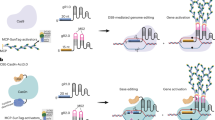
Abstract
Transformation via Agrobacterium tumefaciens is the predominant method used to introduce exogenous DNA into plant genomes1,2. Transfer DNA (T-DNA) originating from Agrobacterium can be integrated as a single copy or in complex concatenated forms3,4, but the mechanisms affecting final T-DNA structure remain unknown. Here we demonstrate that inclusion of retrotransposon (RT)-derived sequences in T-DNA can increase T-DNA copy number by more than 50-fold in Arabidopsis thaliana. These additional T-DNA copies are organized into large concatemers, an effect primarily induced by the long terminal repeats (LTRs) of RTs that can be replicated using non-LTR DNA repeats. We found that T-DNA concatenation is dependent on the activity of the DNA repair proteins MRE11, RAD17 and ATR. Finally, we show that T-DNA concatenation can be used to increase the frequency of targeted mutagenesis and gene targeting. Overall, this work uncovers molecular determinants that modulate T-DNA copy number in Arabidopsis and demonstrates the utility of inducing T-DNA concatenation for plant gene editing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data generated from this study are included within the main figures, extended data and Supplementary Information. Raw sequencing data are deposited at the NCBI SRA (BioProject PRJNA892619). Analysis of the data made use of the TAIR10 Arabidopsis genome (https://www.arabidopsis.org/download/index-auto.jsp%3Fdir%3D%252Fdownload _files%252FGenes%252FTAIR10_genome_release) and the ‘Orthologous gene family’ list sourced from the Dicots PLAZA 5.0 database (https://ftp.psb.ugent.be/pub/plaza/plaza_public_dicots_05/GeneFamilies/genefamily_data.ORTHOFAM.csv.gz). There are no restrictions on data availability. Source data are provided with this paper.
References
Gelvin, S. B. Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 51, 195–217 (2017).
Gelvin, S. B. Plant DNA repair and Agrobacterium T-DNA integration. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22168458 (2021).
Jupe, F. et al. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet. 15, e1007819 (2019).
Pucker, B., Kleinbolting, N. & Weisshaar, B. Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis. BMC Genomics 22, 599 (2021).
Chilton, M. D. et al. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11, 263–271 (1977).
Galbiati, M., Moreno, M. A., Nadzan, G., Zourelidou, M. & Dellaporta, S. L. Large-scale T-DNA mutagenesis in Arabidopsis for functional genomic analysis. Funct. Integr. Genomics 1, 25–34 (2000).
Zambryski, P. et al. Tumor DNA structure in plant cells transformed by A. tumefaciens. Science 209, 1385–1391 (1980).
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A. & Voytas, D. F. DNA replicons for plant genome engineering. Plant Cell 26, 151–163 (2014).
Kralemann, L. E. M. et al. Distinct mechanisms for genomic attachment of the 5′ and 3′ ends of Agrobacterium T-DNA in plants. Nat. Plants 8, 526–534 (2022).
van Kregten, M. et al. T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat. Plants 2, 16164 (2016).
Jacob, Y. et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466, 987–991 (2010).
Davarinejad, H. et al. The histone H3.1 variant regulates TONSOKU-mediated DNA repair during replication. Science 375, 1281–1286 (2022).
Zaratiegui, M. Cross-regulation between transposable elements and host DNA replication. Viruses https://doi.org/10.3390/v9030057 (2017).
Cavrak, V. V. et al. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 10, e1004115 (2014).
Ito, H. et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119 (2011).
Wolter, F., Klemm, J. & Puchta, H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 94, 735–746 (2018).
Bechtold, N., Ellis, J. & Pelletier, G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III 316, 1194–1199 (1993).
Alonso, J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003).
McElver, J. et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763 (2001).
Sessions, A. et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994 (2002).
Kamp, J. A., van Schendel, R., Dilweg, I. W. & Tijsterman, M. BRCA1-associated structural variations are a consequence of polymerase theta-mediated end-joining. Nat. Commun. 11, 3615 (2020).
Jacob, Y. et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253 (2014).
Jacob, Y. et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 16, 763–768 (2009).
Nishizawa-Yokoi, A. et al. Agrobacterium T-DNA integration in somatic cells does not require the activity of DNA polymerase θ. New Phytol. 229, 2859–2872 (2021).
Inagaki, S. et al. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 18, 879–892 (2006).
Hussmann, J. A. et al. Mapping the genetic landscape of DNA double-strand break repair. Cell 184, 5653–5669.e25 (2021).
Budzowska, M. et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 23, 3548–3558 (2004).
Wang, Q. et al. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 33, 862–877 (2014).
Williams, G. J., Lees-Miller, S. P. & Tainer, J. A. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair 9, 1299–1306 (2010).
Fauser, F. et al. In planta gene targeting. Proc. Natl Acad. Sci. USA 109, 7535–7540 (2012).
Scheiffele, P., Pansegrau, W. & Lanka, E. Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J. Biol. Chem. 270, 1269–1276 (1995).
Ward, E. R. & Barnes, W. M. VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 242, 927–930 (1988).
Neale, M. J., Pan, J. & Keeney, S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057 (2005).
Brzezinka, K., Altmann, S. & Baurle, I. BRUSHY1/TONSOKU/MGOUN3 is required for heat stress memory. Plant Cell Environ. 42, 771–781 (2019).
Li, W. et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl Acad. Sci. USA 101, 10596–10601 (2004).
Valuchova, S. et al. Protection of Arabidopsis blunt-ended telomeres is mediated by a physical association with the Ku heterodimer. Plant Cell 29, 1533–1545 (2017).
Heacock, M. L., Idol, R. A., Friesner, J. D., Britt, A. B. & Shippen, D. E. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 35, 6490–6500 (2007).
Heitzeberg, F. et al. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 38, 954–968 (2004).
Samanic, I., Simunic, J., Riha, K. & Puizina, J. Evidence for distinct functions of MRE11 in Arabidopsis meiosis. PLoS ONE 8, e78760 (2013).
Feng, W. et al. Large-scale heterochromatin remodeling linked to overreplication-associated DNA damage. Proc. Natl Acad. Sci. USA 114, 406–411 (2017).
Culligan, K., Tissier, A. & Britt, A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104 (2004).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Puizina, J., Siroky, J., Mokros, P., Schweizer, D. & Riha, K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16, 1968–1978 (2004).
Molina-Risco, M. et al. Optimizing Agrobacterium-mediated transformation and CRISPR-Cas9 gene editing in the tropical japonica rice variety presidio. Int. J. Mol. Sci. https://doi.org/10.3390/ijms222010909 (2021).
Clemente, T. Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). Methods Mol. Biol. 343, 143–154 (2006).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016).
Rouillard, J. M., Zuker, M. & Gulari, E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31, 3057–3062 (2003).
Sage, D. et al. DeconvolutionLab2: an open-source software for deconvolution microscopy. Methods 115, 28–41 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Jovtchev, G., Schubert, V., Meister, A., Barow, M. & Schubert, I. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114, 77–82 (2006).
Ollion, J., Cochennec, J., Loll, F., Escude, C. & Boudier, T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841 (2013).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Cock, P. J. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Lamesch, P. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 (2012).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997v2 (2013).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Van Bel, M. et al. PLAZA 5.0: extending the scope and power of comparative and functional genomics in plants. Nucleic Acids Res. 50, D1468–D1474 (2022).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience https://doi.org/10.1093/gigascience/giab008 (2021).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Jasper, F., Koncz, C., Schell, J. & Steinbiss, H. H. Agrobacterium T-strand production in vitro: sequence-specific cleavage and 5′ protection of single-stranded DNA templates by purified VirD2 protein. Proc. Natl Acad. Sci. USA 91, 694–698 (1994).
Kleinboelting, N. et al. The structural features of thousands of T-DNA insertion sites are consistent with a double-strand break repair-based insertion mechanism. Mol. Plant 8, 1651–1664 (2015).
Feng, W. et al. Marker-free quantification of repair pathway utilization at Cas9-induced double-strand breaks. Nucleic Acids Res. 49, 5095–5105 (2021).
Acknowledgements
We thank all current and former members of the Jacob lab, especially F. Langhammer and G. Villarino, for reagents, discussions and advice; C. Bolick and his staff at Yale for help with plant growth and maintenance; H. Puchta from the Karlsruhe Institute of Technology for his generous gifts of the gene targeting binary plasmids used in this work; and Marco Molina and Mayra Molina (Multi-Crop Transformation Facility, Texas A&M University) for generating the transgenic tobacco plants used in this study. This project was supported by grant R35GM128661 from the National Institutes of Health (Y.J.) and an NIH Director’s New Innovator Award (DP2GM137414 to S.W.).
Author information
Authors and Affiliations
Contributions
Y.J. supervised the study and designed the experiments with L.D. and W.Y. All experiments were performed by L.D. and W.Y., except the targeted mutagenesis work (done by C.L.) and the bioinformatic analyses (done by G.T.). S.W. designed the probes for the FISH experiments. Y.J. and C.L. wrote the paper, with contributions from L.D. and W.Y.
Corresponding author
Ethics declarations
Competing interests
A patent application (patent application no. 63/481,276, US Patent and Trademark Office, 24 January 2023), with Y.J., L.D. and W.Y. as inventors, related to the use of repetitive sequences and genetic mutations to regulate T-DNA copy number and improve gene editing, has been filed. The other authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Avraham Levy, Damon Lisch and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
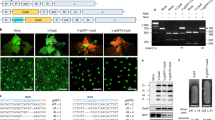
Extended Data Fig. 1 DNA FISH analysis.
a, DNA-qPCR of ALS in Col-0 (untransformed) and T1 plants used for the FISH experiment (panel b and Fig. 1e). Dots represent DNA samples from individual leaves from the same T1 plant (n = 4 for Col-0, ONSEN RT #1 and ONSEN RT #2, n = 3 for No RT). Horizontal bars indicate the mean. SD is shown. b, FISH in leaf nuclei targeting the T-DNA sequence. Nuclei were stained with DAPI and probes for ALS (green) and NPTII (red). The FISH experiment was performed >3 times with similar results.
Extended Data Fig. 2 Whole genome sequencing analysis of T-DNA insertions.
a, DNA-qPCR of ALS in the No RT and ONSEN RT T1 plants used for whole genome sequencing (panel b, Fig. 1f,g and Fig. 3b). b, Locus diagrams for the identified T-DNA insertions. The coordinates for each insertion are based on the TAIR10 annotation and correspond to the Arabidopsis genomic borders surrounding each identified T-DNA. For each insertion, top lines represent unaltered genomic sequence with annotated genes (shades of navy blue). Red arrows represent insertion points. The bottom lines show the borders of the insertion in more detail, with the identified insertion of binary vector (non-T-DNA region) sequence or T-DNA components shown. Dashed lines represent contiguous T-DNA-associated cassettes. Red bars indicate binary vector sequence (non-T-DNA). Dark blue bars (LB) and light gray bars (pea3A(T)) indicate the 5′ end of the T-DNA construct (though it may be in 5′ or 3′ orientation in the plant genome). Darker gray bars adjacent to the red bars are filler sequences, and the teal bar represents NPTII sequence.
Extended Data Fig. 3 Repetitive sgRNA genes do not contribute to T-DNA concatenation.
a, Schematic representation of the sgRNA gene deletion from the No RT vector. b, DNA-qPCR of ALS in Col-0 (untransformed) and in T1 plants transformed with the No RT plasmid and the No RT plasmid with a sgRNA gene deleted. Each dot represents an individual plant (n = 21 for Col-0, n = 26 individual T1 plants for No RT and No RT, sgRNA gene deletions). Horizontal bars indicate the median. PCol-0 = 0.00000002, ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). c. Schematic representation of the sgRNA gene deletion from the ONSEN RT vector. d. DNA-qPCR of ALS in Col-0 (untransformed) and in T1 plants transformed with the ONSEN RT plasmid and the ONSEN RT plasmid with both sgRNA genes deleted. Each dot represents an individual plant (n = 22). Horizontal bars indicate the median. PCol-0 = 0.00000004, ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). e. DNA-qPCR of ALS in tobacco plants transformed by biolistic bombardment with particles coated with No RT or ONSEN RT plasmid DNA. Each dot represents an individual plant. Horizontal bars indicate the median. ns = not significantly different (two-tailed Mann-Whitney U test). ns, P > 0.05; ****, P < 0.0001.
Extended Data Fig. 4 T-DNA junctions support the involvement of TMEJ in T-DNA concatenation.
a, Multiple sequence alignments of junctions between two T-DNA sequences grouped by orientation of the two sequences (RB-LB, LB-LB or RB-RB). Reference sequences were constructed assuming T-DNA molecules began and ended immediately upstream of the endonuclease (VirD2) recognition sequence (5′-CAGGATATATT-3′)63,64. Filler sequences are in red and sequences consistent with microhomology-associated deletions are underlined. If filler sequences have a similar sequence nearby, they (and the nearby sequence) are also underlined. Asterisks indicate identical junctions occurring in independent plants. b, Depiction of T-DNA junctions with another T-DNA or binary vector sequence not immediately internally adjacent to the LB or RB. c. Classification of RB-LB, LB-LB and RB-RB sequences for each T1 line following a procedure previously described65. NHEJ (<4 bp deletions and <5 bp insertions), insertions (≥5 bp with any deletion), non-microhomology (Non-MH; ≥4 bp deletion or <5 bp insertions with microhomologies <2), and microhomology (MH; ≥4 bp deletion with microhomologies ≥2). The latter three are associated with DNA polymerase theta.
Extended Data Fig. 5 T-DNA concatenation is not DNA replication-dependent.
a, DNA-qPCR of ALS in Col-0, atxr5/6, and tsk plants transformed with the ONSEN RT construct. Each dot represents an individual T1 plant (n = 24). Horizontal bars indicate the median. ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). b, DNA-qPCR of ALS in DNA extracted from leaves of T1 plants at 16 days and 30 days after germination.
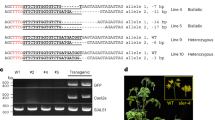
Extended Data Fig. 6 T-DNA concatenation increases the efficiency of targeted mutagenesis and gene targeting.
a. Gene structure of CRY2. Gray bars represent exons, and blue bars represent regions targeted by sgRNAs. b, c. INDEL frequency of CRY2 PCR products amplified from DNA extracted from leaves of individual T1 plants transformed with either No RT or ONSEN RT constructs. Constructs carried either (b) sgRNA #2 or (c) sgRNA #3. d. DNA-qPCR of ALS in T1 plants with no detectable CRY2 indels (No indels) or detectable indels (Indels) using CRY2 sgRNA #3. Each dot represents an individual T1 plant (n = 20). Horizontal bars indicate the median. *P = 0.0350 (two-tailed Mann-Whitney U test) e. DNA-qPCR of ALS in No RT and ONSEN RT T1 plants (gray) in relation to the percentage of true (non-ectopic) gene targeting rates in the T2 generation (red).
Extended Data Fig. 7 Model of T-DNA concatenation.
The model builds on the T-DNA integration model from Kralemann et al., 20226. Chromosomal capture of a T-DNA strand 3′ end is mediated by TMEJ. After conversion to a double-stranded T-DNA intermediate, capture of the T-DNA 5′ end is accomplished by removal of the Agrobacterium protein VirD2 by TDP2 or MRE11. TDP2-mediated removal of VirD2 creates blunt-ended DNA at the T-DNA 5′ end that is ligated to the chromosome by NHEJ. In contrast, MRE11, acting as part of the MRN complex (MRE11-RAD50-NBS1; loaded on DNA by RAD1725), removes VirD2 by cutting the T-DNA internally, generating a staggered end at the T-DNA 5′ end. TDP2/NHEJ activity leads to a single T-DNA copy integration (outcome A), while MRE11/TMEJ activity leads to multiple outcomes (B-E), with the simplest one being chromosomal capture of the T-DNA 5′ end (outcome B). Alternatively, the staggered T-DNA 5′ end can facilitate recruitment of additional T-DNA strands for ligation, leading to concatenation. Capture of the 5′ end of an additional T-DNA strand by TDP2/NHEJ instead of MRE11/TMEJ is more likely to terminate the concatenation cycle. In this model, T-DNA features like DNA repeats may increase concatenation levels by increasing the number of available T-DNA strands for integration, and/or their accessibility. ssDNA: single-stranded DNA. dsDNA: double-stranded DNA. Created with BioRender.com.
Supplementary information
Supplementary Table 1
Supplementary Table 1. List of probes used for FISH analysis and primers.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dickinson, L., Yuan, W., LeBlanc, C. et al. Regulation of gene editing using T-DNA concatenation. Nat. Plants 9, 1398–1408 (2023). https://doi.org/10.1038/s41477-023-01495-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01495-w
This article is cited by
-
The power of repetition
Nature Plants (2023)