reads of individual nuclei were split into separate files based on the cell barcodes. Duplicated reads in each nucleus were removed using FastUniq v1.1 (ref. 70). Reads from each nucleus were aligned to the GRCh38/hg38 assembly using STAR v2.5.3a as described in bulk ATAC-seq data analysis. Genomic alignments of nuclei from the same cell type were merged to generate pseudo-bulk signals for peak calling with MACS2 v2.2.7.1 and visualization. Peaks called from each cell type were merged to generate a master peak set, which was used for Peak2GeneLinkage analysis with ArchR and differentially accessible peak analysis. To define cell type-specific differentially accessible peaks in patients versus controls, we conducted a one-sided Poisson test on the normalized pseudo-bulk reads count and significantly differentially accessible peaks were defined by fold change > 1.5, Padj < 0.01 and peak RPKM signal > 1 in the tested group. For visualization in a heatmap (Fig. 2d), differentially accessible peaks were filtered by pseudo-bulk RPKM > 10 in at least one cell type to retain most accessible peaks. Differentially accessible individual retrotransposons were defined by overlapping retrotransposons with differentially accessible peaks.
Retrotransposon subfamily analysis
We used our SAMA pipeline36 for retrotransposon subfamily quantifications in bulk and single-nucleus RNA-seq and ATAC-seq. Briefly, reads from each nucleus for single-nucleus data or from each sample for bulk data were mapped to the GRCh38/hg38 assembly using STAR v2.5.3a with the parameter --outFilterMultimapNmax 150. Reads with more than one best genomic alignment that is uniquely anchored to the same repeat subfamily were rescued. Rescued multi-aligned reads and uniquely aligned reads were collected for subfamily quantification and differential analysis.
Motif, GO and retrotransposons enrichment analysis
Motif analysis was conducted using HOMER71 (v4.9.1 for ATAC-seq peaks and v4.11 for CUT&Tag peaks) using whole genome as background. Gene ontology analysis was performed using Metascape v3.5 (ref. 72) and GREAT v4.0.4 (ref. 73). Subfamily enrichment of differentially expressed individual retrotransposons was calculated by the ratio of observed over expected counts of elements in each subfamily. The expected numbers were estimated by (n / N) × X, where n is the total number of elements of each subfamily, N is the total number of retrotransposons in the genome and X is the total number of differential retrotransposons. P values were calculated using a hypergeometric test.
ChIP–seq data analysis
For the TSC micro-ChIP–seq (single-end) dataset generated in this study, reads were aligned to the GRCh38/hg38 assembly using Bowtie v1.3.0 (ref. 74) with the parameters ‐v 3 ‐ m 1 ‐‐best ‐‐strata. For the public placenta ChIP–seq dataset (paired-end)41,42, the reads were aligned to the GRCh38/hg38 assembly using Bowtie2 v7.5.0 with the parameters ‐N 1 ‐L 25 ‐X 500 ‐‐no‐discordant ‐‐no‐mixed. Multi-aligned reads were removed. PCR duplicates were removed using Picard MarkDuplicates v2.23.4 for both datasets.
Hi-C data analysis
The TSC Hi-C reads were mapped to the GRCh38/hg38 assembly using Juicer v1.13 (ref. 75) with the parameter -s Arima. Significant interactions were called by Fit-Hi-C at 5-kb resolution with threshold q < 0.05. For virtual 4C analysis, reads that interacted with the bait (LTR8B elements of interest) and their flanking 2-kb regions were extracted and aligned to the genome with BWA v0.7.15 and the RPKM signal was calculated for visualization.
Sensitivity analysis
Sensitivity analysis was conducted on bulk RNA-seq and snRNA-seq to determine the impact of different clinical conditions on transcriptomic data. Patient samples were divided into the following clinical conditions: term (Cov2, Cov3, Cov4 and Cov7) versus pre-term (Cov1, Cov5 and Cov6), Caesarean section (Cov2, Cov3, Cov5, Cov6 and Cov7) versus natural delivery (Cov1 and Cov4) and hypertension (Cov7). For analyses of term versus pre-term and Caesarean section versus natural delivery, we split the samples based on the conditions and performed differential analysis using DESeq2 v1.22.1 (ref. 58) with default settings. For hypertension, we removed Cov7 from the snRNA-seq dataset and repeated differential gene and GO analysis using the same methods mentioned earlier.
Statistics and reproducibility
No statistical methods were used to pre-determine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. The experiments were not randomized. All statistical analyses were done using a one-tailed Student’s t-test, two-tailed Student’s t-test, one-tailed Wilcoxon test, two-tailed Wald’s test, one-tailed hypergeometeric test, one-tailed MAST hurdle model or a one-tailed Poisson test. Details such as the exact P values, statistical tests and experimental replicates are indicated in the figures or figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All sequencing datasets generated in this study have been deposited at ArrayExpress under the accession ID E-MTAB-11749 and at the European Genome-phenome Archive under the accession ID EGAS00001006263. Published trophoblast epigenomic datasets were acquired from JGA under the accession IDs JGA000074 and JGA000117. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Code availability
All code used in this study was previously published and no customized code was used in this manuscript.
References
Narang, K. et al. SARS-CoV-2 infection and COVID-19 during pregnancy: a multidisciplinary review. Mayo Clin. Proc. 95, 1750–1765 (2020).
Villar, J. et al. Maternal and neonatal morbidity and mortality among pregnant women with and without COVID-19 infection: the INTERCOVID multinational cohort study. JAMA Pediatr. 175, 817–826 (2021).
Singh, M., Bansal, V. & Feschotte, C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 32, 108175 (2020).
Ashary, N. et al. Single-cell RNA-seq identifies cell subsets in human placenta that highly expresses factors driving pathogenesis of SARS-CoV-2. Front. Cell Dev. Biol. 8, 783 (2020).
Jamieson, D. J. & Rasmussen, S. A. An update on COVID-19 and pregnancy. Am. J. Obstet. Gynecol. 226, 177–186 (2022).
Mor, G., Aldo, P. & Alvero, A. B. The unique immunological and microbial aspects of pregnancy. Nat. Rev. Immunol. 17, 469–482 (2017).
Ander, S. E., Diamond, M. S. & Coyne, C. B. Immune responses at the maternal–fetal interface. Sci. Immunol. 4, eaat6114 (2019).
Sasaki, Y. et al. Proportion of peripheral blood and decidual CD4+ CD25bright regulatory T cells in pre-eclampsia. Clin. Exp. Immunol. 149, 139–145 (2007).
Hanna, J. et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat. Med. 12, 1065–1074 (2006).
Rebollo, R., Romanish, M. T. & Mager, D. L. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 46, 21–42 (2012).
Mi, S. et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (2000).
Blaise, S., de Parseval, N., Bénit, L. & Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA 100, 13013–13018 (2003).
Blond, J. L. et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74, 3321–3329 (2000).
Chuong, E. B., Rumi, M. A., Soares, M. J. & Baker, J. C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329 (2013).
Dunn-Fletcher, C. E. et al. Anthropoid primate-specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS Biol. 16, e2006337 (2018).
Sun, M. A. et al. Endogenous retroviruses drive lineage-specific regulatory evolution across primate and rodent placentae. Mol. Biol. Evol. 38, 4992–5004 (2021).
Yin, Y., Liu, X. Z., He, X. & Zhou, L. Q. Exogenous coronavirus interacts with endogenous retrotransposon in human cells. Front. Cell. Infect. Microbiol. 11, 609160 (2021).
Lu-Culligan, A. et al. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal–fetal interface. Med 2, 591–610 (2021).
Garcia-Flores, V. et al. Maternal–fetal immune responses in pregnant women infected with SARS-CoV-2. Nat. Commun. 13, 320 (2022).
Sureshchandra, S. et al. Single-cell RNA sequencing reveals immunological rewiring at the maternal–fetal interface following asymptomatic/mild SARS-CoV-2 infection. Cell Rep. 39, 110938 (2022).
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Inoue, A., Jiang, L., Lu, F., Suzuki, T. & Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017).
Zazhytska, M. et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell 185, 1052–1064 (2022).
Yıldırım-Buharalıoğlu, G., Bond, M., Sala-Newby, G. B., Hindmarch, C. C. T. & Newby, A. C. Regulation of epigenetic modifiers, including KDM6B, by interferon-γ and interleukin-4 in human macrophages. Front. Immunol. 8, 92 (2017).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045 (2020).
Gupta, R. K. et al. Blood transcriptional biomarkers of acute viral infection for detection of pre-symptomatic SARS-CoV-2 infection: a nested, case-control diagnostic accuracy study. Lancet Microbe 2, e508–e517 (2021).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Moser, G., Windsperger, K., Pollheimer, J., de Sousa Lopes, S. C. & Huppertz, B. Human trophoblast invasion: new and unexpected routes and functions. Histochem. Cell Biol. 150, 361–370 (2018).
Reynolds, L. P. & Redmer, D. A. Angiogenesis in the placenta. Biol. Reprod. 64, 1033–1040 (2001).
Tran, V. et al. The Angiopoietin–Tie2 axis contributes to placental vascular disruption and adverse birth outcomes in malaria in pregnancy. EBioMedicine 73, 103683 (2021).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Leaños-Miranda, A. et al. Soluble endoglin as a marker for preeclampsia, its severity, and the occurrence of adverse outcomes. Hypertension 74, 991–997 (2019).
Vrijens, K., Tsamou, M., Madhloum, N., Gyselaers, W. & Nawrot, T. S. Placental hypoxia-regulating network in relation to birth weight and ponderal index: the ENVIRONAGE Birth Cohort Study. J. Transl. Med. 16, 2 (2018).
Masoud, G. N. & Li, W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 5, 378–389 (2015).
Karijolich, J., Abernathy, E. & Glaunsinger, B. A. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-κB pathway. PLoS Pathog. 11, e1005260 (2015).
Jiang, Q. et al. G9a plays distinct roles in maintaining DNA methylation, retrotransposon silencing, and chromatin looping. Cell Rep. 33, 108315 (2020).
Bustamante Rivera, Y. Y., Brütting, C., Schmidt, C., Volkmer, I. & Staege, M. S. Endogenous retrovirus 3—history, physiology, and pathology. Front. Microbiol. 8, 2691 (2018).
Moore, T. & Dveksler, G. S. Pregnancy-specific glycoproteins: complex gene families regulating maternal–fetal interactions. Int. J. Dev. Biol. 58, 273–280 (2014).
Arnold, L. L. et al. Pregnancy-specific glycoprotein gene expression in recurrent aborters: a potential correlation to interleukin-10 expression. Am. J. Reprod. Immunol. 41, 174–182 (1999).
Chang, C. L., Chang, C. Y., Lee, D. X. & Cheng, P. J. Characterization of human pregnancy specific glycoprotein (PSG) gene copy number variations in pre-eclampsia patients. Adv. Exp. Med. Biol. 924, 63–65 (2016).
Stunnenberg, H. G., International Human Epigenome Consortium & Hirst, M. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 167, 1145–1149 (2016).
Hamada, H. et al. Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am. J. Hum. Genet. 99, 1045–1058 (2016).
Gao, X. et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 21, 687–699 (2019).
Paul, S., Home, P., Bhattacharya, B. & Ray, S. GATA factors: master regulators of gene expression in trophoblast progenitors. Placenta 60, S61–S66 (2017).
Krendl, C. et al. GATA2/3–TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc. Natl Acad. Sci. USA 114, E9579–E9588 (2017).
Yockey, L. J. & Iwasaki, A. Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity 49, 397–412 (2018).
Buchrieser, J. et al. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science 365, 176–180 (2019).
Burton, G. J. & Jones, C. J. Syncytial knots, sprouts, apoptosis, and trophoblast deportation from the human placenta. Taiwan J. Obstet. Gynecol. 48, 28–37 (2009).
Domizio, J. D. et al. The cGAS–STING pathway drives type I IFN immunopathology in COVID-19. Nature 603, 145–151 (2022).
Farine, T., Parsons, M., Lye, S. & Shynlova, O. Isolation of primary human decidual cells from the fetal membranes of term placentae. J. Vis. Exp., e57443 (2018).
Burton, G. J. et al. Optimising sample collection for placental research. Placenta 35, 9–22 (2014).
Okae, H. et al. Derivation of human trophoblast stem cells. Cell Stem Cell. 22, 50–63 (2018).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930–1935 (2019).
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 12, 323–323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–558 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Xiang, G. et al. S3norm: simultaneous normalization of sequencing depth and signal-to-noise ratio in epigenomic data. Nucleic Acids Res. 48, e43 (2020).
Meers, M. P., Tenenbaum, D. & Henikoff, S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42–44 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Germain, P., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Research 10, 979 (2021).
Jain, A. & Tuteja, G. PlacentaCellEnrich: a tool to characterize gene sets using placenta cell-specific gene enrichment analysis. Placenta 103, 164–171 (2021).
Suryawanshi, H. et al. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788 (2018).
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563, 347–353 (2018).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Xu, H. et al. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS ONE 7, e52249 (2012).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523–1526 (2019).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell. Syst. 3, 95–98 (2016).
Acknowledgements
This work was supported by the Hong Kong Research Grant Council (grants nos. GRF16103721 (D.L.) and CRF C5045-20EF (C.C.W.)), the Hong Kong Epigenome Project (Lo Ka Chung Charitable Foundation (D.L.)), the Croucher Innovation Award (D.L.), the INPer COVID Project 2020-1-32 (G.E.-G.) and Direct Grant (grant no. CUHK 2020.053 (C.C.W. and L.C.P.)). We thank Q. Jiang and V. Ho (HKUST) for their assistance in carrying out the study; L. Chen, T. C. Y. Ma, M. Mak and A. S. T. Tai for their involvement in the study coordination; all participants and their attending obstetricians (T. Ma, F. N. Y. Yu, C. W. Kong, T. K. Lo and P. L. So), nurses and midwives; and the laboratory technicians at all participating hospitals (Queen Elizabeth Hospital, United Christian Hospital, Princess Margaret Hospital and Tuen Mun Hospital, Hong Kong, China) for case recruitment and sample collection. We also thank P. Liu for sharing the TSC cell line used in this study. The schematic in Fig. 1a was created with BioRender.com.
Author information
Authors and Affiliations
Contributions
L.G., V.M., S.K.M.T., X.Z., C.C.W., L.C.P. and D.L. designed the study. G.E.-G., S.M. and L.C.P. collected all clinical samples and data. L.G., V.M., S.K.M.T., X.Z., L.Y.C. and B.W.L. performed all experiments. L.G., V.M., S.K.M.T., X.Z. and M.F.C. performed all data analyses. L.G., V.M., S.K.M.T., X.Z., C.C.W., L.C.P. and D.L. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Fasil Tekola-Ayele, Junyue Cao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Case timelines for the patient samples.
Timeline of SARS-CoV-2 infection for each COVID-19 patient participating in this study. The timelines show the relationship between infection period, hospital admission due to SAR-CoV-2 infection, hospital admission for delivery and postpartum period. Discharge, discharge from hospital; COVID-19, Coronavirus disease 2019; Ab, antibody; IgG, immunoglobulin G; Ag, Antigen; DTS, deep throat saliva; NPS, nasopharyngeal swab; NPA, nasopharyngeal aspiration; TS, throat swab; CS, Caesarean section; EmCS, emergency Caesarean section; RDS, respiratory distress syndrome; Ct, cycle threshold; NSD, normal spontaneous delivery; TTN, transient tachypnea of the newborn; ICU, intensive care unit; NICU, neonatal intensive care unit; +ve, positive finding; −ve, negative finding; PP, postpartum; and GA, gestational age.
Extended Data Fig. 2 Quality control and cell-type analysis for samples and multi-omic datasets.
a, Haematoxylin and eosin (top) histological staining and immunostaining of SARS-CoV-2 nucleocapsid protein (bottom) in patients and control tissue. Scale bars, 50 µm. No replicates were performed for the haematoxylin and eosin or N protein immunostaining. b, Principal component analysis (PCA) on the patient samples (n = 7) used in this study based on gene expression from the bulk RNA-seq. The colour of the sample indicates if the pregnancy was carried out to pre-term or term and the shape represents the mode of delivery—either CS or natural delivery. c, DEGs within patient samples when compared according to term of pregnancy (pre-term/term; top) and mode of delivery (CS/natural; bottom). The significance is presented as adjusted two-tailed P value from multiple testing using the Benjamini and Hochberg method (Padj) from DESeq2. Threshold, Padj < 0.05 and log2(fold change) > 1. d, Expression of top marker genes from the snRNA-seq calculated by Seurat. e, Cell-type distribution in all samples from the snRNA-seq after filtering. f, Expression of known SARS-CoV-2 receptors in each cell type from the snRNA-seq calculated by Seurat (bottom). The size of the bubble indicates the percentage of cells in each cell type that expresses the gene. RT–qPCR results of ACE2 and TMPRSS2 in our patient and control samples (top). Each dot represents a sample (n = 7 for both patient and control); P values were calculated using a one-tailed Wilcoxon test. The centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively. g, Gene score of top marker genes from the snATAC-seq calculated by ArchR. h, Cell-type distribution in all samples from the snATAC-seq after filtering.
Extended Data Fig. 3 Characterization of transcriptional changes in patient samples.
a, DEGs between patient and control samples in all 21 cell types from the snRNA-seq. Upregulated genes (one-tailed MAST model Padj < 0.05 and log2(fold change) > 0.25) and downregulated genes (one-tailed MAST model Padj < 0.05 and log2(fold change) < −0.25) are displayed in red and blue, respectively. b, Immunohistochemistry staining (left) and the corresponding H-score (right) of The DEGs PAPPA and FLNB in placenta and decidua tissue. Scale bars, 50 µm. Each dot in the bar chart represents a sample (PAPPA, n = 7 for patient and n = 5 for control; FLNB, n = 7 for patient and n = 7 for control). Data are presented as the mean with error bars showing the s.e.m.; P values were calculated using a two-tailed Student’s t-test. c, Boxplots showing the bulk RNA-seq FPKM signal of DEGs defined by the snRNA-seq for all samples. Each dot represents a sample (n = 7 for both patient and control); a two-tailed Wald test Padj was calculated from DESeq2. The centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively. d, Expression of IFI6 (top) and VEGFA (bottom) for each cell type from the snRNA-seq calculated by Seurat.
Extended Data Fig. 4 Characterization of epigenomic changes in patient samples.
a, Differential peaks from the H3K27me3 (left) and H3K27ac (right) CUT&Tag data. The fold change and two-tailed Wald test P values were calculated by DESeq2. The H3K27me3 (left) volcano plot shows increased peaks (P < 0.05 and log2(fold change) > 0.5, n = 77) and decreased peaks (P < 0.05 and log2(fold change) < −0.5, n = 502) in red and blue, respectively. The H3K27ac (right) volcano plot shows increased peaks (P < 0.05 and log2(fold change) > 0.5, n = 816) and decreased peaks (P < 0.05 and log2(fold change) < −0.5, n = 2,844) in red and blue, respectively. b, Differential chromatin accessibility between patient and control samples in nine cell types from the snATAC-seq. Increased peaks (one-tailed Poisson test Padj < 0.01, log2(fold change) > 0.585 and RPKM > 1 in Cov) and decreased peaks (one-tailed Poisson test Padj < 0.01, log2(fold change) < −0.585 and RPKM > 1 in Ctrl) are shown in red and blue, respectively. c,d, HOMER motif analysis of both increased (top) and decreased (bottom) peaks from the bulk ATAC-seq (c) and the H3K27ac CUT&Tag (d). P values were calculated using a one-tailed hypergeometric test. e, Genome browser screenshot (left) and violin plot (right) showing the upregulation of the NCOA1 gene in syncytiotrophoblasts (ST). The pseudo-bulk tracks of ST cells from the snRNA-seq are displayed as RPM and the y axes range from 0 to 5 (left). The y axis of the violin plot shows the expression of NCOA1 in ST cells from the snRNA-seq calculated by Seurat (right).
Extended Data Fig. 5 Gene ontology analysis and CellPhoneDB results for bulk and single-nucleus assays showing immune and angiogenesis dysregulation.
a, GO analysis for upregulated (top) and downregulated genes (bottom) from the bulk RNA-seq. The colour scale represents the ratio of dysregulated genes to total genes under each GO term; P values were calculated using a one-tailed hypergeometric test. b,c, GREAT analysis of decreased peaks from the H3K27me3 CUT&Tag (b) and increased (c,top) and decreased (c,bottom) peaks from the bulk ATAC-seq. P values were calculated using a one-tailed binomial test. d, Expression of selected interferon genes from the bulk RNA-seq. The fold change (patient/control) and Padj using the two-tailed Wald test were calculated by DESeq2. Significantly dysregulated genes (|log2(fold change)| > 1 and Padj < 0.05) are marked by red stars. e, RT–qPCR results of selected interferon genes in our patient and control samples. Each dot represents a sample (n = 7 for both patient and control); P values were calculated using a one-tailed Wilcoxon test. The centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively. f, GO analysis of significant patient-specific receptor–ligand interactions, defined by CellPhoneDB, in which the ligand is differentially expressed (|log2(fold change)| > 0.25 and one-tailed MAST model Padj < 0.05) in given cell types from Seurat. The colour scale represents the ratio of dysregulated genes to total genes under each GO term; P values were calculated using a one-tailed hypergeometric test. g, Significant patient-specific receptor–ligand pairs under the GO term regulation of cytokine production defined by CellPhoneDB (see Methods). Only interactions between immune and trophoblast cell types are shown. The mean expression and P values (one-tailed Wilcoxon test) were calculated by CellPhoneDB.
Extended Data Fig. 6 Single-nucleus assays showing angiogenesis dysregulation in fibroblast and endothelial cell types.
a, GO of upregulated genes in villous endothelial (V.Endo; left) and villous fibroblast 1 (V.FB1; right) cells from the snRNA-seq without the Cov7 (hypertension sample). The colour scale represents the ratio of dysregulated genes to total genes under each GO term; P values were calculated using a one-tailed hypergeometric test. b, Genome browser screenshot of the region around ENG. All pseudo-bulk tracks are displayed as RPM with y axes ranging from zero to two. c, Expression of hypoxia-related genes from the bulk RNA-seq. The fold change (patient/control) and Padj (two-tailed Wald test) were calculated by DESeq2. The genes that are significantly dysregulated (|log2(fold change)| > 1 and Padj < 0.05) are marked by a red star.
Extended Data Fig. 7 Dysregulation of retrotransposons due to SARS-CoV-2 infection during pregnancy.
a, Differentially expressed retrotransposon subfamilies in ST (top) and V.Endo (bottom) cells from the snRNA-seq. Upregulated subfamilies (red), P < 0.05 and log2(fold change) > 0.25, n = 6 (ST) and 54 (V.Endo). Downregulated subfamilies (blue), P < 0.05 and log2(fold change) < −0.25, n = 600 (ST) and 57 (V.Endo). P value, two-tailed Student’s t-test. b, Expression of the LTR16A1 subfamily from the snRNA-seq (n numbers in Supplementary Table 9). P value, two-tailed Student’s t-test. c, GREAT analysis of upregulated individual retrotransposons from the bulk RNA-seq. P value, one-tailed binomial test. d, Observed versus expected counts of downregulated individual retrotransposons within subfamilies from the bulk RNA-seq. Subfamilies with observed/expected count > 5 and two-tailed Student’s t-test P < 0.001 are indicated in blue. e, Chromatin accessibility of the LTR5 subfamily from the snATAC-seq (n numbers in Supplementary Table 9). P value, two-tailed Student’s t-test. f, Expression (left) and chromatin accessibility (right) of the HERV17-int subfamily from the snRNA-seq and the snATAC-seq, respectively (n numbers in Supplementary Table 9). P value, two-tailed Student’s t-test. g, Observed versus expected numbers of individual retrotransposons with increased (left) and decreased (right) chromatin accessibility within subfamilies from the bulk ATAC-seq. Subfamilies with observed/expected count > 5 and two-tailed Student’s t-test P < 0.001 are shown in red (left) and blue (right). h, GREAT analysis of individual retrotransposons with increased chromatin accessibility from the bulk ATAC-seq. P value, one-tailed binomial test. i, HOMER motif analysis of individual retrotransposons with decreased chromatin accessibility in ST cells from the snATAC-seq. P value, one-tailed hypergeometric test. j, Transcription factor expression and transcription start site chromatin accessibility called in i. Padj, one-tailed MAST model from Seurat (snRNA-seq) and one-tailed Poisson test (snATAC-seq). For all boxplots in this figure, the centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively.
Extended Data Fig. 8 Downregulation of the expression of PSG genes due to SARS-CoV-2 infection during pregnancy.
a, Expression of PSG2 (top left), PSG3 (top right), PSG5 (middle left), PSG6 (middle right), PSG7 (bottom left) and PSG8 (bottom right) in the trophoblast cell types from the snRNA-seq calculated by Seurat. b, Expression of PSG genes from the bulk RNA-seq. Each dot represents a sample (n = 7 for both patient and control) and the Padj (two-tailed Wald test) was calculated by DESeq2. c, Chromatin accessibility signal of each intronic LTR8B within a PSG gene in the ST cell type from the snATAC-seq. d, Chromatin accessibility signal of intronic LTR8B elements within PSG genes in all nine cell types from the snATAC-seq. Each dot represents one LTR8B element (n = 11); P values were calculated using a one-tailed Wilcoxon test. e, Luciferase assay for promoter activity of individual intronic LTR8B elements within different PSG genes. Data are presented as mean values with error bars showing the s.d. and each dot representing an independent experiment (n = 3). f,g, Correlations between bulk RNA-seq expression levels of PSGs and the bulk ATAC-seq signals (f) as well as the H3K27ac CUT&Tag signals (g) of corresponding intronic LTR8B elements. The y axes represent the FPKM signal from the bulk RNA-seq and the x axes represent the RPKM signal from the respective assays. The error bands in grey represent a 95% confidence level interval for predictions from a linear model; Spearman’s correlation was used to calculate the R value. For all boxplots in this figure, the centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively.
Extended Data Fig. 9 Decreased expression of PSG genes is associated with reduced intronic LTR8B chromatin accessibility.
a, Peak-to-gene linkage analysis of PSG genes and their corresponding intronic LTR8B elements (red boxes). Tracks are displayed as pseudo-bulk signals of each cell type from the snATAC-seq and the colour scale represents the correlation value. b, Genome browser screenshot of the PSG gene cluster with expression and histone modifications tracks of TSCs. The blue shadings highlight the intronic LTR8B elements within PSG genes. All tracks are displayed as RPM, y axes are range from 0 to 2 for RNA-seq, 0 to 80 for H3K27ac, 0 to 40 for H3K4me1 and 0 to 80 for H3K4me3. c, Chromatin accessibility of LTR8B individual elements in all cell types from the snATAC-seq. Each dot represents an LTR8B element; P values were calculated using a one-tailed Wilcoxon test. Elements were filtered for RPKM > 1 in at least one cell type and length > 400 bp (n = 242). d, Chromatin accessibility of all LTR8B individual elements (left; n = 125) and LTR8B elements outside the PSG cluster (right; n = 114). Each dot represents an LTR8B element; P values were calculated using a one-tailed Wilcoxon test. LTR8B elements were filtered by RPKM > 1 in either control or patient sample and length > 400 bp. e, Genome browser screenshot of the STS gene promoter region. The blue shading highlights an LTR8B element. All pseudo-bulk tracks from the snRNA-seq, snATAC-seq and ChIP–seq tracks from the IHEC data repository are displayed in RPM. Significant interactions (one-tailed binomial test Padj < 0.05) predicted by Fit-Hi-C are displayed by purple arcs. The y axes are range from 0 to 15 for snRNA-seq, 0 to 6 for snATAC-seq, 0 to 200 for H3K27ac, 0 to 200 for H3K4me1 and 0 to 500 for H3K4me3. For all boxplots in this figure, the centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively.
Extended Data Fig. 10 Epigenetic regulation of PSG genes in trophoblasts and decreased expression of GATA2 in patient samples.
a, Genome browser screenshot of the PSG gene cluster showing the H3K27ac and H3K27me3 CUT&Tag signals. All tracks are displayed as RPM, with y axes ranging from zero to three for H3K27ac and zero to two for H3K27me3. The blue shadings highlight the intronic LTR8B elements within PSG genes. b, Expression and chromatin accessibility of transcription factors in the ST cell type called in Fig. 8c. The Padj was calculated using the one-tailed MAST model from Seurat for snRNA-seq and a one-tailed Poisson test for snATAC-seq. c, Genome browser screenshot showing expression and chromatin accessibility of GATA2 in the ST cell type from the snRNA-seq and the snATAC-seq. All tracks are displayed as RPM, with y axes ranging from 0 to 40 for snRNA-seq and 0 to 5 for snATAC-seq. d, Chromatin accessibility of GATA2-targeted genes transcriptional start site regions (top; n = 245) and number-matched shuffle control (bottom) between patient and control samples in the ST cell type from the snATAC-seq. Each dot represents a GATA2-targeted gene; P values were calculated using a one-tailed Wilcoxon test. The centre and bounds of boxes indicate the median and quartile of all data points, respectively. The minima and maxima of whiskers indicate quartile 1 − 1.5× the interquartile range and quartile 3 + 1.5× the interquartile range, respectively.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and legends.
Supplementary Tables 1–9
Supplementary Tables 1–9.
Supplementary Data 1
Source data for Supplementary Fig. 1c,d.
Source data
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, L., Mathur, V., Tam, S.K.M. et al. Single-cell analysis reveals transcriptomic and epigenomic impacts on the maternal–fetal interface following SARS-CoV-2 infection. Nat Cell Biol 25, 1047–1060 (2023). https://doi.org/10.1038/s41556-023-01169-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-023-01169-x
This article is cited by
-
Effect of SARS-CoV-2 infection in early pregnancy on placental development
Science China Life Sciences (2024)