Abstract
Hypercholesterolemia is a major risk factor for atherosclerosis and associated cardiovascular diseases. The liver plays a key role in the regulation of plasma cholesterol levels and hosts a large population of tissue-resident macrophages known as Kupffer cells (KCs). KCs are located in the hepatic sinusoids where they ensure key functions including blood immune surveillance. However, how KCs homeostasis is affected by the build-up of cholesterol-rich lipoproteins that occurs in the circulation during hypercholesterolemia remains unknown. Here, we show that embryo-derived KCs (EmKCs) accumulate large amounts of lipoprotein-derived cholesterol, in part through the scavenger receptor CD36, and massively expand early after the induction of hypercholesterolemia. After this rapid adaptive response, EmKCs exhibit mitochondrial oxidative stress and their numbers gradually diminish while monocyte-derived KCs (MoKCs) with reduced cholesterol-loading capacities seed the KC pool. Decreased proportion of EmKCs in the KC pool enhances liver cholesterol content and exacerbates hypercholesterolemia, leading to accelerated atherosclerotic plaque development. Together, our data reveal that KC homeostasis is perturbed during hypercholesterolemia, which in turn alters the control of plasma cholesterol levels and increases atherosclerosis.
Similar content being viewed by others
Introduction
Atherosclerotic cardiovascular disease (ASCVD) and its clinical outcomes such as myocardial infarction are a leading cause of morbidity and mortality worldwide. Multiple lines of evidence have established that hypercholesterolemia, characterized by high concentrations of cholesterol-rich low-density lipoproteins (LDL)1 and their oxidatively damaged forms (oxLDL)2, is a leading risk factor in ASCVD. While the liver and intestine are the major organs regulating plasma cholesterol levels, several lines of evidence suggest the immune system could also participate. Thus, reducing3,4 or increasing5 tissue-resident macrophage (trMac) numbers associated with plasma cholesterol elevation or reduction, respectively. Yet, whether a specific trMac population is primarily involved remains undefined. Though, these models had in common a modulation in the hepatic trMacs, Kupffer cells (KCs), arguing for their potential implication in the control of cholesterolemia.
Located in the hepatic sinusoids, KCs are the largest trMac population in direct contact with the blood and thus with circulating lipoproteins. They originate from various progenitor waves during embryogenesis and maintain by self-renewal independently from circulating monocytes in the steady-state adult liver6,7,8. However, when the embryo-derived KC (EmKC) network is altered during metabolic dysfunction-associated steatohepatitis (MASH)9,10,11,12, excessive intravascular hemolysis13, bacterial14 or parasitic15 infections, blood monocytes can be recruited to the liver and differentiate into KCs. Analysis of how these monocyte-derived KCs (MoKCs) engraft the liver tissue has revealed that KCs closely interact with liver sinusoidal endothelial cells (LSECs), hepatic stellate cells (HSCs) and hepatocytes in order to acquire their tissue-imprinted signature as well as the signals allowing them to self-maintain16,17. Aside their undisputable role as an intravascular immune barrier that constantly filters the blood for pathogens18,19, but also for damaged red blood cells13, KCs have been recently endowed with activities influencing the liver response to metabolic diseases such as obesity20 or MASH10.
Here, we evaluated whether KCs homeostasis was altered in the context of hypercholesterolemia using cholesterol-fed mice deficient for the LDL receptor gene (Ldlr−/−). Our findings reveal a two-phase adaptation of the KC pool to plasma cholesterol elevation. In a first stage, EmKCs adapt to this environmental change by loading large amount of cholesterol and increasing their tissue density. Then, in a second phase, EmKCs are gradually lost over weeks and replaced by MoKCs exhibiting a diminished ability to uptake lipoprotein-derived cholesterol. To infer the functional consequences of EmKCs replacement by MoKCs, we specifically deleted EmKCs. Replacement of EmKCs by MoKCs in hypercholesterolemic mice resulted in perturbed hepatic cholesterol and bile acids homeostasis with decreased activation of the retinoid X receptors (RXR)/liver X receptors (LXR) pathway that associated with increased cholesterol content in the liver tissue and in plasma. Overall, our results support a role for EmKCs in regulating hepatic metabolic adaptation to hypercholesterolemia.
Results
Embryo-derived Kupffer cells markedly increased shortly after the induction of hypercholesterolemia
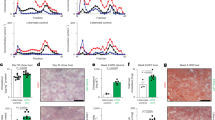
To characterize the potential changes in hepatic leukocyte populations that occur rapidly after the induction of hypercholesterolemia, Ldlr−/− mice were subjected to a chow diet enriched with 1% cholesterol (HC diet). After 4 days of HC diet, cholesterolemia raised by 4 times, reaching approximately 1000 mg/dL (Fig. 1A). We then performed a flow cytometry analysis of common hepatic leukocyte populations in livers perfused in situ with collagenase. Of note, in situ perfusion was critical to retrieve KCs under hypercholesterolemia as classical digestion protocols failed. A t-distributed stochastic neighbor embedding (tSNE) analysis of the cytometry data revealed no major changes between chow and HC diet conditions, except for KCs. KCs, identified by their classical marker VSIG4, distributed very differently between conditions in the tSNE projection (Fig. 1B). To further analyze the impact of short-term HC diet feeding on KCs, RNA sequencing was performed on KCs isolated from mice fed a chow or a HC diet for 4 days. This revealed marked changes in gene expression upon HC diet (Fig. 1C). KEGG pathway analysis on genes up-regulated in the HC-fed condition showed terms associated with lysosomes and cholesterol metabolism (Fig. 1D). Highlighted pathways were also suggestive of intense metabolic activities (metabolism of xenobiotics, arginine biosynthesis, drug metabolism, ribosome). Interrogation of the GSEA Molecular Signatures Database (MSigDB) confirmed such activities (Xenobiotic, Mtorc1 signaling, Glycolysis) and also revealed the induction of genes involved in cell cycling (G2m checkpoint, Mitotic spindle) (Fig. 1D). We then assessed whether the enrichment in pathways linked to cell proliferation was associated with increased KC numbers. Our previous studies have demonstrated that all CLEC2hi leukocytes observed by flow cytometry in the healthy or diseased liver identified KCs, including MoKCs that lack TIMD4 expression10. After 4 days of hypercholesterolemia, CD45+ CLEC2+ KCs displayed, similar to the chow condition, homogeneous cell surface expression for the EmKCs markers CLEC4F, VSIG4 and TIMD410,21 (Fig. 1E). Thus, all KCs remain of embryonic origin after 4 days of HC diet and EmKCs absolute numbers increased by 2–3-fold in the HC-fed condition (Fig. 1F). Increased KC density was confirmed by immunofluorescence microscopy of frozen sections using E-CADHERIN staining to demarcate periportal regions22 and CLEC4F staining to identify KCs (Fig. 1G). This led us to assess the proportion of KCs expressing the proliferation marker KI-67. The results showed approximately a 4-fold increase in KI-67+ EmKCs in mice fed the HC diet for 4 days as compared to mice fed the chow diet (Fig. 1H). A kinetic analysis showed that increased EmKCs proliferation was already noticeable as soon as 3 days after the start of HC diet feeding (Fig. S1). Thus, as suggested by our RNA-Seq data, HC diet-induced hypercholesterolemia increased EmKCs proliferation and density in the liver of HC diet-fed animals. We next wondered what drove EmKCs proliferation and focused on colony-stimulating factor 1 (CSF1) that is known to play a critical role in tissue macrophage maintenance and proliferation16,23. RT-qPCR measurement of Csf1 mRNA expression showed its progressive elevation from day 2 to day 4 in the livers of Ldlr−/− mice fed the HC diet (Fig. 1I). Cell sorting of endothelial (LSECs) and stellate cells (HSCs), the main CSF1-expressing cells within the liver16, showed that HSCs were the cells that expressed more Csf1 4-days post-induction of hypercholesterolemia (Figs. 1J, S2A). DESMIN staining on liver slices also indicated that HSCs density was increased after 4 days of HC diet in Ldlr−/− mice (Figs. 1K, S2B), suggesting KC niche adaptation to sustain EmKCs expansion. Finally, administration of the CSF1R inhibitor PLX3397 to Ldlr−/− mice during the first 2-days of HC diet feeding fully blocked EmKCs proliferation and increase in numbers (Fig. 1L). Thus, EmKCs proliferate and expand shortly after the induction of hypercholesterolemia in a CSF1R signaling-dependent manner and involving HSCs activation to provide CSF1.
A Plasma total cholesterol in Ldlr−/− male mice fed a chow diet (n = 8 mice) or chow diet supplemented with 1% cholesterol (HC) (n = 7 mice) for 4 days (2 independent experiments combined; ****p < 0.0001). B tSNE projections of identified leukocytes by flow cytometry in livers of chow-fed or HC-fed Ldlr−/− mice for 4 days. High expression of VSIG4 identifies KCs. C Volcano plot depicting differentially expressed genes (fold change ≥ 1.3 and adjusted p-value ≥ 0.05 in red) in sorted KCs from livers of chow-fed or HC-fed Ldlr−/− mice for 4 days. D Pathways enriched in KCs in response to hypercholesterolemia. E Flow cytometry analysis of KCs and histograms showing the expression of KC markers CLEC4F, VSIG4 and TIMD4. F KC density in livers of chow- (n = 8 mice) and HC-fed (n = 8 mice) Ldlr−/− male mice for 4 days (2 independent experiments combined; ****p < 0.0001). G Representative microscopy images of livers of chow- and HC-fed (4 days) Ldlr−/− male mice showing expression of CLEC4F (red) and E-CADHERIN (green). Nuclei (DAPI) are shown in blue. Scale bar = 100 μm. Quantification of CLEC4F+ and DAPI+ cells density in HC-fed (4 days, n = 3) and chow-fed (n = 8) Ldlr−/− mice (****p < 0.0001). H Percentage of KI-67+ cells among CLEC2+ KCs of chow (n = 10) or HC-fed (n = 8) Ldlr−/− male mice (2 independent experiments combined; ****p < 0.0001). I Fold-change in Csf1 gene expression in livers of Ldlr−/− male mice fed HC diet for the indicated days (n = 12, 3, 7, 6 and 10 mice for day 0, 1, 2, 3 and 4, respectively). Data are expressed relative to day 0 and p values correspond to statistical differences versus day 0 (1-way ANOVA and Dunnett’s multiple comparison test; 0 vs 3 days: *p = 0.04 and 0 vs 4 days: ****p < 0.0001). J RT-qPCR measurement of Csf1 mRNA expression in sorted HSC and LSEC cells of mice fed HC (n = 3) or chow diet (n = 3). Statistical significance assessed with 2-way ANOVA (chow HSCs vs HC HSCs: p = 0.057). K Quantification of DESMIN+ and DAPI+ cells in HC-fed (4 days, n = 3) and chow-fed (n = 3) Ldlr−/− mice (**p = 0.009). L Percentage of KI-67+ KCs and KC numbers in Ldlr−/− female mice fed a chow or HC diet for 3 days and treated with PLX3397 (CSF1R inhibitor) or vehicle. Statistical significance tested with 1-way ANOVA and Tukey multiple comparison test (n = 10, 8 and 9 mice for chow, HC and HC + PLX respectively; Combined 2 independent experiments; For KI-67+ KCs, chow vs HC and HC vs HC + PLX: ****p < 0.0001 and for KC numbers, chow vs HC: **p = 0.009 and HC vs HC + PLX: *p = 0.02). All data in this figure are presented as mean values ± SEM. Statistical significance has been assessed with a two-sided t-test unless otherwise stated on the corresponding panel legend. Source data are provided as a Source Data File.
Massive cellular accumulation of cholesterol occurs in Kupffer cells following induction of hypercholesterolemia
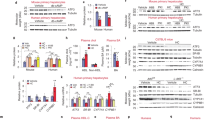
We next aimed to further characterize the cellular changes occurring in EmKCs shortly after the induction of hypercholesterolemia (4 days). As reported above, genes associated with cholesterol metabolism were enriched in KCs isolated from hypercholesteremic animals (Fig. 1D). Thus, we wondered if genes involved in cholesterol biosynthesis were regulated in KCs after the induction of hypercholesteremia. We performed Gene Set Enrichment Analysis (GSEA) using a gene dataset that include all the major genes involved in cholesterol synthesis. The analysis revealed a clear downregulation of this pathway in the HC-fed condition (Fig. 2A). Such expression profile is usually observed in cholesterol-loaded cells, including foamy macrophages24. Foamy macrophages are typically found in atherosclerotic lesions and transcriptomic profiles of intimal foamy and non-foamy macrophages were recently reported25. We thus conducted GSEA using the 250 most up-regulated genes in foamy versus non-foamy macrophages and revealed a specific enrichment of these genes in EmKCs isolated from HC-fed Ldlr−/− mice (Fig. 2A). Foamy macrophages found in atherosclerotic plaques were previously characterized by their high granularity and elevated intracellular lipid levels as revealed by bodipy 493/503 staining25. Flow cytometry analyses also showed that hypercholesterolemia specifically elevated granularity (SSC-A parameter) in KCs (Fig. 2B) that associated with higher bodipy staining (Fig. 2C). Thus, KCs expand and accumulate intracellular lipids shortly after the induction of hypercholesterolemia. Increased granularity and bodipy staining could be observed also in some other tissue macrophage populations, such as spleen and kidney macrophages, but to a lower extent than in KCs and without changes in tissue densities of these macrophage populations (Fig. S3). Targeted lipidomic showed an increase in both free cholesterol (4-fold) and cholesteryl-esters (CE) (15-fold) in KCs isolated from mice fed the HC diet as compared to chow-fed controls (Fig. 2D). All CE species quantified were found elevated in cholesterol-loaded KCs with CE derived from the monounsaturated fatty acid oleate (CE 18:1) being the most prevailing form (Fig. 2D). We then used two-photon laser scanning microscopy combined with Coherent anti-Stokes Raman Spectroscopy (CARS) to detect lipids in situ in the liver of Ldlr−/− mice fed chow and HC diet for 4 days. We observed the presence of numerous lipid droplets in KCs of hypercholesterolemic mice (Fig. 2E) and, as expected, larger lipid droplets were also detectable in hepatocytes under these conditions (Fig. 2E). Lipidomic analysis of the liver revealed that hepatic cholesterol content raised only by 2-fold after 4 days of hypercholesterolemia, and primarily in the form of CE (Fig. 2F). Considering both KC numbers and cholesterol-loading increase following the induction of hypercholesterolemia, we estimated a 25-fold increase in the amount of cholesterol stored by the KC population upon HC feeding (Fig. 2G). Nevertheless, despite this important increase, the amount of cholesterol stored in KCs was only a marginal fraction of the total liver cholesterol content as it was estimated to represent only 0.25%.
A Gene Set Enrichment analysis (GSEA) of the reactome_cholesterol_biosynthesis and foamy plaque macrophages gene signatures in KC from chow- and HC-fed (4 days) Ldlr−/− mice using the Phantasus software. The foamy plaque macrophages gene signature was generated from a previously published dataset (GSE116239). B KCs in HC-fed mice exhibit high granularity (SSC-Ahi) and (C) strongly stain for lipids (bodipy; n = 8 chow and n = 7 HC; combined 2 independent experiments; ****p < 0.0001). D Changes in cholesterol content (*p = 0.0115), in cholesteryl-esters (*p = 0.0115) and various CE species in KCs sorted from chow- and HC-fed (4 days) Ldlr−/− mice (n = 3 chow and n = 4 HC; 1 experiment). E Two-photon laser scanning microscopy images of the liver of chow and HC-fed (4 days) Ldlr−/− mice. Numerous lipid droplets (white spots) are detected by Coherent anti-Stokes Raman Spectroscopy (CARS) in TIMD4+ KCs (red; 3D reconstruction by mask rendering on the right images) and in the hepatocytes (autofluorescence in green) of HC-fed mice as compared to the chow condition. Scale bar = 10 μm. F Calculated amount of total cholesterol (**p = 0.006) and of free cholesterol (FC) and cholesteryl-esters (CE) (****p < 0.0001) present in the liver of chow- (n = 6 mice) and HC-fed (4 days; n = 8 mice; 1 experiment) Ldlr−/− mice. G Calculated amount of total cholesterol (****p < 0.0001) and of FC (**p = 0.003) and CE (**p = 0.003) present in the KC pool of chow- (n = 3 mice) and HC-fed (4 days; n = 5 mice; 1 experiment) Ldlr−/− mice. H Heatmap generated from the RNA-Seq depicting the level of expression of scavenger receptors in KCs from chow- and HC-fed (4 days) Ldlr−/− mice. I Changes in the granularity (SSC-A; ***p = 0.0002) and lipid content (bodipy; **p = 0.005) of KCs in mice fed HC diet overnight and injected in vivo either with anti-CD36 blocking antibodies (n = 8) or isotype control antibodies (n = 9) (2 independent experiments combined). All data in this figure are presented as mean values ± SEM. Statistical significance has been assessed with a two-sided t-test unless otherwise stated on the corresponding panel legend. Source data are provided as a Source Data File.
Considering the phagocytic nature of KCs, we hypothesized that cholesterol loading in KCs would result from the uptake of modified forms of LDL generated upon hypercholesterolemia, as it occurs in atherosclerotic foamy macrophages. Among the different scavenger receptors involved in modified LDLs uptake, Cd3626 and Colec1227 were found to be up-regulated in KCs from HC-fed animals (Fig. 2H). To test the potential involvement of CD36, we administered a CD36 blocking mAbs28 or its isotype control to Ldlr-/- fed the HC diet for 1 day. Blocking CD36 led to decreased KC granularity as well as bodipy staining (Fig. 2I). Thus, cholesterol loading is, at least partially, dependent on CD36.
We next sought to exclude the possibility that the changes observed in KCs were derived from other mechanisms than the hypercholesterolemic state, such as dietary cholesterol-induced gut microbiota dysbiosis29. To do so, Ldlr−/− mice were treated with the selective cholesterol absorption inhibitor ezetimibe while given cholesterol in the diet. A pre-treatment period with ezetimibe before addition of cholesterol was set up to ensure full activity of the inhibitor (Fig. 3A). Ezetimibe treatment fully blocked plasma cholesterol elevation when mice were switched to HC diet (Fig. 3B). As a result, KCs proliferative response (Fig. 3C), expansion (Fig. 3D) and lipid loading (Fig. 3E) were abolished, demonstrating that induction of hypercholesterolemia was the only triggering factor.
A Experimental plan for ezetimibe treatment to block intestinal cholesterol absorption. B Feeding ezetimibe to Ldlr−/− male mice fed HC diet blocked plasma cholesterol elevation (chow vs HC and HC vs HC + Eze: ****p < 0.0001), resulting in (C) no increased proliferation (as determined by KI-67 staining; chow vs HC: ***p = 0.0002 and HC vs HC + Eze: **p = 0.004) (D), no increased pool density (chow vs HC: ***p = 0.0008 and HC vs HC + Eze: **p = 0.002), and (E) no lipid loading (bodipy; chow vs HC and HC vs HC + Eze: ****p < 0.0001) of KCs. (n = 4, 4 and 3 for chow-, HC- and HC + eze-fed mice, respectively). All data in this figure are from one experiment and presented as mean values ± SEM. Statistical significance tested with 1-way ANOVA and Tukey multiple comparison test. Source data are provided as a Source Data File.
Altogether, we show that EmKCs are actively mobilized during hypercholesterolemia and accumulate large amounts of cellular cholesterol, notably through the scavenging of modified forms of cholesterol-rich LDLs from the bloodstream.
Monocyte-derived KC with reduced cholesterol-loading sustain the KC pool that contracts during prolonged exposure to hypercholesterolemia
We next evaluated the consequences of prolonged exposure to hypercholesterolemia on KCs. Female Ldlr−/− mice were fed the HC diet for up to 3 weeks and analyzed at different time points during this period. After the initial expansion of the KC pool observed at day 4, KC numbers then dropped and stabilized at an intermediate level (Fig. 4A). As mentioned above, previous works revealed that TIMD4 expression is essentially absent on monocyte-derived KCs10,21. Here, we observed that the percentage of CLEC2+ TIMD4− cells among KCs significantly raised after 8 days of HC diet feeding (Fig. 4B). By day 21, approximately 50% of KCs were TIMD4- (Fig. 4C), suggesting that MoKCs were generated to sustain the KC pool. Absolute cell quantification during this time course analysis clearly showed the progressive engraftment of MoKCs (Fig. 4D). We next generated Ccr2−/− × Ldlr−/− mice, in which circulating Ly-6Chi monocyte numbers are markedly reduced30, and showed that CLEC2+ TIMD4− MoKCs were not observed in these animals when subjected to hypercholesterolemia for 3 weeks (Fig. 4E). This confirmed the monocytic origin of MoKCs. After the short expansion phase already described above, we found that EmKCs decreased overtime, returning to steady state levels after 3 weeks of HC diet (Fig. 4D). We thus quantified hepatic Csf1 mRNA expression levels to determine whether the decrease in EmKCs could result from diminished maintenance signals. Csf1 mRNA expression progressively increased till day 8 and was then slightly reduced, remaining however elevated by 2-fold as compared to the control chow condition (Fig. 4F). Thus, the decrease in EmKCs numbers after day 4 is unlikely to be the consequence of reduced CSF1 amounts available between day 4 and 8 as Csf1 mRNA expression kept rising during this time frame. As previously noticed in the context of MASH10, MoKCs also presented with a 2–3-fold higher proliferative rate than EmKCs after 3 weeks of HC diet (Fig. 4G). Thus, increased proliferation could confer an advantage to MoKCs as compared to their embryonically-derived counterparts in colonizing the KC niche in the hypercholesterolemic environment. We then asked whether MoKCs generation kinetics would differ between females and males. Thus, changes in KCs homeostasis upon prolonged hypercholesterolemia were also evaluated in male Ldlr−/− mice (Fig. S4A). The contraction of the EmKC pool that occurs after the early proliferative phase was less abrupt in males and only started after day 8 (Fig. S4B). In addition, MoKCs engraftment started only after day 8 and their proportion among the KC pool after 3 weeks of diet was twice as less than that seen in females (Fig. S4C). Overall, the results obtained were comparable to those obtained in females, albeit slight differences were noticeable. Having characterized changes occurring to the KC population, we wondered whether inflammatory monocyte-derived macrophages (MoDMacs) were also generated in the liver upon hypercholesterolemia. To do so, we assessed whether we could identify CD64+ CLEC2− macrophages by flow cytometry. While Ly-6C+ monocytes increased in the liver after 3 weeks of HC diet, very few MoDMacs could be found (Figs. 4H, S4D). Thus, during prolonged exposure to hypercholesterolemia, monocytes are progressively recruited to the liver and differentiate into MoKCs, but only marginally as inflammatory MoDMacs.
A KC numbers before and 4, 8, 13 and 21 days after induction of hypercholesterolemia (0 vs 4 days: ****p < 0.0001 and 0 vs 13 days: *p = 0.04). B Representative flow cytometry of CLEC2 + TIMD4- KCs during prolonged exposure of the mice to hypercholesterolemia. C Frequency of CLEC2 + TIMD4- KCs (0 vs 8 days: **p = 0.0015, 0 vs 13 days: ***p = 0.0001 and 0 vs 21 days: ****p < 0.0001). D EmKCs and MoKCs numbers before and at 4, 8, 13 and 21 days after induction of hypercholesterolemia (Panels A, C and D: n = 9, 8, 5, 4 and 8 mice for experimental points 0, 4, 8,13, and 21 days; 2 independent experiments combined) (1-way ANOVA and multiple comparison test; EmKCs: 0 vs 4 days: ****p < 0.0001; MoKCs: 0 vs 8 days: **p = 0.0013, 0 vs 13 days: ***p = 0.0002 and 0 vs 21 days: ****p < 0.0001). E Representative FACS plots showing TIMD4- cells among CLEC2 + KCs in Ldlr−/− and Ccr2−/− x Ldlr−/− female mice subjected to hypercholesterolemia for 3 weeks. F Fold-change in Csf1 expression (normalized to day 0) in livers of Ldlr−/− mice fed HC diet (n = 5, 4, 4, 4 and 4 mice for day 0, 4, 7, 13 and 21, respectively; 1-way ANOVA and multiple comparison test; 0 vs 4 days: *p = 0.026, 0 vs 8 days: ****p < 0.0001, 0 vs 13 days: *p = 0.043 and 0 vs 21 days: **p = 0.007). G Percentage of KI-67+ cells among EmKCs and MoKCs in HC-fed female Ldlr−/− mice for 3 weeks (***p = 0.0007). H Ly6C+ monocytes and MoDMacs in livers of Ldlr−/− mice fed chow (n = 4) or HC diet (n = 4) for 21 days. (Holm-Sidak multiple comparison test; Chow vs HC: Monocytes **p = 0.002 and MoDMacs **p = 0.006). I Granularity (****p < 0.0001) and (J) lipid content (****p < 0.0001) of EmKCs and MoKCs of mice (n = 9) fed HC diet for 3 weeks. K Lipidomic analysis of sorted EmKCs and MoKCs of mice fed HC diet for 3 weeks (n = 3 mice; Chow vs HC, FC: *p = 0.04 and CE: ***p < 0.001). L Heatmap of scavenger receptors mRNA expression in sorted EmKCs and MoKCs from Ldlr−/− mice after 3 weeks of HC diet (n = 4 mice). M CD36 expression (ΔMFI = CD36 MFI minus MFI of non-stained cells) on EmKCs and on VSIG4− and VSIG4+ MoKCs at 3 weeks of HC diet (n = 4 mice) showing lower expression on VSIG4− MoKCs (**p = 0.009). N FITC MFI (bodipy) for EmKCs and for VSIG4- and VSIG4+ MoKCs after injection of oxidized-LDL labeled with bodipy or not (1-way ANOVA; 2 independent experiments combined; EmKCs vs VSIG4+ MoKCs: *p = 0.031 and EmKCs vs VSIG4− MoKCs: ***p = 0.0008). All data in this figure are presented as mean values ± SEM. Statistical significance has been assessed with a two-sided t-test (or multiple t-tests) unless otherwise stated on the corresponding panel legend. Source data are provided as a Source Data File.
Next, we addressed whether MoKCs properties differed from EmKCs fed the HC diet for 3 weeks. We noticed that MoKCs exhibited reduced granularity (Fig. 4I) and intracellular lipid content (Fig. 4J) as compared to EmKCs in female mice. Similar observations were also made in males (Figs. S4E, S4F). A reduction in both free cholesterol and all the CE species quantified accounted for the diminished lipid loading observed in MoKCs (Figs. 4K, S4G). In addition, RT-qPCR analysis showed decreased expression of the scavenger receptors Cd36, Colec12 and Marco, but unchanged expression for Scarb1 and Msr1, in MoKCs as compared to EmKCs (Fig. 4L). Lower cell surface expression of CD36 protein in MoKCs was confirmed using flow cytometry (Fig. 4M). Noteworthy that the lowest CD36 expression was found on MoKCs that do not express VSIG4 marker, suggesting that CD36 is progressively acquired during MoKCs maturation (Fig. S4H). Thus, we sought to functionally assess whether decreased expression of these scavenger receptors could impact MoKCs ability to uptake modified-lipoproteins. Bodipy-labeled copper-oxidized LDL (oxLDL) were prepared and injected intravenously to evaluate their uptake by KC subsets in vivo using flow cytometry. We observed that both EmKCs and MoKCs phagocytized bodipy-labeled oxLDL. Nevertheless, bodipy-labeled oxLDL uptake was significantly lower in MoKCs than EmKCs (Fig. 4N). This was true for VSIG4+ and VSIG4- MoKCs, although the latter exhibited the lowest capacity to scavenge modified-lipoproteins. This result was consistent with the respective lipid contents of these KC subsets, VSIG4- MoKCs exhibiting the lowest (Fig. S4H).
To consolidate our results demonstrating that KCs homeostasis is perturbated in response to hypercholesterolemia, similar experiments were repeated in a non-genetically Ldlr deficient mouse model using liver-directed recombinant adeno-associated viral (AAV)-mediated gene transfer. Thus, wild-type mice received a single administration of an AAV vector encoding the gain-of-function form of murine PCSK9D377Y. PCSK9D377Y-mediated degradation of hepatic LDL receptors31 resulted in a marked elevation of plasma cholesterol levels when mice were subjected to the HC diet (Fig. S5A). KCs analysis of HC-fed AAV-PCSK9D377Y injected mice provided similar results to those obtained in Ldlr−/− mice. Thus, EmKCs pool markedly increased 4-days post-induction of hypercholesterolemia. This pool expansion was associated with a strong proliferative response of EmKCs. Prolonged exposure to hypercholesterolemia resulted also in the generation of MoKCs that exhibited a lower lipid-loading capacity as compared to EmKCs (Fig. S5A). Analysis of spleen and kidney macrophages also revealed no impact of hypercholesterolemia on their populations, but moderate lipid loading in spleen macrophages (Fig. S5B). Finally, we sought to evaluate whether cholesterol ester transfer protein (CETP), that plays important role in human lipoprotein cholesterol metabolism and that has been shown to be expressed by macrophages and, KCs in particular32,33, could affect our observations. Ldlr−/− mice were bred to human CETP transgenic mice (mice do not naturally express CETP) and fed HC diet for 3 weeks. CETP expression had no major impact, and KC pool homeostasis was similarly remodeled by hypercholesterolemia in Ldlr−/− mice expressing CETP or not (Fig. S6).
Altogether, these results show that hypercholesterolemia triggers a specific adaptive response of KCs with two phases. Following a major EmKCs pool expansion in response to the early induction of hypercholesterolemia, the KC pool is then remodeled with a marked reduction in the number of heavily cholesterol-loaded EmKCs and the engraftment of MoKCs displaying a decreased capacity to scavenge modified-LDLs.
Protection of EmKCs from apoptosis diminished MoKCs engraftment during hypercholesterolemia
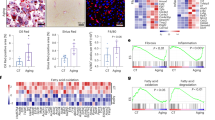
Mitochondrial dysfunction and endoplasmic reticulum stress are important triggers of macrophage foam cell apoptosis34,35,36,37. In particular, modified forms of LDL and accumulation of cellular free cholesterol can induce the production of mitochondrial reactive oxygen species (ROS) that generate cellular oxidative stress and damage35,37,38. We thus sought to determine whether the foamy KCs observed upon hypercholesterolemia underwent mitochondrial stress. KCs were stained with the mitochondria-specific ROS indicator Mitosox and analyzed by flow cytometry. Mitosox fluorescence in EmKCs raised by 1.6- and more than 3-fold when Ldlr−/− mice were fed the HC diet for 4 and 21 days, respectively (Fig. 5A), suggesting progressive accumulation of mitochondrial superoxide in EmKCs. At 21 days, mitochondrial ROS detection in MoKCs demonstrated significantly less signal than that measured in EmKCs (Fig. 5B), which might be linked to the lower amount of intracellular free cholesterol measured in MoKCs (Fig. 4K).
A Generation of mitochondrial ROS determined by mitosox staining in EmKCs of Ldlr−/− female mice fed chow (n = 5) or HC diet for 4 (n = 4) or 21 days (n = 4). Statistical significance tested with 1-way ANOVA and Tukey multiple comparison test (Chow vs HC4: *p = 0.025, Chow vs HC21: ****p < 0.0001 and HC4 vs HC21: ****p < 0.0001). B Comparative mitosox staining between EmKCs and MoKCs of Ldlr−/− female mice fed HC diet 21 days (n = 4 mice, **p = 0.0012). C Experimental strategy to track the fate of monocytes in hypercholesterolemic chimeric mice with apoptosis-resistant EmKCs. D Representative flow cytometry analysis showing the contribution of monocyte-derived (GFP+) cells to EmKCs and MoKCs. E Frequency of MoKCs (***p = 0.0009) and (F) absolute numbers of GFP+ MoKCs (*p = 0.018) in female Ccr2−/− (n = 6 mice) and Cd68-hBcl2 x Ccr2−/− (n = 6 mice) chimeras fed the HC diet for 3 weeks (2 independent experiments combined). All data in this figure are presented as mean values ± SEM. Statistical significance has been assessed with a two-sided t-test unless otherwise stated on the corresponding panel legend. Source data are provided as a Source Data File.
We then wondered whether protecting EmKCs from mitochondrial stress would influence MoKCs generation. We thus used Cd68-hBCL239 mice exhibiting enforced expression of the mitochondrial anti-apoptotic protein BCL2 in the myeloid lineage, including KCs. Cd68-hBCL2 mice crossed in a Ccr2−/− background were submitted to low-dose irradiation (3 Gy) and subsequently transplanted with bone marrow (BM) cells expressing the green fluorescent protein (GFP). BMGFP-transplanted Ccr2−/− mice served as controls. In this model, the fate of bone marrow-derived cells can be traced thanks to GFP expression. These chimeric mice were injected with AAV-PCSK9D377Y to allow induction of hypercholesterolemia upon HC diet feeding of the mice (Fig. 5C). After 3 weeks of HC diet, only a small proportion ( < 3%) of EmKCs expressed GFP (Fig. 5D) in agreement with their embryonic origin, radioresistance and maintenance independent from circulating monocytes. GFP+ EmKCs may correspond to the few MoKCs that acquired TIMD4 expression with time, as previously reported10,21,40. By contrast, more than 60% of MoKCs were GFP+ and thus derived from GFP+ donor cells. The remaining GFP− MoKCs most likely corresponded to the few host-derived MoKCs (5–6% of the KC pool at the steady-state, Figs. 4C, S4C) that proliferated together with the KC pool early after HC diet feeding. The overall frequency of MoKCs was decreased by 2-fold in Cd68-hBCL2 x Ccr2−/− recipient mice (Fig. 5E). This was also observed for absolute numbers of MoKCs, whereas EmKCs numbers were increased (Fig. S7). Finally, quantification of GFP+ MoKCs demonstrated diminished amount in Cd68-hBCL2 x Ccr2−/− recipient mice (Fig. 5F). Altogether, these results lend support to the conclusion that protection of EmKCs from cell death, and potentially as a consequence of mitochondrial oxidative stress, limits the recruitment and engraftment of MoKCs under hypercholesterolemia.
Reducing EmKCs numbers raises cholesterolemia and accelerates atherosclerosis
We finally sought to investigate the potential consequences of decreasing EmKC numbers during hypercholesterolemia. To this aim, we crossed Ldlr−/− mice to Cd207-DTR animals in which diphteria toxin (DT) administration leads to depletion of EmKCs as previously shown10. Mice were injected either with DT or heat (boiled)-inactivated DT (bDT) and then fed the HC diet for 8 days (Fig. 6A). Total KC numbers were similar in both groups after 8 days of HC diet (Fig. 6B). However, DT administration substantially reshaped the KC pool with diminished EmKC counts and a concomitant increase in MoKCs numbers (Fig. 6C). Thus, while few MoKCs were detectable in the bDT group, MoKCs represented about 60% of the KC pool in DT-treated animals (Fig. 6D). As reported above, recruited MokCs were less lipid-loaded than EmKCs in both bDT and DT-treated mice (Fig. 6E). Importantly, both groups exhibited similar counts of Ly-6C+ monocytes and neutrophils (Fig. 6F), confirming previous observations that DT administration did not create chronic inflammation10,21. In parallel to the increased intracellular lipid content observed in the KCs of DT-treated animals (Fig. 6E), we found that plasma cholesterol concentrations were significantly more elevated in these mice (Fig. 6G). This effect could not be explained by increased lipoprotein production (as very low-density lipoproteins, or VLDLs) by the liver of DT-treated mice, as VLDL secretion rates were comparable in both groups (Fig. 6H). However, plasma cholesterol elevation was undoubtedly associated with EmKCs depletion and not DT off-target activity as DT-treated Ldlr−/− mice fed the HC diet for 8 days did not display elevated cholesterolemia (Fig. S8A). We then assessed whether similar observations could be done when the HC diet was administered for 8 days to Cd207-DTR x Ldlr−/− mice that received DT and then underwent a 3 weeks recovery period (Fig. S8B). This set up allows the KC pool to reconstitute before inducing hypercholesterolemia. In this context, EmKC numbers were also found decreased in DT-treated animals and MoKCs counts markedly increased (Fig. S8C). Total KC numbers was similar in both conditions (Fig. S8D) but MoKCs represented 40% of the KC pool in DT-treated mice (Fig. S8E). Again, no overt sign of inflammation was observed upon DT treatment (Fig. S8F). In this scenario, we observed that plasma cholesterol levels were also more elevated in DT-treated animals (Fig. S8G). To evaluate the long-term consequences of EmKCs replacement by MoKCs, Cd207-DTR × Ldlr−/− female mice administered DT or bDT were fed the HC diet for 4 weeks (Fig. 6I). Total KC numbers were slightly diminished in DT-treated mice as compared to bDT-treated controls (Fig. 6J). This was primarily due to a marked decrease in EmKC counts as MoKCs numbers were found comparable to the bDT-treated controls (Fig. 6K). MoKCs represented 60% of the KC pool in bDT-treated Cd207-DTR × Ldlr−/− female mice (Fig. 6L), as observed in Ldlr−/− female mice under prolonged hypercholesterolemia (Fig. 4C). This proportion raised to 84% in DT-treated animals (Fig. 6L). Consistent with our previous observations that macrophage populations other than KCs are not targeted in Cd207-DTR mice10, we observed no change in spleen or kidney macrophage populations 4 weeks after DT administration (Fig. S8H). Blood monocytes and neutrophils also did not differ between DT- or bDT-treated mice (Fig. S8I). Thus, even after several weeks of hypercholesterolemia EmKCs were unable to reconstitute their pool after DT treatment and remain lower in DT-treated mice. Consistent with our findings after 8 days of HC diet feeding, cholesterolemia was also more elevated in DT-treated mice after 4 weeks of diet (1194 mg/dL vs 913 mg/dL in the bDT group, p = 0.0009) (Fig. 6M). As the liver plays a key role in regulating plasma cholesterol levels, RNA-Seq was performed on livers of these bDT and DT-treated mice. Transcription factor enrichment analysis (CHEA) revealed that many of the down-regulated genes (FC ≥ 1.3) were putative target genes of RXR, LXR and PPARα (based on the use of the ChIP-seq liver mouse dataset GSE3526241) (Fig. 6N), all nuclear receptors known for their pivotal roles in the transcriptional control of lipid and bile acid metabolisms. KEGG pathway analysis performed on the differentially expressed genes highlighted bile secretion pathway (Fig. S8J). Thus, lipidomic analysis on liver and feces were conducted and confirmed differences in cholesterol and bile acids contents between the two groups (Fig. 6O-S). Notably, DT-treated mice exhibited elevated content of cholesterol biosynthesis intermediates (squalene, lanosterol, lathosterol and desmosterol) (Fig. 6O). Liver free cholesterol and cholesteryl-esters were also increased in DT-treated animals as compared to bDT-treated controls (Fig. 6O). Then, if liver overall primary and secondary bile acid pools were not significantly different between the two groups of mice, some bile acid species such as cholic acid or α-muricholic acid were found increased in DT-treated mice (Fig. 6P). Noteworthy, that these changes were not associated with more liver suffering as measured by plasma ALT levels (Fig. 6Q). Changes in hepatic cholesterol and bile acids contents in DT-treated mice also associated with increased primary and secondary bile acids excretion in feces (Fig. 6R), but not of cholesterol (Fig. 6S). Altogether, this shows that transcriptional control of cholesterol and bile acids metabolism is rewired in the liver of mice exhibiting decreased EmKCs numbers, affecting cholesterol and bile acids recycling. These results led us to examine whether these changes transposed into increased susceptibility to atherosclerosis. Quantification of atherosclerosis in the aortic sinus revealed a 1.5-fold increase in intimal atherosclerotic lesions in DT-treated Cd207-DTR × Ldlr−/− female mice (Fig. 6T). This experimental protocol was also conducted in male mice and provided similar observations. Notably, DT-treated Cd207-DTR × Ldlr−/− males exhibited significantly reduced EmKC numbers and elevated hypercholesterolemia (Fig. S8K) that associated with larger atherosclerotic lesions (Fig. 6T). Finally, we could demonstrate that these effects were long lasting since similar results were also observed in females after 2 months of HC diet (Figs. 6U, S8L). Thus, loss of EmKCs and their subsequent replacement by MoKCs associates with an altered metabolic adaptation of the liver to hypercholesterolemia that further aggravates it and promotes accelerated development of atherosclerosis.
A Experimental strategy used to deplete EmKCs in the context of hypercholesterolemia (n = 7 mice for bDT and n = 6 mice for DT). B Total KC and (C) EmKCs (****p < 0.0001) and MoKCs (***p = 0.0002) numbers in livers of Cd207-DTR × Ldlr−/− male mice after 8 days of HC diet and injected either with bDT or DT. D Percentage of MoKCs (****p < 0.0001), E bodipy staining of EmKCs (*p = 0.049) and MoKCs (*p = 0.034), F numbers for liver monocytes and neutrophils, and (G) plasma cholesterol concentrations after 8 days HC diet feeding (1 independent experiment; **p = 0.0033). H Hepatic production of VLDLs in Cd207-DTR × Ldlr−/− mice 8 days post-injection of DT (n = 5) or bDT (n = 5) and HC diet feeding (2 independent experiments combined). Plasma triglycerides (TG) were measured overtime after p407 injection (intravascular lipolysis inhibition). I Experimental strategy. J Total KC (*p = 0.044) and (K) EmKCs (**p = 0.0027) and MoKCs numbers in livers of Cd207-DTR × Ldlr−/− female mice after 4 weeks of HC diet and injected either with bDT (n = 9) or DT (n = 9) (2 independent experiments combined). L Percentage of MoKCs (****p < 0.0001) and (M) plasma cholesterol concentrations (***p = 0.0009) after 4 weeks HC diet feeding. N The list of significantly downregulated genes (RNA-Seq) in livers of DT-treated mice as compared to bDT-treated controls was submitted to Enrichr for transcription factor enrichment analysis (ChEA_2022). O Heat map showing fold-changes in cholesterol biosynthesis intermediates in livers of DT-treated mice (n = 9) as compared to bDT-treated animals (n = 9). Differences in cholesterol (*p = 0.048) and cholesteryl-esters liver (*** < 0.001) concentrations between the two-groups are shown. P Liver content in primary (I) and secondary (II) bile acids. Fold-change for specific I and II bile acids species in livers of DT-treated mice (n = 5) as compared to bDT-treated (n = 5) animals provided as a heat map. Q Hepatic damage (ALT) in Cd207-DTR × Ldlr−/− female mice injected with bDT (n = 10) or DT (n = 11) (2 independent experiments combined) after 4 weeks of HC diet feeding. R Bile acids (I BA: *p = 0.043 and II BA: *p = 0.032) and (S) cholesterol contents in feces for both groups (n = 6 in each group) were also determined. T Representative images of atherosclerosis in the aortic root of Cd207-DTR × Ldlr−/− mice after 4 weeks of HC diet and injected either with bDT or DT. Lipid lesions were quantified by ORO staining in female (n = 6 in each group; 2 independent experiments combined; **p = 0.008) and male mice (DT, n = 9 and bDT, n = 11; 4 independent experiments combined; *p = 0.013). U Plasma cholesterol (*p = 0.05) and arterial lipid lesions (*p = 0.05) of Cd207-DTR × Ldlr−/− female mice after 2 months of HC diet and injected either with bDT (n = 7) or DT (n = 7) (2 independent experiments combined). All data in this figure are presented as mean values ± SEM. Statistical significance has been assessed with a two-sided t-test (or multiple comparison t-tests). Source data are provided as a Source Data File.
Discussion
The consequences of hypercholesterolemia on the retention of LDL lipoproteins in focal areas of the arterial tree with their engulfment by macrophages to form lipid-laden foam cells have been studied for decades as an early event in the formation of atherosclerotic lesions. Here, our work demonstrate that hypercholesterolemia also leads to the generation of heavily cholesterol-loaded Kupffer cells in the liver. We also showed that EmKCs homeostasis is perturbated with a first stage of expansion early after the induction of hypercholesterolemia followed by their impaired maintenance and the engraftment of monocyte-derived KCs while hypercholesterolemia becomes chronic. In addition, the present study lends support for a metabolic function of EmKCs in contributing to cholesterol homeostasis and atherosclerosis development in the context of hypercholesterolemia.
The study of the initial response of EmKCs to elevation of plasma LDL-cholesterol concentration demonstrated a remarkable adaptation of the pool that expanded in both male and female mice. Transcriptomic analysis and KI-67 staining revealed that EmKC expansion was due to increased proliferation. We noticed an increase in liver Csf1 mRNA expression and a CSF1R inhibitor blunted EmKCs proliferation, strongly supporting a major role for the CSF1-CSF1R axis. More specifically, increased hepatic Csf1 gene expression was due to hepatic stellate cells (HSCs) that we showed expressed more Csf1 and were more numerous. Thus, the adaptative response of KC to hypercholesterolemia relies also on adaptation of its niche, and notably HSCs with which they are in close contact16. Whether HSCs activation is a direct response to hypercholesterolemia, or more likely, implies a two-cell circuit42 in which HSCs would respond to growth factors produced by activated KCs to secrete more CSF1 and maintain a stable HSC-KC ratio will have to be further explored.
After this rapid increase in EmKCs density, we then observed a progressive contraction of the KC pool. The continuous increase in Csf1 expression overtime did not suggest that EmKCs contraction was due to a limitation in the amount of local CSF1 available to sustain EmKCs maintenance. Rather, excessive cholesterol loading of EmKCs may have contributed to decrease their survival capacity. Our lipidomic analyses revealed that, while liver tissue increased its cholesterol content in the form of non-toxic esterified forms of cholesterol, EmKCs accumulated large amount of both free cholesterol and cholesteryl-esters. Excess free cholesterol in macrophages is a potent inducer of their death. Indeed, macrophage free-cholesterol loading has been reported to induce apoptosis, notably by triggering endoplasmic reticulum (ER) stress43 but also by increasing mitochondrial oxidative stress and dysfunction38. The progressive increase in mitochondrial ROS we observed in EmKCs during HC diet feeding would concur to such scenario. In addition, the involvement of CD36 in EmKCs lipid loading suggested that cholesterol loading is driven by the uptake of modified lipoproteins under hypercholesterolemia. This would notably corroborate a recent study showing KCs avidity for APOB-containing lipoproteins in the context of atherogenic dyslipidemia44. CD36 has been linked to atherosclerosis through its recognition of modified endogenous ligands, including oxidized-LDL45. Macrophage uptake of such atherogenic lipoproteins potently induces ER stress and mitochondrial oxidative stress34,35,36, and can even lead to macrophage apoptosis in a CD36-dependent pathway34. Thus, under hypercholesterolemic conditions, EmKCs could suffer similar cellular stresses than those operant in lesional macrophages in atherosclerotic plaques.
Loss of EmKCs in depletion models10,17,21, in pathological contexts whereby EmKCs are challenged10,11,12,13,14,15 or through genetic invalidation of critical genes for KCs survival17,46 have been shown to create niche availability and the engraftment of monocyte-derived KCs. This also occurs in condition of hypercholesterolemia as MoKCs started to progressively emerge when EmKC numbers diminished after the early proliferative phase. The accelerated and stronger loss of EmKCs in females as compared to males also coincided with a more rapid and larger generation of MoKCs in females. It was proposed that niche accessibility and niche availability were the predominant factors for the engraftment of monocytes in a restricted number of niches per organ47. The fact that almost no MoKCs were generated in Ccr2−/− × Ldlr−/− mice strongly suggests that signals of niche accessibility require an operant CCL2-CCR2 axis in this pathological hypercholesterolemic context. Nonetheless, such signaling can be overcome if the niche is brutally emptied as demonstrated in acute depletion models carried on the Ccr2−/− background16. Protecting cholesterol-loaded EmKCs from death through enforced hBCL2 gene expression, thus limiting niche availability, supported also the proposed niche concept as we observed less recruitment of MoKCs in hypercholesterolemic Cd68-hBCL2 chimeric mice. These data also suggested that loss of cholesterol-loaded EmKCs upon prolonged exposure to hypercholesterolemia was likely linked to mitochondrial-related stress. Such cellular stress could be partially relieved by overexpression of the mitochondrial membrane BCL2 protein that blocks the apoptotic death pathway. With this in mind, reduced cholesterol uptake capacity concomitant to diminished mitochondrial ROS in recruited MoKCs could confer a competitive advantage to these cells over cholesterol-overloaded EmKCs for niche occupancy. Notwithstanding that MoKCs exhibited a higher proliferative state as compared to EmKCs, as previously reported during MASH10.
MoKCs have been shown to activate gene expression patterns and display phenotypic characteristics similar to those of EmKCs16,21. Nevertheless, specific transcriptomic differences have also been reported between the two cell types10,13,21, and functional differences were also highlighted21,48. In the context of MASH, we recently revealed that EmKCs were protective over MoKCs to limit liver damage10. Here, in the context of hypercholesterolemia, our study adds further support to a beneficial role of EmKCs as compared to MoKCs. Indeed, we observed that both hepatic cholesterol content and circulating concentrations of cholesterol were increased when MoKCs replaced EmKCs in HC-fed and DT-treated Cd207-DTR × Ldlr−/− mice. On the basis of our transcriptomic analysis, reduced LXR and RXR signaling in livers of DT-treated mice provides a potential explanation for such effects. Indeed, liver-specific deletion of either Lxra49 or Rxra50 in mice results in altered processing and elimination of dietary cholesterol with marked cholesteryl-esters accumulation in their livers and high plasma cholesterol levels when challenged with cholesterol-rich diets. Moreover, excess dietary cholesterol is normally managed by increasing bile acid production and subsequent fecal elimination, a process in which LXR plays an important role51. We observed specific modulation of hepatic bile acids pool composition, while fecal bile acids content, but not fecal cholesterol, were increased in HC-fed and DT-treated mice, suggesting altered bile acid and cholesterol recycling. LXRs (LXRa and LXRb) form obligate heterodimers with RXR and function as important regulators of cellular sterol homeostasis52. LXR/RXR heterodimer can be activated by ligands for either partner, and notably oxysterols and desmosterol for LXRs. Thus, decreased activation of LXR/RXR target genes in the liver of DT-treated mice with less EmKCs and more MoKCs could imply diminished hepatic availability of such ligands (in particular oxysterols as desmosterol is poorly active in hepatocytes53). Noteworthy that uncoupling of LXR activation with reduced oxysterols availability in a context of increased hepatic cholesterol content has been reported earlier in condition of hepatic regeneration54. Here, whether EmKCs contribute to hepatic cholesterol metabolism adjustment to hypercholesterolemia by providing LXR ligands to hepatocytes and whether MoKCs, with a diminished capacity to scavenge circulating oxidized-lipoproteins, might not be as effective as EmKCs to provide such ligands, remain to be established.
Our data also highlight the possibility that the actual development of nanoparticle-based therapeutic strategies to target macrophages within atherosclerotic lesions may also present a great deal of interest to preserve KCs homeostasis and function in the liver of hypercholesterolemic patients. In that respect, it would be highly relevant to assess in preclinical models the beneficial actions on KCs of nanoparticle-mediated delivery of functional miRNA55,56 or synthetic LXR agonists57,58 that have been shown to promote cholesterol efflux from foamy macrophages, or alternatively, nanoparticle-based approaches to scavenge ROS59.
In summary, we have demonstrated that EmKCs are progressively lost and replaced by MoKCs during hypercholesterolemia, which in turn may further exacerbate hypercholesterolemia and atherosclerosis development.
Methods
Mice and treatments
Animal procedures were reviewed and approved by the Charles Darwin ethics committee on animal experimentation (project approval #16734 & 46344) and were performed in accordance with the French ministry of agriculture. This investigation conformed to the European directive 2010/63/EU revising directive 86/609/EEC on the protection of animals used for scientific purposes.
Animals were on a C57BL/6 J background. UBC-GFP (C57BL/6-Tg(UBC-GFP)30Scha/J) mice, Ccr2−/− (B6.129S4-Ccr2tm1Ifc/J) mice, CETP transgenic (B6.CBA-Tg(CETP)5203Tall/J), homozygous Cd207-DTR (B6.129S2-Cd207tm3(DTR/GFP)Mal/J) mice and Ldlr−/− (B6.129S7-Ldlrtm1Her/J) mice were from the Jackson Laboratory. Ldlr−/− were bred in-house to Ccr2−/−, CETP and Cd207-DTR animals. Cd68-hBCL2 mice previously generated by our team39 were bred in-house to Ccr2−/− animals. The mice were housed in standard cages with a 12-h light-dark cycle, at 21 °C ambient temperature and with ad libitum access to water and food. Hypercholesterolemia was induced in Ldlr−/− mice by feeding the animals with a chow diet (SAFE A04, Augy, France,) containing 1% cholesterol (HC diet). For studies with ezetimibe, the animals were fed with powdered chow diet (SAFE A04) supplemented or not with 0.005% ezetimibe for 4 days and then switched to powdered chow or HC diets supplemented with 0.005% ezetimibe (Bertin Pharma, France) for 4 days. For confirmatory studies in hypercholesterolemic mice via overexpression of PCSK9, C57BL/6 J mice were administered intravenously 1011 vector genome copies of a recombinant adeno-associated virus pAAV9-TBG_D377YmPcsk9 encoding the gain-of-function form of murine PCSK931 under the liver specific control of the TBG promoter (virus production by VectorBuilder Inc, USA). For CSF1R blocking experiments, 100 mg/kg of PLX3397 or vehicle (0.5% HPMC, 1% Tween 80, 2.5% DMSO) was given to mice by gavage every day from day 0 to day 2 of HC diet feeding. Mice were euthanized and analyzed on day 3. For hepatic VLDL-triglycerides (TG) production experiments, mice were fasted for 4 h and injected i.p. with Poloxamer-407 (Sigma) in saline at the optimal dose of 1 g/kg60. Immediately prior to injection, and at 0.5, 1, 2, and 4 h following injection, blood samples were drawn for plasma TG concentration measurements by commercial kits (Diasys).
Plasma, cell-sorted and tissue lipid analyses
Blood was collected on anesthetized mice by retro-orbital bleeding in tubes containing EDTA. Plasma was stored at −80 °C. Plasma alanine aminotransferase (ALT) activity was determined on a Indiko™ Plus Clinical Chemistry Analyzer (Thermo Fisher Scientific) using a commercial kit (ALAT/GPT; Thermo Fisher Scientific). Plasma total and free cholesterol levels were determined using commercial kits (Diasys). Quantification of cholesterol and cholesteryl ester species in sorted cells or liver tissue was performed by LC-ESI/MS/MS using a Prominence UFLC and a QTrap 4000 mass spectrometer (ICANalytics core facility of the institute of Cardiometabolism and Nutrition (IHU-ICAN, ANR-10-IAHU-05). Cell pellets or liver tissues resuspended in methanol 70% and homogenized were supplemented with internal standards: CE(18:1)_d7 and cholesterol_d7. Lipids were extracted according to a modified Bligh and Dyer method in methanol/CHCl3 (2:1) and HCl 0.01 N. Phase separation was triggered by addition of CHCl3 and H2O. The lower phase was dried and resuspended in LC/MS compatible solvent. Samples were injected to a Ascentis C18 column. Mobile phase A consisted of ACN/H2O (60:40), 10 mM ammonium formate, 0.1% formic acid and mobile phase B of ISP/ACN (90:10), 10 mM ammonium formate, 0.1% formic acid. Lipid species were detected using scheduled multiple reaction monitoring (sMRM). For bile acids quantification, dried feces or liver samples were homogenized in water and, then volumes equivalent to 10 mg of sample were supplemented with deuterated internal standards. 7 volumes of cold acetonitrile were added for protein precipitation. After centrifugation, the upper phases were collected, dried and resuspended in methanol/water (50:50). Samples were injected to an Ascentis Express C18 column and bile acids analyzed by LC-ESI/MS/MS using a Prominence UFLC and a QTrap 4000 mass spectrometer. Mobile phases consisted of water and acetonitrile containing 0.1% formic acid. Bile acids were detected using sMRM in the negative-ion mode. Cholesterol content in feces was measured by GC-MS. Homogenized feces in water were supplemented with deuterated sterols. Esterified sterols were saponified with KOH (0.5 N) in methanol for 20 mins at 60 °C. After addition of H2O, sterols were extracted with hexane. Total sterols were derivatized with BSTFA/TMCS (90:10) for 1 h at 80 °C. For GC-MS analysis, samples were injected on a 0.25 µm DB-5MS capillary column (250 °C in split mode). Cholesterol quantification was done on a single quadripole using electronic ionization.
Cell suspensions preparation for leukocytes analysis
Mice were euthanized by cervical dislocation. Immediately after, livers were perfused through the portal vein with 1 mL of PBS, followed by 2 mL (1.5 ml/min) perfusion with HBSS containing collagenase D (0.2 mU/mL, Sigma). Livers were removed and incubated for 30 min at 37 °C under gentle agitation with HBSS containing collagenase D (0.2 mU/mL, Sigma) and fetal bovine serum (3%). Cell suspensions were passed through a 100 µm cell strainer before staining. All subsequent procedures were performed on ice. Spleens, lungs and kidneys were harvested from PBS-perfused animals and digested in HBSS containing collagenase D (0.2 mU/mL, Sigma). For isolation of lamina propria leukocytes, freshly harvested intestines were quickly washed in PBS, opened, and cut into smaller pieces. To remove epithelial cells, samples were placed into 40 mL of PBS (without calcium and magnesium) containing glucose (1 g/L), HEPES (10 mM), EDTA (5 mM), fetal bovine serum (5%), and dithiothreitol (0.5%), and incubated for 30 min at 37 °C under agitation. After cells were washed (5 times), samples were chopped with scissors and placed in a digestion solution of HBSS (with calcium and magnesium), fetal bovine serum (3%), collagenase D (1.25 mg/mL, Sigma-Aldrich) and DNase (10 U/mL, Sigma-Aldrich). After 30 min at 37 °C, cell suspensions were passed through a 18 G needle before filtration on a 70 μm filter before staining.
Flow cytometry
Cell suspensions were first pre-incubated with anti-mouse CD16/32 antibody (93, biolegend) for 20 mins to block Fc receptors. Cells were stained for 30 min on ice: antibodies used were Ly-6C (HK1.4; # 128012, BioLegend), Ly-6G (1 A.8, #127648, BioLegend), VSIG4 (NLA14, #25-5752-82, ThermoFisher Scientific), CLEC4F (Polyclonal Goat IgG, #AF2784, R&D systems), TIMD4 (RMT4-54, #130008, BioLegend), CLEC2 (17D9, #146104, BioLegend), KI-67 (B56, #561277, BD Biosciences), TCRb (H57-597, # 17-5961-81, ThermoFisher Scientific, CD8a (53-6.7, #61-0081-82, ThermoFisher Scientific), B220 (RA3-6B2, #103206, Biolegend), MHC-II (M5/114.15.2, #107608, Biolegend), Nk1.1 (PK136, #108704, Biolegend), CD4 (RM4-5) BV711, Cat#100557, Biolegend), CD115 (AFS98, 135505, Biolegend), F4/80 (BM8, #123110,Biolegend), CD170 (S17007L, #155506, Biolegend), CD73 (TY/11.8, #127220, Biolegend), CD140a (APA5, #135910, Biolegend), CD61 (2C9.G2, # 104322, BioLegend), CD31(390, #102418, BioLegend), CD64 (X54-5/7.1, #139303 and #139306, BioLegend), CD45 (30-F11, #103154, BioLegend), CD11c (N418, #117334, and #117310, BioLegend), CD11b (M1/70, #48-0112-82, ThermoFisher Scientific), and CD36 (CRF D-2712, #565094, BD Biosciences). Draq7 (BioLegend) was used to exclude dead cells. Intracellular KI-67 staining was performed using the Foxp3 staining kit from Thermo Fisher Scientific. Bodipy 493/503 staining was performed on cells fixed using the Cytofix/CytopermTM kit from BD Biosciences. Mitosox staining was performed before extracellular staining according to manufacturer’s instructions (MitoSOX™ Red Mitochondrial Superoxide Indicator, Thermo Fisher Scientific). To determine absolute cell counts, a fixed quantity of non-fluorescent beads (10,000 10-μm polybead carboxylate microspheres from Polysciences) was added to each sample tube. The following formula was applied: number of cells = (number of acquired cells × 10,000) / (number of acquired beads). The final cell counts were expressed as the number of cells per mg of tissue. Data were acquired on a BD LSRFortessa flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star). The dimensionality reduction algorithm tSNE (t-Distributed Stochastic Neighbor Embedding) was run using the plugin integrated in FlowJo. KCs (EmKCs CD45+CLEC2+TIMD4+CD31− and MoKCs CD45+CLEC2+TIMD4−CD31−) cell sorting were performed on a BD FACSAria II cell sorter. For cell sorting of hepatic stellate cells (HSCs, CD45−CLEC2−CD31−UVhighCD73+PDGFRa+) and of liver sinusoidal endothelial cells (LSECs, CD45−UV−CLEC2+CD31+CD61+), livers were cut into small pieces with scissors, homogenized using a gentelMACS dissociator (Miltenyi Biotec), incubated with collagenase D (0.2 mU/mL, Sigma) at 37 °C for 30 mins with gentle shaking and further homogenized with the gentelMACS dissociator. Cell suspensions were filtered through 70 µm cell strainers before sorting on a BD FACSAria II sorter.
Microscopy
Following liver removal, tissue slices measuring 3–5 mm were fixed in 4% paraformaldehyde (PFA) for 24 h at 4 °C, then washed in PBS and incubated in 30% sucrose for another 24 h. The samples were embedded in Tissue-Tek OCT compound (Sakura Finetek) and frozen using isopentane and liquid nitrogen. Sections 8 µm thick were cut on a Leica CM 1900 cryostat, rehydrated in PBS for 5 min, and incubated with 0.5% Triton and 3% bovine serum albumin for 30 min at room temperature. The tissue sections were then labeled overnight at 4 °C in a humid chamber with goat anti-mouse CLEC4F (AF2784, R&D Systems) and rat anti-mouse/human CD324 (DECMA-1, BioLegend) antibodies. After washing with PBS, they were further incubated for 1 h at room temperature with Cy3 AffiniPure F(ab’)2 fragment rabbit anti-goat IgG and Alexa Fluor 647 AffiniPure donkey anti-rat IgG secondary antibodies (Jackson ImmunoResearch Europe Ltd). For DESMIN staining, liver cryosections were incubated overnight at 4 °C with recombinant rabbit monoclonal anti-DESMIN antibody (ARC0235, ThermoFisher Scientific). After several PBS washes, the samples were incubated with an Avidin/Biotin blocking kit (Abcam) for 15 min, followed by incubation with biotinylated goat anti-rabbit IgG antibody (Vector Laboratories) for 1 h at room temperature. Signal amplification was carried out using the Tyramide SuperBoost kit (ThermoFisher Scientific), according to the manufacturer’s instructions. Finally, the slides were mounted using Vectashield mounting medium with DAPI (Vector Laboratories) and imaged with a Zeiss AxioImager M2 microscope (Carl Zeiss) using Zen software. For lipid imaging of liver tissue, Ldlr−/− mice fed a chow diet or HC diet for 4 days were anesthetized using Isoflurane and injected intravenously with 5 µg anti-mouse TIM4-APC antibody (RMT4-54, BioLegend). They were then euthanized 5 min after injection by cervical dislocation and perfused through the portal vein with PBS and then with 4% ice-cold PFA. Livers were harvested, lobes separated and incubated overnight at 4 °C in a fixation/permeabilization buffer (BD Biosciences) diluted 1:4. Tissue samples were washed thoroughly with PBS before imaging. The two-photon laser-scanning microscopy (TPL SM) set-up used was a 7MP (Carl Zeiss) coupled to a Ti: Sapphire Crystal multiphoton laser (ChameleonU, Coherent), which provides 140-fs pulses of near-infrared light, selectively tunable between 680 and 1050 nm and an optical parametric oscillator (OPO-MPX, Coherent) selectively tunable between 1050 and 1600 nm. The NLO and the OPO beams were spatially aligned and temporally synchronized using a delay line (Coherent) allowing Coherent anti-stoke Raman Scattering (CARS) imaging approach. The excitation wavelength was 820 nm for the NLO beam and 1070 nm for the OPO beam to detect the vibrational signature of lipid rich structures at a frequency of 2850 cm-1 with an emission wavelength at 665 nm. The system included a set of external nondescanned detectors in reflection with a combination of a LP-600-nm dichroic mirror (DM) followed by a LP-645-nm DM with 624-/40-nm emission filter (EF) and a LP-462-nm DM with 417-/60-nm emission filter (EF), LP-500-nm DM with 480-/40-nm EF, LP-550 nm DM with 525-/50-nm and 575/50 nm EFs. Images were performed directly on the whole liver lobe with a water immersion objective, plan apochromat × 20 (numerical aperture = 1). Mask rendering and treatment were done using Imaris software (Bitplane).
Generation of bone marrowGFP-transplanted Ccr2−/− mice
Bone marrow cells were harvested from femurs of UBC-GFP donor female mice. 10 million cells were injected (i.v.) into 3 Gy irradiated Cd68-hBCL2 × Ccr2−/− or Ccr2−/− female mice and a 3-week period was allowed to warrant engraftment of donor cells and reconstitution of blood monocytes. To ensure induction of hypercholesterolemia, mice were then administered intravenously 1011 vector genome copies of pAAV9-TBG_D377YmPcsk9. Mice were fed 7 days later with HC diet for three weeks before sacrifice. Cell suspensions were prepared from livers and analyzed by flow cytometry as described above. Absolute counts for GFP-expressing cells were normalized to liver monocyte chimerism (94% and ∼84% in Ccr2−/− and Cd68-hBCL2 × Ccr2−/− recipients, respectively).
Diphteria toxin (DT) mediated depletion of KCs in Cd207-DTR mice
Kupffer cells were depleted following two intraperitoneal DT (Sigma) injections (2 × 1 µg, 8 to 10 h apart) in homozygous Cd207-DTR × Ldlr−/− mice. Heat-inactivated DT (bDT, boiled 25 mins) was used as control.
In vivo lipid uptake blocking by CD36 neutralizing antibodies
Chow fed Ldlr−/− mice were fasted during the day and administered i.v. 50 µg of anti-CD36 monoclonal antibody (MF3, thermoFisher Scientific) or 50 µg of rat IgG2a isotype (thermoFisher Scientific) before given access to HC diet. Antibody injections were repeated the day after, 1 h before sacrifice.
In vivo oxidized-LDL uptake
LDL were isolated from human plasma by sequential ultracentrifugation in the density range of 1.019 < d < 1.063 g/ml, dialyzed against PBS and filter-sterilized. Oxidation was initiated by dialysis of the LDL preparation against a 5 µM copper sulfate solution at 37 °C. After an overnight incubation, the oxidation reaction was stopped by extensive dialysis against PBS-EDTA 0.1 mM. Changes in the electrophoretic mobility of LDL lipoproteins after oxidation was monitored on agarose gel (hydrogel LIPO + Lp(a) K20, Sebia). Oxidized-LDL (oxLDL) were then incubated with BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentaméthyl-4-bora-3a,4a-diaza-s-indacène, ThermoFischer Scientific) (100 µM final) at 37 °C for 30 mins and then overnight at 4 °C. Free bodipy was removed by passing the oxLDL preparation through a PD-10 column (Pharmacia Biotech, Uppsala, Sweden) using PBS as buffer. Finally, bodipy-labeled oxLDL were concentrated using Spin-X UF concentrator (Corning) to approximately 2 mg protein/mL. 150 µL of the preparation was injected i.v. into Ldlr−/− male mice fed 3 weeks HC diet and switched for one week on chow diet to reduce the cellular fluorescence background generated by KCs lipid loading. oxLDL were injected in control mice. Animals were euthanized 1 h after injection to measure bodipy content of Kupffer cells using flow cytometry.
qPCR analysis
For gene expression analysis on liver tissue, total RNA preparation was performed using the NucleoSpin RNA Plus kit (MACHEREY-NAGEL). cDNA was synthesized using random hexamer and SuperScript III (Thermo Fisher Scientific). For gene expression analysis on sorted cells, total RNA was prepared from 20,000 cells using the RNeasy Plus micro kit (QIAGEN). RNA was reverse transcribed using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). Quantitative PCR analyses using Sybr (LightCycler 480 SYBR Green I Master, Roche) were performed using a LightCycler 480 real-time PCR system and dedicated software (Roche). Initial differences in mRNA quantities were controlled using reference mouse genes 18 s, Hprt, Rpl13a, and Nono. All primer sequences are available upon request.
RNA-Seq
Total RNA preparation was performed from 20,000 sorted KCs using the RNeasy Plus Micro Kit (Qiagen) and Nucleospin RNA plus kit for liver samples. cDNA libraries for sorted KCs were generated using Next Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs). RNA-Seq libraries were sequenced on an Illumina NovaSeq 6000 (40 million reads per sample). RNA-Seq analysis was completed using the Eoulsan pipeline. The STAR index was used to map raw reads to the genome and data normalization was performed with DESeq2. A LIMMA analysis was conducted to select differentially expressed genes with a 1.3-fold change cutoff between at least two conditions. Adjusted p value for multiple gene testing were used. Annotated genes with a count mean over 100 in at least one condition and a coefficient of variation of more than 0.5 between at least two conditions were retained. GSEA analysis was performed using the Phantasus web platform. The R packages EnhancedVolcano and EnrichR were used for volcano plotting and pathway analyses.
Analysis of aortic lipid lesions
Atherosclerosis was assessed using serial cross-sections of the aortic root. Mice were anesthetized with 2% isoflurane, euthanized via cervical dislocation, and perfused with PBS. The hearts were then collected and fixed in 4% PFA for 30 min, followed by overnight incubation in a 20% sucrose phosphate-buffered solution at 4 °C. Next, the hearts were embedded in Tissue-Tek OCT compound (Sakura Finetek). Sections 10 μm thick were cut through the proximal aorta, covering the three cusps of the aortic valves. Three sections, spaced 40 μm apart around the valves, were fixed and stained with oil red O (0.3% in triethylphosphate) for 30 min, followed by counterstaining with Mayer’s hematoxylin for 1 min. Images were captured with a Zeiss AxioImager M2 microscope, and plaque area was quantified using AxioVision Zeiss software.
Statistical analyses
Statistical significance of differences was performed using GraphPad Prism (GraphPad Software). Two-tailed Student’s t test was used to assess the statistical significance of the difference between means of two groups. Experiments were repeated at least twice. Graphs depicted the mean ± SEM. One-way ANOVA and Tukey’s or Dunnett post hoc analyses were used for multiple comparison tests. Statistical significance is represented as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data supporting the findings described in this manuscript are available in the article, in the Supplementary Information and from the corresponding author upon request. Source data are provided with the paper. The RNA-Seq data generated in this study were deposited under the accession number E-MTAB-12611 and E-MTAB-12744. Source data are provided with this paper.
References
Ference, B. A. et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 38, 2459–2472 (2017).
Que, X. et al. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558, 301–306 (2018).
Shearn, A. I. U. et al. Bcl-x inactivation in macrophages accelerates progression of advanced atherosclerotic lesions in Apoe(−/−) mice. Arterioscler. Thromb. Vasc. Biol. 32, 1142–1149 (2012).
Demetz, E. et al. The haemochromatosis gene Hfe and Kupffer cells control LDL cholesterol homeostasis and impact on atherosclerosis development. Eur. Heart J. 41, 3949–3959 (2020).
Bouchareychas, L. et al. Promoting macrophage survival delays progression of pre-existing atherosclerotic lesions through macrophage-derived apoE. Cardiovasc. Res. 108, 111–123 (2015).
Sawai, C. M. et al. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity 45, 597–609 (2016).
Hoeffel, G. et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015).
Liu, Z. et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525.e19 (2019).
Devisscher, L. et al. Non-alcoholic steatohepatitis induces transient changes within the liver macrophage pool. Cell. Immunol. 322, 74–83 (2017).
Tran, S. et al. Impaired Kupffer cell self-renewal alters the liver response to lipid overload during non-alcoholic steatohepatitis. Immunity 53, 627–640.e5 (2020).
Remmerie, A. et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity 53, 641–657.e14 (2020).
Seidman, J. S. et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity 52, 1057–1074.e7 (2020).
Theurl, I. et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat. Med. 22, 945–951 (2016).
Bleriot, C. et al. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 42, 145–158 (2015).
Lai, S. M. et al. Organ-specific fate, recruitment, and refilling dynamics of tissue-resident macrophages during blood-stage malaria. Cell Rep. 25, 3099–3109.e3 (2018).
Bonnardel, J. et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity 51, 638–654.e9 (2019).
Sakai, M. et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain Kupffer cell identity. Immunity 51, 655–670.e8 (2019).
Zeng, Z. et al. CRIg functions as a macrophage pattern recognition receptor to directly bind and capture blood-borne gram-positive bacteria. Cell Host Microbe 20, 99–106 (2016).
Gola, A. et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136 (2021).
Blériot, C. et al. A subset of Kupffer cells regulates metabolism through the expression of CD36. Immunity 54, 2101–2116.e6 (2021).
Scott, C. L. et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016).
Doi, Y. et al. Development of complementary expression patterns of E- and N-cadherin in the mouse liver. Hepatol. Res. 37, 230–237 (2007).
Cecchini, M. G. et al. Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development 120, 1357–1372 (1994).
Spann, N. J. et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151, 138–152 (2012).
Kim, K. et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ. Res. 123, 1127–1142 (2018).
Endemann, G. et al. CD36 is a receptor for oxidized low density lipoprotein. J. Biol. Chem. 268, 11811–11816 (1993).
Cheng, C. et al. Recognition of lipoproteins by scavenger receptor class A members. J. Biol. Chem. 297, 100948 (2021).
Wong, H. S. et al. Chemokine signaling enhances CD36 responsiveness toward oxidized low-density lipoproteins and accelerates foam cell formation. Cell Rep. 14, 2859–2871 (2016).
Zhang, X. et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 70, 761–774 (2021).
Serbina, N. V. & Pamer, E. G. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 7, 311–317 (2006).
Bjørklund, M. M. et al. Induction of atherosclerosis in mice and hamsters without germline genetic engineering. Circ. Res. 114, 1684–1689 (2014).
Van Eck, M. et al. Important role for bone marrow–derived cholesteryl ester transfer protein in lipoprotein cholesterol redistribution and atherosclerotic lesion development in LDL receptor knockout mice. Circ. Res. 100, 678–685 (2007).
Wang, Y. et al. Plasma cholesteryl ester transfer protein is predominantly derived from Kupffer cells. Hepatology 62, 1710–1722 (2015).
Seimon, T. A. et al. Atherogenic lipids and lipoproteins trigger CD36-TLR2-dependent apoptosis in macrophages undergoing endoplasmic reticulum stress. Cell Metab. 12, 467–482 (2010).
Asmis, R. & Begley, J. G. Oxidized LDL promotes peroxide-mediated mitochondrial dysfunction and cell death in human macrophages: a caspase-3–independent pathway. Circ. Res. 92, e20–e29 (2003).
Sanda, G. M. et al. Aggregated LDL turn human macrophages into foam cells and induce mitochondrial dysfunction without triggering oxidative or endoplasmic reticulum stress. PLoS ONE 16, e0245797 (2021).
Wang, Y., Wang, G. Z., Rabinovitch, P. S. & Tabas, I. Macrophage mitochondrial oxidative stress promotes atherosclerosis and nuclear factor-κB–mediated inflammation in macrophages. Circ. Res. 114, 421–433 (2014).
Yao, P. M. & Tabas, I. Free cholesterol loading of macrophages is associated with widespread mitochondrial dysfunction and activation of the mitochondrial apoptosis pathway. J. Biol. Chem. 276, 42468–42476 (2001).
Gautier, E. L. et al. Macrophage apoptosis exerts divergent effects on atherogenesis as a function of lesion stage. Circulation 119, 1795–1804 (2009).
Beattie, L. et al. Bone marrow-derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J. Hepatol. 65, 758–768 (2016).
Boergesen, M. et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell Biol. 32, 852–867 (2012).
Zhou, X. et al. Circuit design features of a stable two-cell system. Cell 172, 744–757.e17 (2018).
Devries-Seimon, T. et al. Cholesterol-induced macrophage apoptosis requires ER stress pathways and engagement of the type A scavenger receptor. J. Cell Biol. 171, 61–73 (2005).
Di Nunzio, G. et al. Kupffer cells dictate hepatic responses to the atherogenic dyslipidemic insult. Nat. Cardiovasc. Res. 3, 356–371 (2024).
Kunjathoor, V. V. et al. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J. Biol. Chem. 277, 49982–49988 (2002).
Scott, C. L. et al. The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity 49, 312–325.e5 (2018).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
David, B. A. et al. Combination of mass cytometry and imaging analysis reveals origin, location, and functional repopulation of liver myeloid cells in mice. Gastroenterology 151, 1176–1191 (2016).
Zhang, Y. et al. Liver LXRα expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J. Clin. Invest. 122, 1688–1699 (2012).
Wan, Y.-J. Y. et al. Hepatocyte-specific mutation establishes retinoid X receptor α as a heterodimeric integrator of multiple physiological processes in the liver. Mol. Cell Biol. 20, 4436–4444 (2000).
Peet, D. J. et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell 93, 693–704 (1998).
Tontonoz, P. Transcriptional and posttranscriptional control of cholesterol homeostasis by liver X receptors. Cold Spring Harb. Symposia Quant. Biol. 76, 129–137 (2011).
Muse, E. D. et al. Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proc. Natl Acad. Sci. USA 115, E4680–E4689 (2018).
Lo Sasso, G. et al. Down-regulation of the LXR transcriptome provides the requisite cholesterol levels to proliferating hepatocytes. Hepatology 51, 1334–1344 (2010).
Nguyen, M.-A. et al. Delivery of microRNAs by Chitosan nanoparticles to functionally alter macrophage cholesterol efflux in vitro and in vivo. ACS Nano 13, 6491–6505 (2019).
Li, C. et al. Site‐specific microRNA‐33 antagonism by pH‐responsive nanotherapies for treatment of atherosclerosis via regulating cholesterol efflux and adaptive immunity. Adv. Funct. Mater. 30, 2002131 (2020).
Guo, Y. et al. Synthetic high-density lipoprotein-mediated targeted delivery of liver X receptors agonist promotes atherosclerosis regression. EBioMedicine 28, 225–233 (2018).
Yu, M. et al. Targeted nanotherapeutics encapsulating liver X receptor agonist GW3965 enhance antiatherogenic effects without adverse effects on hepatic lipid metabolism in Ldlr −/− mice. Adv. Healthc. Mater. 6, 1700313 (2017).
Wang, Y. et al. Targeted therapy of atherosclerosis by a broad-spectrum reactive oxygen species scavenging nanoparticle with intrinsic anti-inflammatory activity. ACS Nano 12, 8943–8960 (2018).
Millar, J. S., Cromley, D. A., McCoy, M. G., Rader, D. J. & Billheimer, J. T. Determining hepatic triglyceride production in mice: comparison of poloxamer 407 with Triton WR-1339. J. Lipid Res. 46, 2023–2028 (2005).
Acknowledgements
This work was supported by grants to T.H. from the Agence Nationale de la Recherche (ANR-17-CE14-0044, ANR-21-CE14-0067-01), from the Fondation de France (#00096295) and Sorbonne Université Emergence programme (AAP 2021).
Author information
Authors and Affiliations
Contributions
R.F. provided intellectual input, designed and performed experiments, analyzed and interpreted data. S.D., C.B., M.B., F.L. L.P., C.B., A.G., M.D., M.B., C.L., M.M. performed experiments and analyzed data. A.B. designed and performed experiments, analyzed and interpreted data. E.L.G. provided intellectual input, conceptualized and edited the manuscript. T.H. conceptualized and supervised the study, designed experiments, analyzed and interpreted data, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Menno Hoekstra, Frank Tacke, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fima, R., Dussaud, S., Benbida, C. et al. Loss of embryonically-derived Kupffer cells during hypercholesterolemia accelerates atherosclerosis development. Nat Commun 15, 8341 (2024). https://doi.org/10.1038/s41467-024-52735-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52735-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.