Abstract
Enzymes are crucial in numerous biological processes, with the Enzyme Commission (EC) number being a commonly used method for defining enzyme function. However, current EC number prediction technologies have not fully recognized the importance of enzyme active sites and structural characteristics. Here, we propose GraphEC, a geometric graph learning-based EC number predictor using the ESMFold-predicted structures and a pre-trained protein language model. Specifically, we first construct a model to predict the enzyme active sites, which is utilized to predict the EC number. The prediction is further improved through a label diffusion algorithm by incorporating homology information. In parallel, the optimum pH of enzymes is predicted to reflect the enzyme-catalyzed reactions. Experiments demonstrate the superior performance of our model in predicting active sites, EC numbers, and optimum pH compared to other state-of-the-art methods. Additional analysis reveals that GraphEC is capable of extracting functional information from protein structures, emphasizing the effectiveness of geometric graph learning. This technology can be used to identify unannotated enzyme functions, as well as to predict their active sites and optimum pH, with the potential to advance research in synthetic biology, genomics, and other fields.
Similar content being viewed by others
Introduction
Enzymes play an essential role in various biological processes by catalyzing numerous reactions1,2. Identifying enzyme functions is crucial for the study of metabolism3 and diseases4. Enzyme Commission (EC) number5 is commonly utilized to formulate the enzyme function as a four-digit structure, which provides a unified scheme and expedites advancements in the field of enzyme engineering. However, the experimental determination6 of EC numbers is time-consuming and costly. The development of computational approaches for identifying EC numbers has become imperative.
The computational approaches can be categorized into homology-based7,8, structure-based9,10, and machine learning-based11,12,13 approaches. Homology-based approaches, assuming that highly similar enzymes have similar functions, were proposed to annotate the enzyme function with alignment tools14,15. These methods rely heavily on sequence similarity, which limits their coverage while lacking similar sequences. To improve the coverage, structure-based approaches9,16 scanned structurally similar protein templates to identify consensus functions. For instance, COFACTOR10 compared the query structure to proteins with known structures and functions in the BioLiP library17 for function annotation. Despite the improvement of these methods, difficulties remain due to a lack of high-quality templates. To alleviate the constraints of similar sequences and templates, machine learning-based approaches have been developed. The initial machine learning-based approaches18,19 first extracted vital features before utilizing machine learning algorithms to identify the corresponding EC numbers. The performance of these machine learning algorithms is greatly influenced by the manually crafted features, which are not adapted to rapidly expanding enzyme sequences.
Recently, deep learning methods11,20 have achieved success in enzyme function annotation. To avoid manual feature extraction, DEEPre21 employed CNN and RNN components to capture convolutional and sequential features. ProteInfer12 utilized a dilated convolutional network to establish a mapping between protein space and enzyme function space. Utilizing the InterPro signatures as domain information, GrAPFI22 performed label propagation on a weighted undirected graph. For ECPICK23, the protein sequence was encoded using one-hot embedding, which was subsequently employed to compute the posterior probabilities of around 5000 EC numbers through convolutional and hierarchical layers. CLEAN11, another deep learning method that learned abundant embeddings through contrastive learning24, achieved better accuracy and EC coverage for EC number identification. Nevertheless, these methods still suffer from two limitations. Firstly, they only used protein sequences without incorporating protein structures, thus losing the crucial features implied by the structures. Secondly, the crucial information on enzyme active sites was not employed in the analysis of enzyme function.
Due to the lack of native structures, present methods don’t fully exploit the information from protein structures. AlphaFold225 has made a breakthrough in protein structure prediction, with the predicted structures confirmed to be useful in DNA-binding site prediction26,27, antibiotic discovery28, and the study of intrinsically disordered proteins29. Regrettably, the high computational demand of AlphaFold2 limits its applicability for genome-wide use. To address this issue, Lin et al.30 proposed a pre-trained language model ESMFold for precise and quick structure prediction, attaining comparable accuracy to AlphaFold2 while significantly reducing inference time by up to 60 times. The high efficiency of ESMFold enables the analysis of protein structures in metagenomics31, which has shown remarkable achievements in nucleic-acid-binding site prediction32 and drug discovery33. With the aid of predicted structures, geometric graph learning34, a technique that has proven beneficial in protein design35,36 and docking37, can extract structural information efficiently. To augment geometric graph learning, some studies32,38 have attempted to incorporate informative sequence embeddings using unsupervised language models (ProtTrans39 and ESM-1b40).
On the other side, enzyme active sites are typically located on the surface of enzymes and play an important role in catalyzing reactions or binding substrates41. They exhibit a high level of conservation in the process of evolution and significantly determine the function of enzymes42,43. So obviously, it would be highly beneficial to consider the active sites of enzymes when assigning the EC numbers. Meanwhile, current methods for predicting enzyme active sites mainly rely on templates or hand-crafted features, which are unable to keep up with the rapidly growing data. This highlights the need for a fast and accurate enzyme active site predictor. Besides active sites, a label diffusion algorithm44 has been developed for protein function prediction, which can transfer functionally relevant data and aid in identifying EC numbers.
In this work, we proposed GraphEC (geometric Graph learning-based EC number annotation), an accurate network for enzyme function prediction based on predicted protein structures and enzyme active sites. Specifically, the enzyme active sites were identified first, as they play a critical role in predicting enzyme function. With the guidance of active sites, GraphEC was trained through geometric graph learning with the protein structures predicted by ESMFold. To improve the model performance, informative sequence embeddings were generated via a pre-trained language model (ProtTrans) to augment the node features. In addition, a label diffusion algorithm was employed to further enhance the prediction using homology information. Considering that enzyme-catalyzed reactions require specific environmental conditions, we further extended the model to enzyme optimum pH prediction, which can assist in experimental procedures. Through comprehensive comparisons on several independent tests, our model outperformed all the state-of-the-art methods in the predictions of active sites, EC number, and optimum pH. Additional analysis demonstrated that GraphEC is able to learn functional information from enzyme structures, further emphasizing the effectiveness of geometric graph learning.
Results
The overview of the model
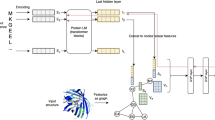
GraphEC, an accurate EC number predictor based on geometric graph learning, incorporates the enzyme active sites and predicted protein structures into enzyme function prediction (Fig. 1). Given a protein sequence, its structure is predicted by ESMFold and used to construct the protein graph. Geometric features were extracted through the predicted structures, which are enhanced by sequence embeddings calculated through a pre-trained language model (PtrotTrans). These features are fed into a geometric graph learning network for learning geometric embeddings, which are utilized in the prediction of active sites, EC number, and optimum pH. Here, enzyme active sites are first predicted by GraphEC-AS, assigning weight scores to each residue. Guided by the weight scores, the initial prediction of the EC number is computed with the attention and pooling layers, which is further improved through a label diffusion algorithm by extracting homologous information. Finally, the model is extended to optimum pH prediction through attention pooling for better representing the reaction conditions (GraphEC-pH).
Given protein sequences, ESMFold was employed to predict the protein structures, which were then utilized to construct the protein graph and extract geometric features. To augment the features, informative sequence embeddings were calculated using a pre-trained language model (ProtTrans). The prepared features were then input into a geometric graph learning network to learn geometric embeddings. These embeddings were then used to predict enzyme active sites (GraphEC-AS), with each residue being assigned a weighted score. Guided by the weight scores of GraphEC-AS, the initial pred of EC number was predicted with the attention and pooling layers. To improve the prediction, a label diffusion algorithm is employed to account for the overlapping communities of enzymes with correlative functions. In addition, the model is further extended to optimum pH prediction through attention pooling for better representing the practical situation (GraphEC-pH).
Enzyme active site prediction (GraphEC-AS)
We first evaluated GraphEC-AS for enzyme active site prediction based on residue using the independent test TS124 (details shown in “Methods”). Figure 2A displays an AUC (area under the receiver operating characteristic curve) of 0.9635 for GraphEC-AS on five-fold cross-validation and 0.9583 for TS124, demonstrating the robustness of the model. Six competing methods (PREvaIL_RF45, PREvaIL_LR, CRpred (residues with coordinates)46, CRpred (all residues), HA (residue identity filter)47, and HA (combination filter) are located between the ROC curves of GraphEC-AS and BiLSTM (the method excluding structural information), indicating the importance of geometric information. In terms of MCC (Matthews correlation coefficient), recall, and precision (Fig. 2B), our method consistently performed the best. The second-best method (PREvaIL_RF) achieved 0.2939, 0.6223, and 0.1487, lower than GraphEC-AS by 40.9, 14.5, and 57.1%, respectively. Source data are provided as a Source Data file. In addition, the F1 score for GraphEC-AS on TS124 is 0.4698 (Supplementary Table S1), while the second-best method, PREvaIL_RF, achieves a score of 0.240, reflecting a decrease of 48.9% relative to GraphEC-AS. The PREvaIL needs the calculation of time-consuming evolutionary profiles using PSI-BLAST48, whereas GraphEC-AS can identify the enzyme active sites rapidly and accurately. Source data are provided as a Source Data file.
A The receiver operating characteristic curves of GraphEC-AS and the geometrically agnostic baseline BiLSTM, as well as their comparison with other state-of-the-art methods. The error band of 5-fold cross-validation represents the standard deviation. B Evaluation of GraphEC-AS’s performance using three metrics (MCC, recall, and precision). Six methods were compared, where PREvaIL_RF and PREvaIL_LR represent the PREvaIL model using random forest and logistic regression algorithms; Crpreda and Crpredb represent the CRpred model using residues with coordinates and all residues; and HAc and HAd represent the HA model using residue identity filter and combination filter. C, D Visualization of the raw ProtTrans embeddings and geometric embeddings learned by GraphEC-AS. E The three-dimensional structure of one example (cis-muconate cyclase, P38677) annotated by BiLSTM and (F) GraphEC-AS. Source data are provided as a Source Data file.
The superiority of GraphEC-AS was further illustrated by its learned embeddings on TS124. The ProtTrans embeddings (Fig. 2C) are scattered while the geometric embeddings learned by GraphEC-AS (Fig. 2D) distinguished active sites from non-active sites clearly. This demonstrates the capability of geometric graph learning to identify the crucial distinctions between them. We further evaluated the impact of the quality of ESMFold-predicted structures using TM-align49 on TS124. More than 85% of proteins had TM-scores greater than 0.8 (Supplementary Fig. S1), which reflects the high quality of the ESMFold-predicted structures. The AUC values increased with TM-scores (Supplementary Fig. S2), which indicates the necessity of high predicted structure quality and emphasizes the importance of employing ProtTrans to enhance the feature embeddings. Figure 2E, F compares the three-dimensional structures of an example (cis-muconate cyclase) predicted by BiLSTM and GraphEC-AS. GraphEC-AS identified all four active sites, whereas BiLSTM only detected H149 due to the absence of local structure characteristics. Compared to H149, the remaining active sites were located far in sequence (more than 20 residues apart) but close in structure (less than 16 Å). These results indicate the capability of GraphEC-AS to learn the local structure information. Additional cases can be seen in Supplementary Fig. S3.
Enzyme EC number identification (GraphEC)
With the guidance of predicted active sites, GraphEC was proposed to identify enzyme EC numbers. GraphEC was evaluated on two independent tests: NEW-392 and Price-149, where NEW-392 comprises 392 enzyme sequences covering 177 different EC numbers, and Price-149 is an experimental dataset validated by Price et al.50. In comparison to four state-of-the-art EC number predictors (i.e., CLEAN, ProteInfer, DeepEC20, ECPred51, GrAPFI, and ECPICK), GraphEC exhibited superior performance in various metrics. Figure 3A illustrates that GraphEC achieved an AUC, recall, precision, and F1 of 0.8404, 0.6908, 0.6132, and 0.6131 on Price-149, surpassing the second-best method (CLEAN) by 14.6, 47.9, 4.9, and 23.9%, respectively. On NEW-392, GraphEC achieved optimal values in AUC (0.8910), recall (0.7988), and F1 (0.5910) (Supplementary Fig. S4). Source data are provided as a Source Data file. As shown in Supplementary Table S3, GraphEC is able to achieve high EC number coverage (5106 EC numbers) while maintaining high performance. Benefiting from the contrastive learning-based representation, CLEAN achieved high precision, but its recall and F1 were 39.8% and 15.6% lower than those of GraphEC, respectively. Relying on the label propagation on a protein domain similarity graph, GrAPFI22 achieved acceptable performance, with AUC values of 0.5095 and 0.5407 on Price-149 and NEW-392 (Supplementary Table S2). ECPICK23 attained the third-best performance through the implementation of a convolutional neural network and hierarchical module, achieving the AUC values of 0.5888 and 0.6502 on Price-149 and NEW-392 (Supplementary Table S2), respectively. Source data are provided as a Source Data file.
A The comparison between GraphEC and several state-of-the-art methods using AUC, recall, precision, and F1 on Price-149. B The model’s precision varies depending on the frequency of the EC number in the training set. C The analysis of GraphEC and three methods (CLEAN, ProteInfer, and DeepEC) at four different levels. D The method ablation focused on the label diffusion algorithm, active site guidance, predicted protein structures, and ProtTrans embeddings. E Three embeddings were visualized on NEW-392, including the GraphEC embeddings, which represent the geometric embeddings learned by GraphEC, as well as the One-hot embeddings and ProtTrans embeddings, which represent the one-hot vector and ProtTrans vector, respectively. F, G The three-dimensional structures of Acyl-protein thioesterase 2 (O95372) and Proline racemase (E3PTZ4) were visualized, with the highlighted portion indicating higher attention scores. Source data are provided as a Source Data file.
GraphEC was further evaluated on different levels of EC numbers and the frequency of each EC number in the training set. Considering the potential impact of EC number frequency in the training set on model performance, precision on NEW-392 was evaluated based on the number of times that the EC number appeared in the training set. (Fig. 3B). More than 66.0% of enzymes have less than ten occurrences, and only 8.9% of enzymes have more than 100 occurrences, demonstrating the challenge of the dataset. As expected, predicting EC numbers with low frequency proved to be difficult. However, GraphEC consistently exhibited higher precision at different occurrences of EC numbers compared to other methods, highlighting the superior performance of our model. The four digits of the EC number correspond to different levels of enzyme functional classification, with the first to fourth digits indicating a hierarchical breakdown. The recall of GraphEC on NEW-392, compared to CLEAN, improved by 1.1%, 1.7%, 3.4%, and 66.0% from the first level to the fourth level, with values of 0.9468, 0.9116, 0.8945, and 0.7988 (Fig. 3C). The superiority of GraphEC becomes more apparent as the level increases, indicating the effectiveness of our model. Source data are provided as a Source Data file.
Considering the utilization of active sites in EC number prediction, we have evaluated the impact of mutations in the active sites. We first identified the active sites of enzymes on NEW-392 and Price-149 based on the predicted results (score > 0.5). Subsequently, these active sites were mutated to Alanine (A), and the predicted scores for true EC numbers were compared before and after the mutation. After mutation, the predicted scores for true EC numbers have decreased (Supplementary Fig. S6), demonstrating the influence of mutations in the active sites on the prediction of EC numbers. Among the mutated enzymes, 59.1% can be identified as non-enzymes, such as L-2-hydroxyglutarate dehydrogenase (Uniprot ID: A0A011QK89) and Farnesyl pyrophosphate synthase (Uniprot ID: B4YA15) (more cases can be seen in Supplementary Table S4). Source data are provided as a Source Data file. Furthermore, the predicted scores for active sites before and after the mutation were compared, discovering a reduction in predicted scores for active sites after mutation (Supplementary Fig. S7). This indicates a reduced focus of the model on the mutated active sites. In addition, we have compared the average computational time per protein of different methods on Price-149. The average inference time for GraphEC is 0.26 seconds (s), while CLEAN, ProteInfer, and DeepEC have inference times of 1.28, 0.21, and 0.14 s, respectively (Supplementary Fig. S8). Source data are provided as a Source Data file. Due to the considerable time needed to compute the pairwise distances between the query sequence and each EC number cluster center in CLEAN, GraphEC’s inference speed is 392.3% faster than that of CLEAN. By combining the time required for ESMFold to compute protein structures (11.44 s) with the inference time of GraphEC (0.26 s), a total of 11.7 s is necessary for each enzyme. In this case, computing the functions of 1000 enzymes requires just 3.25 h, thereby meeting the need for high-throughput analysis.
The ablation studies of GraphEC
The ablation studies of GraphEC were conducted to investigate the contribution of each module. When removing label diffusion, the AUC values slightly decreased (Fig. 3D) likely because of the ability of GraphEC to learn homology information. The removal of active site guidance resulted in a decrease of 2.8% and 3.5% in AUC on NEW-392 and Price-149, demonstrating its great importance. For evaluating the impact of ESMFold-predicted structures, a geometrically agnostic baseline (BiLSTM) was constructed. Without structural information, the AUC decreased by 4.8% and 2.1% on NEW-392 and Price-149, indicating the crucial role of predicted structures. The ProtTrans embeddings were used to enhance the node features, and the removal of them led to a decrease in AUC by 6.6% and 2.8%. The PortTrans embeddings used here are residue-level representations, which are different from the protein-level ESM-1b representations (mean representations) used in CLEAN (Supplementary Fig. S9). Source data are provided as a Source Data file. In addition, we have evaluated the effects of physicochemical properties in reference to previous studies52,53. The incorporation of these physicochemical properties failed to further improve the performance of GraphEC (Supplementary Table S5), suggesting that the geometric features and language model embeddings used in this study may have already inherently captured the physicochemical properties. Source data are provided as a Source Data file.
As shown in Fig. 3E, the learned geometric embeddings (GraphEC embeddings) were compared with ProtTrans embeddings and one-hot embeddings on NEW-392. Among the ten most frequent EC numbers, the one-hot embeddings exhibited limited discriminative capacity. The ProtTrans embeddings can roughly distinguish these EC numbers, yet they cannot cluster the categories to which 3.1.2.22 and 4.2.1.113 belong. In contrast, GraphEC embeddings can clearly separate these EC numbers, demonstrating their strong expressive ability for different EC numbers. Similarly, on Price-149, the one-hot embeddings lacked the ability to distinguish, while the ProtTrans embeddings can provide a basic distinction, and the GraphEC embeddings were able to further differentiate them (Supplementary Fig. S10). Source data are provided as a Source Data file.
To evaluate the importance of predicted structures, we replace the ESMFold-predicted structures with those predicted by AlphaFold2. Utilizing the AlphaFold2-predicted structures, the AUC, recall, precision, and F1 on NEW-392 are 0.9004, 0.8267, 0.5745, and 0.6044, respectively (Supplementary Table S6), slightly higher than those of using ESMFold-predicted structures. On Price-149, comparable performance was obtained when utilizing AlphaFold2-predicted and ESMFold-predicted structures. These results indicate that ESMFold can generate structures with comparable accuracy in much less time than AlphaFold2. In addition, we also evaluated the impact of various cut-off distances (8 Å, 12 Å, and 14 Å) relative to 10 Å on model performance. When the distance is 8 Å, the AUC, recall, precision, and F1 of the model are 0.8761, 0.7729, 0.5577, and 0.5459 on NEW-392 (Supplementary Table S7), lower by 1.7%, 3.2%, 2.3%, and 7.6% when the distance is 10 Å. This may be due to the decreased distance, which reduces the number of neighbor nodes associated with each node, ultimately causing some information loss. When the distance is 12 Å, and 14 Å, the AUC of the model is 0.8876 and 0.8753 on NEW-392, respectively, 0.4% and 1.8% lower than when the distance is 10 Å (0.8910). This might be because a larger distance allows each node to have more edges, resulting in excessive aggregation of information from neighbor nodes during the iterative process, which eventually reduces the node specificity. Similar results on Price-149 are presented in Supplementary Table S7. Source data are provided as a Source Data file.
GraphEC captures the functional regions of enzymes
To verify whether GraphEC can identify functional regions, we studied the connections between predicted enzyme active sites, multi-head attention scores, and true active sites. As shown in Fig. 3F, the true active sites of Acyl-protein thioesterase 2 are S122, D176, and H210, which were correctly predicted through GraphEC-AS and used to guide the EC number prediction. The multi-head attention scores tended to be higher near the true active sites, suggesting that the model can focus on the functional regions. Similarly, the enzyme active sites of Proline racemase were accurately identified, and the muti-head attention scores were prominent when approaching the true active sites (Fig. 3G). Additional cases can be seen in Supplementary Fig. S11. These results indicate that GraphEC could capture the functional regions of enzymes.
The prediction of enzyme optimum pH
Since enzyme pH values are important for enzyme functions, we have also included enzyme optimum pH predictions. To train the model, we have curated a new dataset constructed from the Brenda database (released in January 2023)54 (Supplementary Fig. S12), including 4110 proteins with sequence identity of < 25%. The dataset was divided into a training set (Brenda-train, 3297 enzymes) and an independent test set (Brenda-test, 813 enzymes) with a ratio of 4:1 according to the deposit time. As shown in Fig. 4A, GraphEC-pH achieved an AUPR (area under the precision-recall curve) of 0.9321 for five-fold cross-validation and 0.9170 on the test, indicating the model’s robustness. By removing the structural information, the AUPR of GraphEC-pH w/o structures decreased by 1.4%. In comparison, the two latest methods, EpHod55 and EpHod_SVR, achieved lower performance with points located below the precision-recall curve of GraphEC-pH. Correspondingly, the F1, recall, and precision of GraphEC-pH were 0.8487, 0.8672, and 0.8461, surpassing the second-best method (EpHod) by 9.2%, 16.5%, and 0.09%, respectively (Fig. 4B). Source data are provided as a Source Data file. These results have demonstrated the superior performance of our model. We then evaluate the model’s ability to discern differences among 289 homologous enzyme pairs searched by DIAMOND in the Brenda test. More than 87.9% (254 pairs) of the homologous enzyme pairs have the same type of optimum pH (i.e., “acidic” - “acidic” and “non-acidic” - “non-acidic”), and GraphEC-pH can correctly identify 95.7% of them (243 pairs). Only 35 pairs of enzymes exhibit different optimal pH types (i.e., “acidic” - “non-acidic”), with GraphEC-pH correctly distinguishing 14 pairs (Supplementary Table S8), which is 75% more than EpHod (8 pairs). These results indicate that GraphEC-pH can discern the differences among homologous enzymes to some extent. Source data are provided as a Source Data file.
A The precision-recall curves of GraphEC-pH on Brenda test, compared with 5-fold cross-validation, geometrically agnostic baseline (GraphEC-pH w/o structures), and two of the latest methods (EpHod and EpHod_SVR). The error band of 5-fold cross-validation represents the standard deviation. B F1, recall, and precision were compared for GraphEC-pH, EpHod, and EpHod_SVR. Source data are provided as a Source Data file.
GraphEC learns functional information from enzyme structures
To discover new enzyme functions, a total of 570,830 protein sequences were collected from Swiss-Prot (January 2024 release). After removing the proteins with sequence identity greater than 25% and those with identity above 25% to the training dataset, 52,037 proteins without EC number annotations remained. These proteins were annotated by GraphEC and CLEAN, with over 21% of them including the same EC number annotations. For each protein, the predicted EC number was obtained, and the TM scores were calculated with proteins sharing the same EC number in the training set. Subsequently, the maximum TM scores of proteins were further used to analyze. GraphEC generally has a higher score, with over 82% of the proteins found by Foldseek56 showing a higher TM score compared to CLEAN. When comparing the number of enzymes whose maximum TM-scores exceeded various thresholds (Fig. 5A), GraphEC surpassed CLEAN by 158, 136, 128, and 128% at thresholds of 0.5, 0.7, 0.8, and 0.9, respectively. Source data are provided as a Source Data file. Compared to CLEAN, the newly discovered enzyme functions identified by GraphEC with maximum TM-scores surpassing 0.8 are listed in Supplementary Dataset 1 partially. Despite low sequence similarity, GraphEC can learn functional information from enzymes with high structural similarity (Fig. 5B). Even when the TM-score is low, the enzyme pocket (details shown in “Methods”) around the enzyme active sites can still be aligned (Fig. 5C), demonstrating the capacity of GraphEC to learn critical functional information from enzyme structures. In addition, an example (Q9NWA0) with disorder regions was found to be aligned to the enzyme pocket of Q980B8 in the training set (Supplementary Fig. S13F), which indicates the potential of our method for identifying disordered protein functions. More cases are available for reference in Supplementary Fig. S13. Source data are provided as a Source Data file.
A Comparison of the number of enzymes whose maximum TM-scores exceeded various thresholds. For each protein, the predicted EC number was obtained, and the TM scores were calculated with proteins sharing the same EC number in the training set. Subsequently, the maximum TM-score was further used to compare. The “w/o structures” represents the baseline model (MLP) that only uses ProtTrans embeddings without structures. GraphEC has a higher TM-score compared to CLEAN in over 82% of the proteins found by Foldseek. B The alignment of ESMFold-predicted structures with low sequence similarity, where Q6GIA3 represents the enzyme in the training set, and P96284 represents the protein from Swiss-Prot with less than 25% identity to the training set. Despite low sequence similarity, GraphEC has the ability to learn the functional information from enzymes with high structural similarity. C Despite a low TM-score, the enzyme pocket around the enzyme active sites can still be aligned (the highlighted area represents the enzyme pocket), demonstrating that GraphEC is able to learn functional information from structures even with low structural similarity. Q9GZX3 and O29655 represent the proteins in the training set and Swiss-Prot, respectively. Source data are provided as a Source Data file.
Discussion
GraphEC is a geometric graph learning-based EC number predictor based on the enzyme active sites and predicted structures. The predicted active sites can guide the learning because of their crucial role in enzyme function. Based on the ESMFold-predicted structures, geometric graph learning can efficiently extract structural information, which is especially necessary when lacking homology information. In addition, a label diffusion algorithm and ProtTrans embeddings are able to improve the model performance. For an enzyme, the EC number, active sites, and optimum pH can be analyzed comprehensively.
Despite the essential role of EC numbers, current EC number prediction technologies have not fully recognized the importance of enzyme active sites and structural characteristics. The enzyme active sites represent the chemical reaction regions, which we first predict and use to guide subsequent learning. Due to the limitations of native structures, current methods for EC number prediction don’t fully exploit the information from protein structures. Benefiting from the rapid and precise structure prediction of ESMFold, GraphEC utilizes geometric graph learning to extract important structural information and surpass state-of-the-art methods. Experiments demonstrate the efficacy of our model in predicting active sites, EC numbers, and optimum pH. Furthermore, GraphEC is proven to be able to extract functional information from enzyme structures even in the absence of homology information, emphasizing the effectiveness of geometric graph learning.
Although GraphEC has shown great performance, there is still room for improvement in several aspects. Considering the impact of predicted structure quality, we can explore enhancing the stability of the model by either improving the structural quality or incorporating additional sequence features. In addition, as large language models continue to advance, we can utilize them to extract essential information from textual descriptions and enhance our model’s predictions.
In summary, we have developed an accurate and fast EC number predictor, GraphEC. Researchers can use it to accurately predict enzyme function solely from the enzyme sequences. For specific enzymes, we can further analyze their functional regions (active sites) and determine their reaction conditions (pH), which will be helpful for experimental investigations.
Methods
Dataset construction
To predict the enzyme active sites, we collected eight enzyme datasets and constructed new training and test sets from them. The eight datasets, namely NN57, PC58, HA superfamily47, EF family59, EF superfamily, EF fold, T-37, and T-12446, collectively contain a total of 987 proteins. T-124, containing 124 proteins, was used as the test set (TS124), while the remaining 863 proteins were utilized as a training set. For excluding the sequences with high identity, the chains in the training set that share > 25% identity with TS124 were removed using MMseqs260, resulting in 588 sequences in the training set (Train588). For EC number prediction, referring to CLEAN11, more than 220, 000 enzyme sequences were extracted from UniProt61, and a training set of size 74, 487 for enzyme EC number identification was constructed through 70% clustering. Two independent test sets were used to evaluate the model performance. The first is NEW-392, which collected data from Swiss-Prot released after April 2022. In NEW-392, 392 enzyme sequences were included, encompassing a total of 177 EC numbers. The second is Price-149, an experimental dataset of 149 enzyme sequences described by Price et al.50. For predicting the enzyme optimum pH, 11383 enzymes were collected from BRENDA (released in January 2023)54, which provides the experimental optimum pH for enzyme-catalyzed reactions. After removing the similar sequences with > 25% identity, 4110 enzymes remained and were ranked by the released time. The latest 813 sequences (about 20%) were utilized as the test set (Brenda-test), while the remaining were used as the training set (Brenda-train).
The architecture of the model
As shown in Fig. 1, protein structures are predicted using ESMFold to construct the protein graph, and sequence embeddings are extracted via ProtTrans, which are then fed into a featurizer layer to obtain node and edge features. These features are employed to obtain geometric embeddings through geometric graph learning. Based on the embeddings, enzyme active sites are predicted, and a weighted score is assigned to every residue. Using these weight scores, enzyme EC numbers are identified with an attention layer and label diffusion. In addition, for better determining the reaction conditions, the model is subsequently expanded to optimum pH prediction by incorporating attention pooling.
Featurizer layer
A protein is represented as a radius graph constructed by the \({c}_{\alpha }\) atoms of residues, where the radius defaults to 10 Å. The protein graph comprises the adjacency matrix, as well as node and edge features, which are derived from a local coordinate system. The \({{\rm{C}}}_{{\rm{\alpha }}},{\rm{C}},\) and N atoms of residue \(i\) are employed to build the coordinate system \({Q}_{i}=[{b}_{i},\,{n}_{i},\,{b}_{i} \times {n}_{i}]\). Formally, we define:
Based on the local coordinate system, the node and edge features are defined as follows:
(i) Node features. Given two atoms \(A\in \{{C}_{i},\,{C}_{{\alpha }_{i}},\,{N}_{i},\,{O}_{i},\,{{R}}_{i}\}\) and \(B\in \{{C}_{i},\,{C}_{{\alpha }_{i}},\,{N}_{i},\,{O}_{i},{{R}}_{i}\}\), where \({C}_{i}\), \({C}_{{\alpha }_{i}}\), \({N}_{i}\), and \({O}_{i}\) represent four atoms of residue \(i\) and \({{R}}_{i}\) denotes the centroid of sidechain atoms. By analyzing the characteristics between A and B, the distance, direction, and angle features are computed for each residue. The distance features are \({RBF}(\parallel A-B\parallel )\), where \(A\ne B\) and \({RBF}\) is a radial basis function. The direction features are regulated as \({Q}_{i}^{T}\frac{A-{C}_{{\alpha }_{i}}}{\parallel A-{C}_{{\alpha }_{i}}\parallel }\), indicating the direction of other atoms relative to \({C}_{{\alpha }_{i}}\). For adequately reflecting the geometrical information of the backbone, the torsion angles (\({\phi }_{i},\,{\psi }_{i},\,{\omega }_{i}\)) and bond angles (\({\alpha }_{i},\,{\beta }_{i},{\gamma }_{i}\)) have been exploited and their sine and cosine values are applied as angle features.
To enhance the node features, a pre-trained language model (ProtTrans) was utilized to extract informative protein embeddings from sequences. ProtTrans is a transformer-based pre-trained language model with 3B parameters, trained on BFD and fine-tuned on UniRef50 using the BERT’s denoising objective. Besides the sequence, we also attempted to extract more information from structures. DSSP was used to compute valuable structural properties, including one-hot secondary structure profile and relative solvent accessibility, which were used to further enhance the node features.
(ii) Edge features. For atom pairs \(A\in \{{C}_{i},\,{C}_{{\alpha }_{i}},\,{N}_{i},\,{O}_{i},\,{{R}}_{i}\}\) and \(D\in \{{C}_{j},\,{C}_{{\alpha }_{j}},\,{N}_{j},\,{O}_{j},{{R}}_{j}\}\) representing residues \(i\) and \(j\) respectively, the edge features are defined similarly, including distance, direction, and orientation features. The distance features between residues \(i\) and \(j\) are \({RBF}(\parallel A-D\parallel )\), indicating the distance characteristics of given residue pairs. The direction features are defined as \({Q}_{i}^{T}\frac{D-{C}_{{\alpha }_{i}}}{\parallel D-{C}_{{\alpha }_{i}}\parallel }\), denoting the direction of atoms in residue \(j\) to \({C}_{{\alpha }_{i}}\). To represent the relative rotation between the local coordinate systems, \({q}({Q}_{i}^{T}{Q}_{j})\) is computed as orientation features, where \(q\) represents a quaternion encoding function62.
Geometric graph learning
The node and edge features obtained from featurizer layer were fed into several GNN layers for geometric graph learning. To learn the multi-scale residue interactions, node update, edge update, and global context attention modules were employed at node, edge, and global context levels, respectively.
(i) Node update. Due to the transformer’s reputation as a powerful model for both sequence and graph data63,64, we employed its multi-head attention mechanism for efficient message passing. The feature vectors of node \(i\) and edge \(j\to i\) in layer \(l\) were represented as \({h}_{i}^{l}\) and \({e}_{{ji}}^{l}\), which were transformed into a \(d\)-dimensional space before the GNN operation. To update node \(i\) in layer \(l\), we execute the message passing in the following manner:
the attention weight \({\alpha }_{{ji}}^{l}\) is computed as follows:
Where the \({W}_{Q}^{l}\), \({W}_{K}^{l}\), and \({W}_{V}^{l}\) are three weight matrices utilized to convert the node vectors to query, key, and value representations, respectively. The key and value representations are further supplemented by edge vectors using weight matrices \({W}_{E}^{l}\). \({{NB}}_{i}\) represents the neighbors of the node \(i\). The queries, keys, and values are translated multiple times, with parallel attention functions being performed before concatenating them together.
(ii) Edge update. The edge features are updated through the neighbor nodes to enhance the model performance.
where \({EdgeMLP}\) denotes the MLP operation for edge updates and \(\parallel\) represents the concatenation operation.
(iii) Global context attention. Although local interactions are crucial for learning residue representations, global information has also been shown to be beneficial in enhancing method performance. However, the increased computational overhead in calculating global attention poses a major challenge. To reduce the complexity, an alternative is proposed to calculate a global context vector before employing it for node representations with gate attention36.
where \(n\) represents the quantity of residues in a protein, \(\sigma\) is the sigmoid function, \({\odot}\) is the element-wise product operation and \({GateMLP}\) denotes the MLP for gated attention.
Enzyme active site prediction (GraphEC-AS)
Due to the important role of enzyme active sites in enzyme function, we first predict the active sites before identifying the EC numbers. The geometric embeddings obtained from the geometric graph learning were fed into an MLP layer to assign a score to each residue, indicating its likelihood of belonging to an active site. Using these scores, each residue was assigned a weight to represent its level of importance.
The identification of EC numbers (GraphEC)
Under the guidance of weight scores generated by GraphEC-AS, an EC number predictor was proposed. The previously generated geometric embeddings were further input to an attention layer, where the attention functions were performed in parallel with the multi-head attention mechanism. By integrating the multi-head attention and weight scores, the residue-level information was aggregated to the protein level through a pooling layer. After pooling, the initial prediction was obtained, and a label diffusion algorithm was employed to enhance the prediction using DIAMOND. The label diffusion algorithm was used to extract homologous information, as referenced by S2F44. Following the label diffusion, the final pred was generated to identify the EC numbers as a multilabel classification task.
Enzyme optimum pH prediction (GraphEC-pH)
Since enzymes require certain environmental conditions to exert their catalytic activity, we further predicted the optimal pH of the enzyme. The pH values were categorized into three groups: acidic (less than 5), neutral (between 5 and 9), and alkaline (greater than 9). To get the characterization for predicting the enzyme's optimum pH, multi-head attention was utilized to process the geometric embeddings derived from the geometric graph learning. Then an MLP layer was used to predict the optimum pH. By combining the previous identification of enzyme function with the current prediction of pH, a more effective method can be provided to guide actual experiments.
Hierarchy of catalytic functions
The Enzyme Commission (EC) number is a numerical system used to classify enzymes according to the reactions they catalyze. Each EC number comprises four digits, which hierarchically categorize enzymes based on their catalytic reaction types and specific substrates65 (e.g., EC: 1.3.1.32 represents the maleylacetate reductase). In this study, we collected 5106 EC numbers from the training set and defined a label of length 5106, where each position corresponds to a specific EC number.
The protein language model (ProtTrans)
The informative sequence embeddings were generated through a pre-trained language model ProtT5-XL-U50 (ProtTrans39). ProtTrans is a transformer-based autoencoder known as T566, which has been pre-trained on UniRef5067 to facilitate the prediction of masked amino acids. The features derived from the final layer of the ProtTrans encoder were employed to enhance the node representations.
Protein structure prediction using a language model (ESMFold)
ESMFold30 is a large language model with up to 15B parameters, developed on the premise that language models can capture evolutionary patterns across millions of sequences. Achieving accurate and fast structure prediction, ESMFold reduces inference time by as much as 60 times compared to the state-of-the-art method. Benefiting from its high efficiency, the first evolutionary scale structural characterization of a metagenomic resource has been presented. In this study, we employed ESMFold to predict the protein structures, which were then applied in subsequent geometric graph learning.
Label diffusion algorithm
To enhance the initial predictions of EC numbers, a label diffusion algorithm44,68 was applied during the testing phase. First, the sequences in the training set similar to the test sequences were found using DIAMOND15. Second, based on the sequence identity of protein pairs, a homology network \(M\in {R}^{T\times T}\) was constructed (\(T\) represents the sum of the number of proteins in the test set and the number of hits in the training set). Then, to measure the degree to which a pair of proteins belongs to the same community within the homology network, a Jaccard similarity matrix was defined as follows:
For a target EC number \(x\), the \({x}^{{th}}\) column of the final annotation matrix \(S\) (\({S}_{x}\)) was learned by minimizing the cost function \(P({S}_{x})\):
Where \(\varepsilon\) represents the regularization parameter. The first term serves to preserve the initial labels (\({Y}_{{ix}}\)), and the consistency of the labels of adjacent nodes is accounted for through the second term. And \(\frac{1}{{d}_{i}}\) is defined as:
Furthermore, we define \({M}^{1}\) as:
its Laplacian matrix \(L\) is:
where \({DM}\) is the diagonal degree matrix of \({M}^{1}\). The closed-form solution that minimizes \(P({S}_{x})\) can be converted to:
where \(S\) is the updated annotation matrix, \(I\in {R}^{T\times T}\) indicates an identity matrix, and \(Y\) represents the combination of the training set labels along with the initial predictions for the test set.
Constructing the enzyme pocket from predicted enzyme active sites
The construction of the enzyme pocket involved two steps. First, the predicted enzyme active sites were clustered (k-means), with k set to 2 empirically. To eliminate false positives, we removed the isolated points that were classified separately. Second, using the \({c}_{\alpha }\) coordinates, the enzyme pocket is defined as the area within 10 Å of the cluster center.
Implementation and evaluation
Five-fold cross-validation was performed on training data, where each time, the model was trained on four folds and validated on the remaining one-fold data. This operation was repeated five times, with the best model saved at each iteration. After training, several independent tests were used to test the model performance on different tasks. In enzyme active prediction, TS124 was employed to compare the GraphEC-AS to other methods. The performance of GraphEC in predicting the EC numbers was evaluated on NEW-392 and Price-149. In order to test the accuracy of GraphEC-pH in predicting the enzyme optimum pH, a new independent test (Brenda-test) was built and two of the latest methods were evaluated on it. During testing, the average predictions of the five models from the cross-validation were utilized as the final predictions. Specifically, Pytorch 1.13.1 was used to construct the geometric graph network, which consists of a 3-layer GNN with 256 hidden units. The attention layer of GraphEC employed multi-head attention with 8 attention heads. Based on the binary cross-entropy loss, the Adam optimizer was employed to optimize the model. The training process was limited to a maximum of 35 epochs, and an early stopping with the patience of 4 was implemented, along with a dropout value of 0.1 to prevent overfitting. To comprehensively evaluate model performance, AUC, AUPR, recall, precision, F1-score (F1), and Matthews correlation coefficient (MCC) were utilized, as defined in detail in Supplementary Evaluation metrics.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The enzyme function data is obtained from a previous study (CLEAN), which is available on GitHub (https://github.com/tttianhao/CLEAN/tree/main/app/data). The data about enzyme active sites is derived from a preceding work (CRpred), which is available at http://biomine.cs.vcu.edu/datasets/CRpred/CRpred.html. The data on enzyme optimal pH is curated newly from the Brenda database (https://www.brenda-enzymes.org/), which is available at https://github.com/biomed-AI/GraphEC/tree/main/Optimum_pH/data/datasets. A figshare version is also available at https://doi.org/10.6084/m9.figshare.25714305. Source data are provided with this paper.
Code availability
The source code of GraphEC is available at https://github.com/biomed-AI/GraphEC. A Zenodo version is also available at https://doi.org/10.5281/zenodo.1337527569.
References
Kohli, R. M. & Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013).
Makrydaki, E., et al. Immobilized enzyme cascade for targeted glycosylation. Nat. Chem. Biol. 20, 732–741 (2024).
Finley, S. D., Broadbelt, L. J. & Hatzimanikatis, V. Computational framework for predictive biodegradation. Biotechnol. Bioeng. 104, 1086–1097 (2009).
Hoffmann, B. et al. Nature and prevalence of pain in Fabry disease and its response to enzyme replacement therapy—a retrospective analysis from the Fabry Outcome Survey. Clin. j. Pain 23, 535–542 (2007).
Nomenclature, E., Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes (Academic, New York 1992).
Goddard, J.-P. & Reymond, J.-L. Enzyme assays for high-throughput screening. Curr. Opin. Biotechnol. 15, 314–322 (2004).
Desai, D. K., et al. ModEnzA: accurate identification of metabolic enzymes using function specific profile HMMs with optimised discrimination threshold and modified emission probabilities. Adv. Bioinform. https://doi.org/10.1155/2011/743782 (2011).
Kumar, N. & Skolnick, J. EFICAz2. 5: application of a high-precision enzyme function predictor to 396 proteomes. Bioinformatics 28, 2687–2688 (2012).
Roy, A., Yang, J. & Zhang, Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40, W471–W477 (2012).
Zhang, C., Freddolino, P. L. & Zhang, Y. COFACTOR: improved protein function prediction by combining structure, sequence and protein–protein interaction information. Nucleic Acids Res. 45, W291–W299 (2017).
Yu, T. et al. Enzyme function prediction using contrastive learning. Science 379, 1358–1363 (2023).
Sanderson, T. et al. ProteInfer, deep neural networks for protein functional inference. Elife 12, e80942 (2023).
Zou, H.-L. & Xiao, X. Classifying multifunctional enzymes by incorporating three different models into Chou’s general pseudo amino acid composition. The. J. Membrane Biol. 249, 551–557 (2016).
Altschul, S. F. et al. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Yang, J., Roy, A. & Zhang, Y. BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res. 41, D1096–D1103 (2012).
Volpato, V., Adelfio, A. & Pollastri, G. Accurate prediction of protein enzymatic class by N-to-1 Neural Networks. BMC Bioinformatics 14, 1–7 (2013).
Wang, Y.-C. et al. Prediction of enzyme subfamily class via pseudo amino acid composition by incorporating the conjoint triad feature. Protein Peptide Lett. 17, 1441–1449 (2010).
Ryu, J. Y., Kim, H. U. & Lee, S. Y. Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers. Proc. Natl. Acad. Sci. 116, 13996–14001 (2019).
Li, Y. et al. DEEPre: sequence-based enzyme EC number prediction by deep learning. Bioinformatics 34, 760–769 (2018).
Sarker, B., Ritchie, D. W. & Aridhi, S. GrAPFI: predicting enzymatic function of proteins from domain similarity graphs. BMC Bioinformatics 21, 1–15 (2020).
Han, S.-R. et al. Evidential deep learning for trustworthy prediction of enzyme commission number. Brief. Bioinform. 25, bbad401 (2024).
Heinzinger, M. et al. Contrastive learning on protein embeddings enlightens midnight zone. NAR Genom. Bioinform. 4, lqac043 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Yuan, Q. et al. AlphaFold2-aware protein–DNA binding site prediction using graph transformer. Brief. Bioinform. 23, bbab564 (2022).
Yidong, S., Qianmu, Y. & Yuedong, Y. Application of deep learning in protein function prediction. Synth. Biol. J. 4, 488 (2023).
Wong, F. et al. Benchmarking AlphaFold‐enabled molecular docking predictions for antibiotic discovery. Mol. Syst. Biol. 18, e11081 (2022).
Ruff, K. M. & Pappu, R. V. AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 433, 167208 (2021).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Handelsman, J. et al. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5, R245–R249 (1998).
Song, Y. et al. Accurately identifying nucleic-acid-binding sites through geometric graph learning on language model predicted structures. Brief. Bioinform. 24, bbad360 (2023).
Bal, R., Xiao, Y. & Wang, W. PGraphDTA: Improving drug target interaction prediction using protein language models and contact map. In Advances in Neural Information Processing Systems (2023).
Jing, B. et al. Learning from protein structure with geometric vector perceptions. In International Conference on Learning Representations (2020).
Dauparas, J. et al. Robust deep learning–based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Gao, Z. et al. PiFold: Toward effective and efficient protein inverse folding. In International Conference on Learning Representations (2023).
Stärk, H., et al. Equibind: Geometric deep learning for drug binding structure prediction. In International conference on machine learning. (2022).
Yuan, Q., Tian, C. & Yang, Y. Genome-scale annotation of protein binding sites via language model and geometric deep learning. ELife 13, RP93695 (2024).
Elnaggar, A. et al. Prottrans: Toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2021).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. 118, e2016239118 (2021).
Kahraman, A. & Thornton, J. M. Methods to characterize the structure of enzyme binding sites. Comput. Struct. Biol. Methods Appl. 1, 189–221 (2008).
Torrance, J. W. and J. M. Thornton, Structure‐Based Prediction of Enzymes and Their Active Sites. Prediction of Protein Structures, Functions, and Interactions, (2008).
Roche, D. B., Brackenridge, D. A. & McGuffin, L. J. Proteins and their interacting partners: An introduction to protein–ligand binding site prediction methods. Int. J. Mol. Sci. 16, 29829–29842 (2015).
Torres, M. et al. Protein function prediction for newly sequenced organisms. Nat. Mach. Intell. 3, 1050–1060 (2021).
Song, J. et al. PREvaIL, an integrative approach for inferring catalytic residues using sequence, structural, and network features in a machine-learning framework. J. Theor. Biol. 443, 125–137 (2018).
Zhang, T. et al. Accurate sequence-based prediction of catalytic residues. Bioinformatics 24, 2329–2338 (2008).
Chea, E. & Livesay, D. R. How accurate and statistically robust are catalytic site predictions based on closeness centrality? Bmc Bioinform. 8, 1–14 (2007).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Price, M. N. et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509 (2018).
Dalkiran, A. et al. ECPred: a tool for the prediction of the enzymatic functions of protein sequences based on the EC nomenclature. BMC Bioinform. 19, 1–13 (2018).
Meiler, J. et al. Generation and evaluation of dimension-reduced amino acid parameter representations by artificial neural networks. Mol. Model. Annu. 7, 360–369 (2001).
Chen, J. et al. Structure-aware protein solubility prediction from sequence through graph convolutional network and predicted contact map. J. Cheminform. 13, 1–10 (2021).
Schomburg, I. et al. The BRENDA enzyme information system–From a database to an expert system. J. Botechnol. 261, 194–206 (2017).
Gado, J. E., et al. Deep learning prediction of enzyme optimum pH. Preprint at bioRxiv https://doi.org/10.1101/2023.06.22.544776 (2023).
Van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Gutteridge, A., Bartlett, G. J. & Thornton, J. M. Using a neural network and spatial clustering to predict the location of active sites in enzymes. J. Mol. Biol. 330, 719–734 (2003).
Petrova, N. V. & Wu, C. H. Prediction of catalytic residues using Support Vector Machine with selected protein sequence and structural properties. BMC Bioinformatics 7, 1–12 (2006).
Youn, E. et al. Evaluation of features for catalytic residue prediction in novel folds. Protein Sci. 16, 216–226 (2007).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021).
Huynh, D. Q. Metrics for 3D rotations: Comparison and analysis. J. Math. Imaging Vis. 35, 155–164 (2009).
Ingraham, J., et al. Generative models for graph-based protein design. Advances in Neural Information Processing Systems (2019).
Song, Y., et al. Fast and accurate protein intrinsic disorder prediction by using a pretrained language model. Brief. Bioinform. 24, https://doi.org/10.1093/bib/bbad173 (2023).
Cornish-Bowden, A. Current IUBMB recommendations on enzyme nomenclature and kinetics. Perspect. Sci. 1, 74–87 (2014).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J Mach Learn Res. 21, 5485–5551 (2020).
Suzek, B. E. et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288 (2007).
Yuan, Q. et al. Fast and accurate protein function prediction from sequence through pretrained language model and homology-based label diffusion. Brief. Bioinform. 24, bbad117 (2023).
Song, Y., et al. Accurately predicting enzyme functions through geometric graph learning on ESMFold-predicted structures. Zenodo https://doi.org/10.5281/zenodo.13375275 (2024).
Acknowledgements
This study has been supported by the National Key R&D Program of China (2022YFF1203100) and the National Natural Science Foundation of China (T2394502).
Author information
Authors and Affiliations
Contributions
Y.S. and Y.Y. conceived and supervised the project. Y.S. and Q.Y. contributed to implementing the GraphEC algorithm. Y.S. and Y.Y. wrote the manuscript. S.C., Y.Z., H.Z., and Y.Y. participated in the discussion and proofreading.
Corresponding author
Ethics declarations
Competing interests
The authors declare that no competing interests exist.
Peer review
Peer review information
Nature Communications thanks Sabeur Aridhi and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Song, Y., Yuan, Q., Chen, S. et al. Accurately predicting enzyme functions through geometric graph learning on ESMFold-predicted structures. Nat Commun 15, 8180 (2024). https://doi.org/10.1038/s41467-024-52533-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52533-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.