Abstract
In addition to the ubiquitous loss of the VHL gene in clear cell renal cell carcinoma (ccRCC), co-deletions of chromatin-regulating genes are common drivers of tumorigenesis, suggesting potential vulnerability to epigenetic manipulation. A library of chemical probes targeting a spectrum of epigenetic regulators is screened using a panel of ccRCC models. MS023, a type I protein arginine methyltransferase (PRMT) inhibitor, is identified as an antitumorigenic agent. Individual knockdowns indicate PRMT1 as the specific critical dependency for cancer growth. Further analyses demonstrate impairments to cell cycle and DNA damage repair pathways upon MS023 treatment or PRMT1 knockdown. PRMT1-specific proteomics reveals an interactome rich in RNA binding proteins and further investigation indicates significant widespread disruptions in mRNA metabolism with both MS023 treatment and PRMT1 knockdown, resulting in R-loop accumulation and DNA damage over time. Our data supports PRMT1 as a target in ccRCC and informs a mechanism-based strategy for translational development.
Similar content being viewed by others
Introduction
The clear cell subtype of renal cell carcinoma (ccRCC) is the most common malignancy to arise in the kidney and accounts for the majority of renal cancer-related deaths1. If detected early, localized tumors are surgically resected with the intent to cure2. However, many patients present with disseminated disease at the time of diagnosis3, and even with surgery disease recurrence is common4. The prognosis for metastatic ccRCC is poor. Five-year survival rates hover near 12%5 and, thanks to an insidious resistance to both radiation and cytotoxic chemotherapeutic agents, treatment options remain limited. It has been known for some time that biallelic inactivation of the von Hippel Lindau gene (VHL) gene occurs in most cases of ccRCC, leading to complete loss of pVHL and a constitutive activation of the cell’s hypoxia response6. Although this appears to be a necessary and near universal driver event, evidence suggests that it alone is insufficient to initiate tumorigenesis7,8,9. Recent, large-scale genomic analyses have revealed additional, frequent loss-of-function mutations in several key genes including PBRM1, SETD2, BAP1, KDM5C, KDM6A, MLL2, ARID1A and ARID1B10,11. There exists a surprising and common mechanistic theme to these additional putative tumor suppressor genes: their protein products are all involved in epigenomic regulation.
Given the frequency of gene mutations impacting epigenetic regulatory proteins, and considering recent evidence from genetically engineered mouse models (GEMMs) that substantiate the importance of epigenetic regulators like PBRM1 and BAP1 in ccRCC tumor establishment12,13, we sought to identify epigenetic vulnerabilities that may be exploited to develop new therapies. To accomplish this, we performed an in vitro proliferative screen across a panel of patient-derived ccRCC models14. We used a library of validated chemical probes15,16 that selectively target a spectrum of epigenetic regulatory proteins. The results of our screen identified some previously characterized ccRCC targets, including the enhancer of zeste homolog 2 (EZH2), as well as a newly identified regulator of ccRCC growth: protein arginine methyltransferases (PRMTs).
The methylation of arginine residues on histones and non-histone proteins is a prevalent post-translational modification (PTM) and an important regulator of multiple cellular processes17. PRMTs are the only known family of enzymes responsible for catalyzing the transfer of methyl groups from the S-adenosylmethionine (SAM) cofactor to the terminal nitrogen atoms of the guanidino group of arginine residues18. All nine identified PRMTs catalyze the transfer of one methyl group to produce mono-methylarginine (MMA), and are further classified according to the final methylarginine species they generate: type I members transfer an additional group to the same guanidino nitrogen-producing asymmetric dimethylarginine (aDMA), while type II members transfer an additional group to the other guanidino nitrogen to make symmetric dimethylarginine (sDMA), and the sole type III enzyme, PRMT7, only catalyzes the formation of MMA18. Although arginine methylation is a relatively understudied PTM, emerging evidence has linked the activity of PRMTs to a number of cellular mechanisms important for the development and growth of cancer including epigenetic-mediated gene expression, RNA metabolism, DNA damage response, stem cell function, and the immune response19. Accordingly, the PRMT family has garnered significant attention as a potential therapeutic target and clinical evaluation of type I PRMT inhibitors and PRMT5 selective inhibitors are underway20,21.
In this study, we identify inhibition of type I PRMTs as a vulnerability in ccRCC and using orthogonal genetic approaches, we validate PRMT1 as the specific type I enzyme mediating growth arrest. PRMT1 is the canonical member of the type I family responsible for the majority of all aDMA species produced in the cell18. As such, we employ transcriptomic and proteomic approaches to investigate its role as a mediator of ccRCC growth and survival. Our data suggests that type I PRMT inhibition leads to a pronounced downregulation of the cell cycle, compromised DNA damage repair (DDR) pathways and an accumulation of double-strand breaks (DSBs). Consistent with other reports, proteomic analysis confirms PRMT1’s central role as a regulator of proteins involved in RNA metabolism18,22,23 including targets connected to mRNA splicing and other mRNA metabolic activities. Using cell line and patient-derived xenografts (PDXs), we further validate the inhibition of type I PRMTs and PRMT1 specifically as a viable in vivo strategy for attenuating ccRCC tumor growth. Together, our data argues for the potential translational benefit of PRMT1 inhibition as a clinical therapeutic strategy in ccRCC.
Results
In vitro chemical probe screen identifies Type I PRMTs as mediators of ccRCC cell proliferation
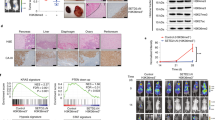
Our lab has previously developed an efficient methodology for establishing patient-derived cell line models of ccRCC from primary tumor tissues14. To ensure faithful representation of the conserved evolutionary subtypes seen clinically9, a panel of seven patient-derived models and one commercially available and widely used ccRCC cell line (786-0) were selected to facilitate the chemical probe screen (Supplementary Fig. 1a). These models underwent targeted sequencing to confirm the presence of relevant evolutionary subtype epigenetic driver mutations including VHL, PBRM1, SETD2, and BAP1. Using a cell permeable, far-red DNA fluorescent dye (DRAQ5TM) and the LI-COR imaging system, growth of these models was interrogated after seven days exposure to a collection of 36 epigenetic chemical probes from the Structural Genomics Consortium (SGC: https://www.thesgc.org/chemical-probes)15,16. This library of small molecule ‘epiprobes’ selectively targets key epigenetic regulatory proteins, including several targets with compounds currently in clinical development (Supplementary Table 1). The results of our screen (Fig. 1a) identified a total of 10 compounds that significantly reduce cell proliferation across the aggregate of all cell line models tested by a minimum of 50% (Supplementary Fig. 1b). Included in this list was UNC1999, an inhibitor of the Polycomb group H3K27me3 lysine methyltransferase EZH2, which has been previously characterized as a target in ccRCC24,25,26. Additionally, GSKJ4, an inhibitor of the lysine demethylases that act on methylated H3K27 (KDM6A and KDM6B) also registered as a hit in our screen, underscoring the importance of the H3K27me3 epigenetic mark in maintaining ccRCC growth. The BET inhibitor JQ1 was also identified as a strong hit; however, early clinical trials have shown that achieving a therapeutic window where an anti-tumor effect can be achieved without undue adverse events due to on-target effects may be challenging with this class of agents27,28. Intriguingly, MS023, an inhibitor of all type I PRMTs, significantly repressed cell proliferation while the more specific type I inhibitors, MS049 (targeting PRMT4 (CARM1) & PRMT6), and SGC707 (targeting PRMT3) did not. The type II PRMT inhibitor GSK591 (targeting PRMT5) also demonstrated significant growth inhibitory effects across our disease models. Follow up dose–response experiments comparing the inhibition of type I PRMTs by MS023 to the activity of a chemically similar but inactive compound, MS094, demonstrated high specificity and potency for type I PRMT inhibition (Fig. 1b and Supplementary Fig. 2a–f). MS023 IC50 values ranged from 0.4 µM to 6 µM across our cell line panel. A second type I PRMT inhibitor, GSK3368715 was subsequently evaluated to corroborate our results with MS023 and similar inhibition profiles were noted (Supplementary Fig. 2g, h).
a Heat map showing the average cell proliferation values in the presence of the indicated epigenetic chemical probe after seven days exposure in eight ccRCC cell lines (data shown as mean of n = 3 technical replicates). b Dose–response curves and MS023 IC50 values across a number of ccRCC models (red). MS094 is the negative control probe for MS023 (blue). Data are presented as the mean ± SD calculated from 3 technical replicates for each cell line. Data presented at day 5 for 786-0, Day 7 for RCC243, day 8 for RCC407, day 12 for RCC22 and day 14 for RCC323 (determined based on the time at which control-treated cells reached confluence). c Western blot analysis of asymmetric dimethylarginine (aDMA) changes in 786-0 and RCC243 cells after MS023 treatment using the ASYM25 antibody for indicated period of time. d Western blot analysis of H4R3me2a changes in acid histone extractions of 786-0 and RCC243 following MS023 treatment for indicated period of time. H4 serves as the gel loading control. Source data are provided as a Source Data file.
To validate MS023’s on-target activity, we treated cell lines RCC243 and 786-0 for a period of 7 days with 5 µM MS023 and using an aDMA-specific antibody (ASYM25), we observed a global downregulation of aDMA species over time (Fig. 1c). Additionally, since MS023 shows high potency against PRMT1 activity29, the type I enzyme solely responsible for the histone mark H4R3me2a30, we also used this post translational modification (PTM) as a proxy for PRMT1 inhibition. MS023 treatment followed by histone extraction and immunoblotting with an H4R3me2a-specific antibody resulted in a noticeable depletion of the PTM in both RCC243 and 786-0 (Fig. 1d).
While type I PRMT inhibition has recently been attracting attention as a potential therapeutic tool in cancer treatment21,31, to our knowledge only one study to date has suggested a role for type I PRMTs in ccRCC32, and no in-depth mechanistic investigations have been reported for these targets in kidney cancer. As such, we selected MS023 as our lead compound and sought to further characterize type I PRMT inhibition in ccRCC.
The PRMT1 enzyme is the critical dependency among type I PRMTs in ccRCC
MS023 is known to have specific activity against all type I PRMT enzymes including PRMT1, PRMT3, PRMT4 (CARM1), PRMT6 and PRMT829. However, no decrease in cell proliferation was noted in our screen for the compounds MS049 and SGC707 that specifically target PRMT4 (CARM1)/PRMT6 and PRMT8, respectively. Transcription levels for each type I enzyme were compared across our previously sequenced cell line models14 and PRMT1 was found to have the highest relative expression (Fig. 2a). Conversely, transcripts for PRMT8, a known membrane bound, neuronal specific protein33, were very low to undetectable. To confirm the functional importance of each type I PRMT enzyme for proliferation and viability, we employed a CRISPR-Cas9, GFP-drop out methodology (adapted from Lin et al.34; Fig. 2b).
a Type I PRMT mRNA expression in ccRCC patient-derived cell lines (each dot represents an individual cell line) shown as fpkm, fragments per kilobase of transcript per million mapped reads. b Schematic representation of the CRISPR-Cas9 competition assay used to determine the functional importance of each type I PRMT enzyme for proliferation and viability in ccRCC cells. c Mean fold-change values (± SEM, n = 3 technical replicates) in the percentage of GFP+ cells, relative to the percentage of GFP+ cells at passage 0 in cell line RCC243 for RPA1 sgRNAs (positive control), ROSA26 gRNAs (negative control) and the indicated type I PRMT sgRNAs (targeting PRMT1, PRMT3, PRMT4/CARM1 and PRMT6). d Mean fold-change values (± SEM, n = 3 technical replicates) in the percentage of GFP+ cells, relative to the percentage of GFP+ cells at passage 0 in cell lines RCC364 and 786-0 for PRMT1 and control sgRNAs. Source data are provided as a Source Data file. Figure (b) Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Briefly, cell line RCC243 was engineered to constitutively express a functional, humanized Cas9 nuclease (RCC243Cas9, Supplementary Fig. 3a). The presence of functional Cas9 was verified by transduction with a construct expressing both GFP and a GFP-targeting gRNA and observation of decreased GFP expression only in Cas9-expressing cells, indicating functional CRISPR editing in these cells (Supplementary Fig. 3b, c). RCC243Cas9 cells were then transduced with GFP-expressing guide RNA (gRNA) vectors targeting individual type I PRMTs at a low multiplicity of infection. This created a mixed population of GFP + /gRNA+ and GFP-/gRNA- cells. Four gRNAs were selected from the Broad Institute’s ‘Brunello Library’35,36 for each of PRMT1, PRMT3, PRMT4 (CARM1) and PRMT6 (Supplementary Table 2). PRMT8 was excluded due to its negligible expression levels in our ccRCC cell lines. Four additional guides were included that target each of a negative control (the human ROSA26 locus – a genetic safe harbor37) and a positive control (RPA3 - an essential replication protein). Following gRNA transduction and for four subsequent passages (cells passaged at a 1:4 ratio every 4 days), the GFP+ population was monitored via analytical flow cytometry and the ratio of GFP+ to GFP- cells was calculated. As expected, cells with gRNAs targeting the human ROSA26 locus persisted in culture while cells with gRNAs targeting RPA3 were depleted by magnitudes of 2.4 to 81.7-fold (Fig. 2c). Conversely, over the course of four passages, the fold depletion for cells harboring gRNAs that target PRMT3, PRMT4 (CARM1) and PRMT6 were negligible and consistent with the negative control guides. However, RCC243Cas9 cells transduced with gRNAs targeting PRMT1 were noticeably depleted in culture with dropout rates ranging from 7.7 to 81.9-fold. These results demonstrate that repeated mutations in PRMT1 induced by Cas9 nuclease led to decreased cell fitness and a resulting drop-out phenotype from the overall population, while mutations in other type I PRMTs were tolerated and did not impact proliferation or viability.
To extend these results, we engineered and functionally validated two additional cell lines to constitutively express Cas9 nuclease (RCC364Cas9 and 786-0Cas9, Supplementary Fig. 3) and repeated the experiment with gRNAs directed against PRMT1 and the relevant negative and positive controls (Fig. 2d). As with RCC243Cas9, gRNA directed mutagenesis of PRMT1 resulted in reduced cell fitness and GFP + /GFP- fold-decreases comparable to those of the positive control protein RPA3 in both cell lines. We thus conclude that PRMT1 is the critical dependency among type I PRMTs in ccRCC.
Knockdown of PRMT1 in ccRCC phenocopies MS023 treatment and overexpression of two major PRMT1 isoforms results in drug-resistant phenotypes
To supplement our CRISPR-Cas9 results, we also engineered two ccRCC cell line models to express doxycycline (dox)-inducible short hairpin RNAs (shRNAs) targeting either PRMT1 (PRMT1-shRNA) or a ‘non-targeting’ control directed against luciferase (NT-shRNA). Upon addition of dox, we observed strong depletion of PRMT1 at day 3 and day 6 in both RCC243PRMT1-shRNA and 786-0PRMT1-shRNA cells (Fig. 3a). As with MS023 treatment, we also noted a marked decrease in the levels of general aDMA species in PRMT1-depleted cells relative to NT-shRNA controls, no-dox treated controls and parental non-transduced cell lines (Fig. 3a). To assess the impact of PRMT1-specific depletion on the growth of each cell line, proliferation was measured over a period of 10 days using the Incucyte® Live-Cell Analysis System. PRMT1 depletion significantly inhibited in vitro cell growth relative to NT-shRNA and non-induced controls (Fig. 3b).
a Western blot analysis of PRMT1 expression (top) and asymmetric dimethylarginine (aDMA) changes using the ASYM25 antibody (bottom) in 786-0 and RCC 243 cells. Cell lines were engineered to express doxycycline (Dox)-inducible PRMT1 targeting or non-targeting (NT) shRNAs and treated with or without 1.0 μg/mL Dox for 3 and 6 days. b Cell line growth curves (confluence measured in Incucyte® Live-Cell Analysis System) of 786-0 and RCC243 cells expressing PRMT1 targeting or NT shRNAs, with or without Dox. Data are presented as the mean of n = 3 technical replicates ± SEM and p-values are calculated by 2-way ANOVA with repeated measures and Sidak’s multiple comparisons test. c Western blot analysis of RCC243 cell line engineered to overexpress PRMT1 isoforms PRMT1v1 (ENST00000391851.8) and PRMT1v2 (ENST00000454376.7). d Cell line growth curve of RCC243 cells engineered to overexpress PRMT1 isoforms. Data are presented as the mean of n = 3 technical replicates ± SEM. e Cell line growth curves of RCC243 cells engineered to overexpress PRMT1 isoforms in the presence of 2.5 μM, 5 μM and 10 μM MS023. Data are presented as the mean of n = 3 technical replicates ± SEM. Source data are provided as a Source Data file.
To further validate ccRCC’s dependence on PRMT1, we engineered patient-derived cell line RCC243 to constitutively overexpress two predominant isoforms of the PRMT1 enzyme: PRMT1v1 (ENST00000391851.8) and PRMT1v2 (ENST00000454376.7). After verifying overexpression via immunoblotting (Fig. 3c) and confirming that said over expressions do not impact the growth dynamics of transduced cell lines relative to parental counterparts (Fig. 3d), we exposed each cell line to increasing doses of MS023. Overexpression of PRMT1v1 and, to a lesser extent, PRMT1v2 restored the aggressive proliferative phenotype seen in parental lines even in the presence of high MS023 doses up to 10 µM (Fig. 3e). The differences in rescue effect between the isoforms are likely due to their characteristic N-terminal sequences that are known to affect both substrate specificity and methyltransferase activity38. Despite this difference, the complete rescue of proliferation at 5 µM MS023 by overexpression of both isoforms further supports PRMT1 as the specific dependency responsible for growth arrest. Taken together, these data indicate that PRMT1 is a targetable therapeutic vulnerability in ccRCC.
MS023 treatment results in downregulation of cell cycle and DNA damage repair (DDR) pathways
To better understand the observed cellular response to both MS023 treatment and the genetic knockdown of PRMT1, our next step was to profile transcriptomic changes via RNA-seq analysis in cell lines RCC243 and 786-0. Following three days of treatment with 5 µM MS023, each cell line was analyzed in duplicate relative to controls and a differential gene expression analysis (DGEA) was performed on the aggregate of both cell line replicates to look for common differentially expressed genes (Fig. 4a). Similarly, RCC243PRMT1-shRNA and 786-0PRMT1-shRNA were induced via dox exposure for four days and analyzed in triplicate relative to non-induced controls (Fig. 4a). Following confirmation of PRMT1 knockdown via Western blotting (Supplementary Fig. 4), DGEA was performed on the aggregate of the induced vs non-induced PRMT1-targeting cell lines to assess resulting transcriptomic changes (Fig. 4a). Intriguingly, in the drug-treated condition, more genes were significantly upregulated (FDR ≤ 0.01, log2(FC) ≥ 1) than down regulated (FDR ≤ 0.01, log2(FC) ≤ −1) − 1005 vs 664 respectively. However, only 9 significant Gene Ontology (GO) biological process pathway overrepresentations (fold change of at least 2.0, FDR ≤ 0.05 and filtered to the most specific subclass) were detected in the upregulated list (Supplementary Fig. 5). Additionally, the majority of genes in this upregulated list (629/1005) did not map to any known GO biological process pathways and consist primarily of microRNAs (miRNAs), long non-coding RNAs (lncRNAs), pseudogenes and novel genes/transcripts. (see Supplementary Data 1a for gene lists). Conversely, for the 664 genes found to be significantly down regulated by day 3 of MS023 treatment (FDR ≤ 0.01, log2(FC) ≤ −1) a total of 20 GO biological process pathways filtered to the most specific subclass were detected with an overrepresentation of at least 2 fold and a FDR of ≤ 0.05 (Supplementary Fig. 5). A more muted effect was noted in the PRMT1-knockdown cell lines where 255 significantly upregulated genes (FDR ≤ 0.01, log2(FC) ≥ 1) and 205 significantly down-regulated genes (FDR ≤ 0.01, log2(FC) ≤ −1) were identified after 4-days of dox induction (Fig. 4a, Supplementary Data 1b). No statistically overrepresented pathways were detected in either of these lists (Supplementary Fig. 6), but a less stringent analysis of all significantly downregulated genes (FDR ≤ 0.01, log2(FC) < 0) revealed overrepresentation of 49 GO biological process pathways, while a similar analysis of all significantly upregulated genes FDR ≤ 0.01, log2(FC) > 0 failed to detect any pathway overrepresentations (Supplementary Fig. 6).
a Volcano plots of log2fold-change for significantly downregulated (red, left) or upregulated (red, right) genes following 3 days of 5 μM MS023 treatment and in cell lines expressing PRMT1 targeting shRNAs treated with or without 1.0 μg/mL Dox for 4 days. Specific mitotic and DNA damage genes of interest explicitly labeled in plot. b Venn diagram highlighting the 165 common significant (FDR ≤ 0.01) downregulated (log2(FC) ≤ 0) genes across MS023 treated and PRMT1 knockdown conditions. c Overrepresentation analysis for gene ontology (GO) biological processes on 165 common significantly down regulated genes across MS023 treatment and PRMT1 knockdown conditions as described in (b). 151/165 genes mapped. Analysis conducted using Fisher’s exact test and PANTHER tool; number of genes in down regulated list per GO biological process listed above each respective bar. GO terms filtered to most specific subclass. d Normalized gene counts (DESeq2’s median of ratios normalization) for mitotic checkpoint protein BUB1B and centromere proteins CENPA and CENPI in RCC243 and 786-0 cells following 5μM MS023 treatment for 3 days (left, n = 2 technical replicates per cell line), and for RCC243 and 786-0 cells expressing PRMT1 targeting shRNAs treated with or without 1.0 μg/mL Dox for 4 days (right, n = 3 technical replicates per cell line). e Normalized gene counts (DESeq2’s median of ratios normalization) for DNA damage proteins FANCM, BRCA2, RAD51AP1 and FANCD2 in RCC243 and 786-0 cells following 5μM MS023 treatment for 3 days (left, n = 2 technical replicates per cell line), and for RCC243 and 786-0 cells expressing PRMT1 targeting shRNAs treated with or without 1.0 μg/mL Dox for 4 days (right, n = 3 technical replicates per cell line).
When comparing these two data sets, we note a total of 165 genes that are significantly down regulated (FDR ≤ 0.01, log2(FC) < 0) across both experimental conditions (Fig. 4b). Pathway overrepresentation analysis on this list of 165 common down regulated genes reveals a statistical overrepresentation of 21 GO biological process pathways filtered to the most specific subclass (Fig. 4c). Prominent among these was a consistent downregulation of pathways related to the cell cycle, particularly genes involved in mitotic progression. Overrepresentations greater than 25-fold were noted for mitotic specific GO annotations including mitotic spindle midzone assembly (GO:0051256), protein localization to kinetochore (GO:0034501) and mitotic spindle assembly checkpoint signaling (GO:0007094). Specific transcripts in these overrepresentations include those encoding the mitotic checkpoint protein BUB1B and a suite of centromere proteins (CENPs) that play a critical role in kinetochore formation, mitotic progression, and chromosome segregation, including CENPA and CENPI (Fig. 4d).
Also notable among the enriched pathways were those related to DNA damage repair (DDR). GO annotations for DNA repair (GO:0006281), DNA duplex unwinding (GO:0032508) and nuclear DNA replication (GO:0033260) were detected at overrepresentation levels greater than 4-fold (Fig. 4c). Included among the proteins in these lists were important players in homologous recombination (HR) including the breast cancer type 2 susceptibility protein (BRCA2) and the RAD51-associated protein (RAD51AP1). Additionally, the Fanconi anemia group M and D2 proteins (FANCM, and FANCD2), key players in the regulation of DNA interstrand cross-link (ICL) repair, were significantly downregulated (Fig. 4e).
Type I PRMT inhibition results in cell cycle arrest, decreased expression of DDR proteins, impaired DDR functions and an accumulation of DSBs
To further assess the apparent transcriptomic downregulation of cell cycle-related pathways, immunoblotting for specific mitotic proteins was performed after treatment with various doses of MS023 for 3 days. Additionally, we assessed the relative cell population actively undergoing division in a time course experiment using an anti-phospho-Histone H3 (Ser10) antibody (Anti-H3S10p), a well characterized marker of mitosis39. An MS023 dose-dependent reduction in levels of BUB1B, CENPA and CENPI was detected after three days of treatment (Fig. 5a) and immunoblotting with Anti-H3S10p in RCC243 following 3, 5 and 7 days of 5 µM MS023 exposure confirmed a decline in the relative cell population actively undergoing mitosis relative to controls (Fig. 5b). However, a DNA content analysis of RCC243 cells over a treatment period of 9 days revealed no accumulation in any specific phase of the cell cycle with prolonged exposure to 5 µM MS023 (Fig. 5c). After 5 days of treatment, a population of cells with sub-2N DNA content (i.e. cells with DNA fragmentation) began to appear, while the number of cells in other phases of the cell cycle began to lag control conditions. By day 9, the sub-2N population became dominant with few cells remaining in other cell cycle phases (Fig. 5c). Overall, MS023 treatment appears to induce a cytostatic phenotype that precedes cell death.
a Western blot analysis of mitotic checkpoint protein BUB1B and centromere proteins CENPA and CENPI in RCC243 cells treated with 0 μM, 0.1 μM, 5 μM, and 10 μM of MS023 for 3 days. b Western blot analysis of phospho-histone H3 in RCC243 cells treated with 0.1 μg/mL Nocodazole (beta-tubulin disruptor and mitotic staller), 100 μM etoposide (DNA damaging agent) and 5 μM MS023 for 3, 5 and 7 days. c Flow cytometric histograms showing cell cycle progression of RCC243 cells in response to 5 μM MS023 inhibition over 9 days. Source data are provided as a Source Data file.
Similarly, to assess the impact of MS023 treatment on DDR proteins, we performed specific immunoblotting in RCC243 cells for the down regulated targets identified in our transcriptomic analysis: BRCA2, RAD51AP1, FANCD2 and FANCM (Fig. 6a). We corroborated these changes in RCC243PRMT1-shRNA cells (Fig. 6b). For all targets assessed, a dose-dependent reduction in protein levels was noted with 3 days of MS023 treatment, and a similar reduction in protein levels was noted after 4 days of PRMT1 knockdown induction in RCC243 cells for all targets except RAD51AP1. These results are consistent with the reduced transcript levels noted in Fig. 4e indicating a potential compromise in the integrity of these critical repair pathways.
a Western blots for indicated proteins in RCC243 cells treated with indicated doses of MS023 for 3 days. b Western blots for indicated proteins in RCC243 cells with dox-inducible PRMT1-shRNAs treated with or without Dox for 4 days. c RCC243 cells were treated with indicated doses of MS023, DNA-PK-inhibitor NU7441 or DMSO followed by 10 Gy of irradiation and allowed to recover for the indicated times. Cells were analyzed by neutral comet assay. Average comet tail moments ± SEM of three independent experiments are shown. P-values are calculated by 2-way ANOVA with repeated measures and Sidak’s multiple comparisons tests. Representative images pictured above. d, e Scatter plots of FANCD2 foci in RCC243 cells treated with and without MS023 for 3 days (d) or with dox-inducible PRMT1-shRNAs treated with or without Dox for 4 days (e) followed by Mitomycin C for 24 h. Data are presented as mean foci counts ± SEM. P-values are calculated by one-way ANOVA with Sidak’s multiple comparisons test. Representative images are shown to the right. ns, not significant. n = 103 cells (DMSO), 104 cells (MMC) and 107 cells (MMC + MS023) (d). n = 111 cells (DMSO), 110 cells (MMC) and 101 cells (MMC + dox) (e). f, g Scatter plots of γH2AX foci in RCC243 cells treated with and without MS023 (f) or with dox-inducible PRMT1-shRNAs treated with or without Dox (g) for indicated times. Data are presented as mean foci counts ± SEM. P-values are calculated by two-tailed unpaired t-tests with Welch’s correction. Representative images from Day 5 (f) or Day 6 (g) are shown to the right. ns, not significant. n = 45 vs 44 cells (Day 1 DMSO vs MS023), 60 vs 43 cells (Day 3 DMSO vs MS023), 81 vs 27 cells (Day 5 DMS0 vs MS023) and 147 vs 16 cells (Day 7 DMOS vs MS023) (f). n = 128 vs 132 cells (Day 3 No Dox vs Dox) and 217 vs 132 cells (Day 6 No Dox vs Dox) (g). Source data are provided as a Source Data file.
To investigate the integrity of DNA DSB repair, we analyzed DNA repair efficiency following ionizing radiation (IR) exposure using the comet assay. This assay was performed in RCC243 cells under neutral conditions to specifically quantify the removal of DSBs. Relative to controls, we observed that cells treated with MS023 repaired DNA breaks more slowly, signaling a defect in the cells’ DSB repair mechanisms (Fig. 6c). This deficit was comparable to that seen with the use of a DNA-dependent protein kinase (DNAPK) inhibitor, a well-characterized disruptor of DSB repair. To assess ICL repair, we performed immunofluorescent staining for FANCD2 foci, a hallmark of a functional FA pathway40. RCC243 cells were treated for 3 days with 5 µM MS023 and then exposed to the interstrand cross linking agent mitomycin C (MMC). Consistent with the observed reduction in levels of FA proteins, treatment with MS023 significantly reduced the number of FANCD2 foci following exposure to MMC relative to controls (Fig. 6d). Similarly, induced RCC243PRMT1-shRNA cells exposed to MMC also showed significantly reduced numbers of FANCD2 foci relative to controls (Fig. 6e). Together, this indicates a compromised ability to handle such damage when type I PRMTs and PRMT1 specifically are inhibited.
To further evaluate the potential for a DDR-compromised phenotype in cell line RCC243, we performed immunofluorescent staining for γH2AX foci, a marker of DSBs. A significant increase in the number of γH2AX foci with 5 µM MS023 treatment was noted over time relative to control conditions indicating an accumulation of unresolved DSBs (Fig. 6f). This result was corroborated in MS023-treated 786-0 cells (Supplementary Fig. 7). A similar accumulation of unresolved DSBs was noted after 4 days of induction in RCC243PRMT1-shRNA cells (Fig. 6g). Taken together our data suggests that the type I PRMTs and PRMT1 specifically coordinate the expression of key DDR genes and that inhibition of these enzymes leads to impaired DDR pathways, accumulation of DSBs, cell cycle arrest and eventual cell death.
Bio-ID reveals RNA-binding proteins (RBPs) as the predominant PRMT1 interactors
To elucidate the molecular mechanisms driving PRMT1-mediated inhibition of ccRCC growth, we employed a proximity-labeling (PL) methodology using miniTurbo41, an adaptation of the BioID technique42. The advantage of miniTurbo relative to the legacy BioID system is the relative speed at which the PL reaction takes place41. Although PRMT1 interactome data has previously been published22,23,43,44, these studies have relied on standard affinity purification of PRMT1 interacting partners coupled with mass spectrometry (AP-MS), or analysis of differentially methylated proteins via AP-MS or isotope labeling techniques such as methyl-SILAC. The establishment of differentially methylated states for substrate interactors of PRMT1 has traditionally been achieved through chemical inhibition of type I PRMTs, which lacks specificity for PRMT1, or via genetic depletion of the enzyme, which is prone to off-target effects. Standard AP-MS techniques also yield a number false negatives for low abundance proteins and for interactors that are difficult to solubilize, such as chromatin-associated proteins, nuclear matrix and other insoluble cellular compartments42.
For our PL method, a genetic fusion was created between the PRMT1v1 isoform, a FLAG-tag and an engineered, promiscuous mutant of the Escherichia coli-derived biotin ligase BirA known as miniTurbo. This fusion was genetically cloned into a dox-inducible construct that was transduced via lentiviral delivery into cell lines RCC243 and 786-0 (Supplementary Fig. 8). Following induction and expression of the fusion protein, exogenous biotin was added to the cell culture to initiate the covalent tagging action of the miniTurbo enzyme (Supplementary Fig. 8). Thus, any protein interactors of PRMT1 that are brought within a few nanometers of the fusion receive a biotin tag. Because of the irreversible nature of the biotin labels, the cells can then be lysed under harsher buffer conditions to maximize solubilization. Labeled proteins were purified by high-affinity streptavidin precipitation and identified by mass spectrometry. With this technique, we were able to define the PRMT1 interactome in ccRCC cells in a specific manner under physiological conditions and over a short window of time (90 min with miniTurbo).
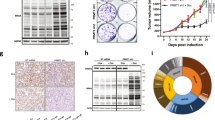
A total of 59 high-confidence PRMT1 interactors (log2(FC) ≥ 1 and ≥ 20 total counts) were identified across both RCC243 and 786-0 (Fig. 7a). The agreement between these data sets was high with a total of 41/59 interactors common to both cell lines. GO biological process and molecular function pathway enrichment analysis of the common list revealed that the majority of identified PRMT1 interactors are proteins involved in RNA metabolism and processing (Fig. 7b). This is consistent with previously published reports and underscores a crucial role for PRMT1 as a regulator of RNA effector proteins involved in splicing, translation, and other RNA regulatory activities22,23,45,46.
a Venn diagram of the 59 total high confidence PRMT1 interactors identified in mini-Turbo Bio-ID experiment (log2(FC) ≥ 2 and ≥20 total counts) in RCC243 and/or 786-0. b Summary diagram of the 41 PRMT1 interactors identified in both cell lines. Protein names were imported into Cytoscape 3.9.1 for visual representation and enrichment analysis was carried out using the STRING Enrichment app using the categories GO biological process, GO molecular function and COMPARTMENTS. Circle size corresponds to fold-change in peptide counts between PRMT1-expressing vs control-miniTurbo cells. The majority of PRMT1 interactors correspond to RNA-binding proteins (blue circles), with the remainder corresponding to other proteins that do not fall into any significantly enriched categories (orange circles). c Quantification of nuclear R-loops via immunostaining with the anti-RNA-DNA hybrid S9.6 monoclonal antibody in indicated cell lines and conditions. Data are presented as mean nuclear intensity ± SEM. P-values are calculated by one-way ANOVA with Sidak’s multiple comparisons test. n = 119 nuclei (15uM CPT, RCC243), n = 148 nuclei (3d DMSO, RCC243), n = 154 (3d 5uM MS023, RCC243), n = 153 nuclei (6d No Dox, RCC243PRMT1-shRNA), n = 149 nuclei (6d Dox, RCC243PRMT1-shRNA). Representative images shown on the right. CPT = camptothecin. d Volcano plots of Alternative Splice Events (ASEs) significantly (FDR < 0.01) downregulated (red, left, ΔPSI < −0.1) or upregulated (red, right, ΔPSI > 0.1) following 3 days of 5 μM MS023 treatment or 4 days of doxycycline induction. Bar graphs of significant ASEs by event type (SE = skipped exons, A5 = alternative 5’ splice site, A3 = alternative 3’ splice site, MXE = mutually exclusive exons, RI = retained introns) are shown below the matching volcano plots. Source data are provided as a Source Data file.
Intriguingly, among the top interactors in both RCC243 and 786-0 were Bcl2-associated transcription factor (Btf or BCLAF1) and the thyroid hormone receptor associated protein 3 (THRAP3). These two proteins have been identified as core members of a DNA damage-induced BRCA1-mRNA splicing complex that promotes the pre-mRNA splicing and subsequent transcript stability of a subset of genes involved in cellular DDR pathways47,48. A subsequent study showed that BCLAF1 and THRAP3 promote RNA splicing and export, respectively, of Fanconi anemia (FA) and HR DDR pathway transcripts independently of DNA damage, including FANCL, FANCD2, BRCA2 and RAD5148. To determine the methylation status of BCLAF1, immunoprecipitation (IP) was done using a BCLAF1 specific antibody followed by aDMA specific immunoblotting. With 5 µM MS023 treatment for 3 days the methylation level for this PRMT1 interactor was markedly decreased, verifying BCLAF1 as a PRMT1 substrate (Supplementary Fig. 9a).
Given the role of BCLAF1 and THRAP3 in the pre-mRNA splicing and nuclear export of genes involved in DDR pathways48, we selected a subset of disrupted DDR genes from our previously generated RNA-seq and Western blot assays and carried out qRT-PCR assays on isolated cytoplasmic and nuclear RNA using primers spanning exon-exon and exon-intron boundaries to quantify levels of pre-spliced and post- spliced RNA in each cellular compartment (Supplementary Fig. 9b). Efficiency of RNA fractionation in these experiments was confirmed by assaying levels of the nuclear RNA, MALAT1 (Supplementary Fig. 9c). qRT-PCR confirmed the overall decrease in mRNA levels, both pre-spliced and post- spliced, of all of the tested genes within 3 days of MS023 treatment. In addition, there is a general trend towards accumulation of pre-spliced transcripts in the nucleus that first decreases but then increases over time, as illustrated by increasing pre-spliced to post-spliced ratios in the nuclear fraction on days 5 and 7 (Supplementary Fig. 9d). The nuclear:cytoplasmic ratio of post-spliced RNA also shows an increase over time for some of the tested genes (Supplementary Fig. S9d). This pattern is most consistent for the FANCD2 gene, in line with our FANCD2 foci data (Fig. 6d, e). This data suggests that MS023 treatment impacts nucleocytoplasmic transport and RNA splicing of these genes, possibly through regulation of the BCLAF/THRAP3 complex. However, this is likely not sufficient to fully explain the observed effects of MS023 on overall transcript levels and the observed DNA damage effects.
Given the enrichment of many additional RNA binding proteins in PRMT1’s interactome, we next sought to explore the downstream effects of MS023 treatment and PRMT1-knockdown on two phenomena relevant to RNA binding proteins: R-loop accumulation and alternative splice events across the transcriptome. R-loops refer to the stable DNA:RNA hybrids generated during transcription. As RNA polymerase progresses along the DNA double helix, newly transcribed RNA hybridizes with the transiently accessible template strand while the displaced non-template DNA forms a loop structure. Although these loops have some biological relevance in the regulation of gene expression, a delicate homeostasis is maintained between formation and resolution of R-loops49. It is now widely accepted that excessive or mis-regulated R-loops are extremely deleterious to the cell and can lead to replication-transcription collisions, DNA damage including double strand breaks, blocks in transcription and genomic instability49,50. Research suggests that nascent RNA is protected from excessive R-loop formation during transcription by RNA binding proteins including splicing machinery components51. For this reason, we sought to quantify R-loops via immunofluorescence using the anti-RNA-DNA hybrid S9.6 monoclonal antibody in our cell line RCC243 to determine if PRMT1 inhibition results in their accumulation. A significant increase in R-loops was detected in RCC243 cells after 3 days of exposure to 5 µM MS023 relative to control conditions (Fig. 7c) and after 6 days of PRMT1 knockdown (Fig. 7c). This suggests that PRMT1-inhibition may compromise the ability of the RNA-binding proteins within the PRMT1 interactome to prevent deleterious R-loop accumulation. Additionally, an in silico analysis of alternative splicing events (ASEs) in RCC243 and 786-0 after 3 days of treatment with 5 µM MS023 revealed massive alterations in the constitutive splicing activities relative to controls. Using the rMATS ‘junction counts”/”exon counts” (JCEC) counting methodology, a total of 2442 significant (FDR ≤ 0.01) ASEs with a delta percent spliced in (PSI) of ≤−0.1 were detected and a total of 2575 significant (FDR ≤ 0.01) ASEs with a delta PSI of ≥ 0.1 were detected after MS023 treatment (Fig. 7d). In both lists of significant ASE’s, the majority of events detected were described as skipped exon events, with fewer but still significant numbers of altered alternative splice site usage events, mutually exclusive exons, and retained introns also occurring. A similar disruption to constitutive splicing, although more muted in magnitude, was noted with the knockdown of PRMT1 in RCC243 and 786-0 (Fig. 7d). Taken together, the accumulation of R-loops and the observed broad splicing changes indicate disruption of the function of RNA-binding proteins as a result of PRMT1 inhibition.
Type I PRMT inhibition and PRMT1 genetic knockdown suppresses tumor growth in vivo
To determine if the anti-proliferative effects of type I PRMT inhibition in ccRCC cell lines translate to antitumor activity in vivo, we evaluated the efficacy of MS023 treatment in ccRCC xenografts. To establish a maximum tolerated dose, intraperitoneal (IP) injections ranging from 40 to 160 mg/kg of MS023 were administered, 5 days per week over a course of 3 weeks and 80 mg/kg was selected as the maximum tolerable dose (≤10% body weight loss over the treatment period). Subsequent pharmacokinetic analysis of drug distribution in serum, kidney, liver, and tumor tissues demonstrated that after three consecutive days of IP injections at 80 mg/kg, residual amounts of the drug could be detected in tumor tissues up to four days later (Supplementary Fig. 10). Thus, to improve tolerability parameters, a dose of 80 mg/kg was selected for efficacy evaluation with an administration schedule of 3 days on/4 days off.
RCC243 and 786-0 cells were implanted subcutaneously, and after tumor establishment, mice were randomized to receive either control (vehicle solution) or 80 mg/kg of MS023 via IP injection in our predetermined dosing schedule. Treatment was continued until controls reached the humane endpoint. Significant inhibition of tumor growth was noted for both RCC243 and 786-0 cell line xenografts relative to controls (Fig. 8a). Global inhibition of the aDMA mark was also noted in xenograft tumor tissue as evaluated by immunoblotting in RCC243 (Fig. 8b) suggesting that tumor growth inhibition coincides with type I PRMT inhibition. To extend these results, we also tested the tumor inhibition potential of MS023 in two patient derived xenograft (PDX) models (generated by direct implantation of patient-derived tumor tissues into immunocompromised mice). Similar to the cell line derived models, growth of RCC63 and RCC243 PDX tumors were significantly reduced by MS023 administration relative to controls (Fig. 8c).
a Tumor growth curves of RCC243 and 786-0 cell line xenograft models treated with MS023 at 80 mg/kg or vehicle control, QD 3 on/4 off. Data are presented as the mean ± SEM and p-values are calculated by 2-way ANOVA with repeated measures and Sidak’s multiple comparisons test. n = 10 mice/group (RCC243) and n = 5 mice/group (786-0). b Western blot analysis of aDMA in RCC243 tumor lysates on day 30. n = 3 mice/group. c Tumor growth curve of RCC63 and RCC243 patient-derived xenograft (PDX) models treated with MS023 at 80 mg/kg or vehicle control, QD 3 on/4 off. Data are presented as the mean ± SEM and p-values are calculated by 2-way ANOVA with repeated measures and Sidak’s multiple comparisons test. n = 5 mice/ group (RCC243 PDX), n = 10 mice/group (RCC63 PDX). d Tumor growth curves of RCC243 and 786-0 cell line xenografts expressing Dox-inducible PRMT1-targeting or NT control shRNAs. n = 5 mice/group (RCC243_NT shRNA (+)Dox, RCC243_PRMT1 shRNA (+)Dox and 786-0_PRMT1 shRNA (+)Dox) and n = 3 mice/group (RCC243_PRMT1 shRNA (-)Dox, 786-0_NT shRNA (+)Dox and 786-0_PRMT1 shRNA (-)Dox). Mice were randomized to either Dox supplemented water (1 mg/mL dox in water) or normal water upon tumor establishment (100–200 mm3). Data are presented as the mean ± SEM and p-values are calculated by 2-way ANOVA with repeated measures and Sidak’s multiple comparisons test. Source data are provided as a Source Data file.
Finally, to further verify that PRMT1 is the specific type I enzyme responsible for tumor growth inhibition, we evaluated the effect of PRMT1 genetic depletion on tumor growth in vivo. RCC243PRMT1-shRNA/RCC243NT-shRNA and 786-0PRMT1-shRNA/786-0NT-shRNA cells were implanted subcutaneously, and tumors were established. Mice were then randomized to receive a normal diet, or one supplemented with dox to induce genetic depletion. Like MS023 treatment, depletion of PRMT1 in mice bearing tumors from both cell lines resulted in significantly inhibited tumor growth relative to NT-shRNA and non-induced controls (Fig. 7d). These results from preclinical models suggest that PRMT1 inhibition holds promise as a potential therapeutic strategy in the treatment of ccRCC.
Discussion
A defining feature of ccRCC tumors is their stubborn resistance to radiation and traditional chemotherapeutics, a hallmark that has left few treatment options beyond surgical resection. Detailed genomic analyses of this cancer have revealed a complex and heterogenous molecular picture, underpinned by two common themes: a near universal dependence on distorted hypoxia signaling and altered chromatin regulation as key drivers of tumorigenesis. In this study, we sought to exploit the latter of these dependencies using a targeted, epigenetic focused chemical screening approach in well characterized patient-derived models of ccRCC. Through this unbiased methodology, we identified the type I PRMT inhibitor MS023 as an attenuator of cell growth. Using orthogonal genetic techniques, we further identified PRMT1 as the specific dependency responsible for growth arrest among the type I enzymes. We show that MS023 administration and PRMT1 inhibition result in widespread loss of aDMA species including the histone mark H4R3me2a. Our transcriptomic data demonstrates pronounced downregulation of key mitotic and DDR proteins that coincide with marked inhibition of the cell cycle, compromised DDR pathways, and an accumulation of DSBs. These observations are consistent with previous reports that also show cell cycle defects and DNA damage resulting from PRMT1 deficiencies. Encouragingly, our preclinical in vivo models also demonstrate significant tumor growth inhibition following MS023 administration and PRMT1 genetic depletion, suggesting this enzyme represents a viable therapeutic target for potential clinical development. The ability of MS023 to impede DDR suggests potential avenues for combination therapies with DNA damage-inducing agents, and warrants future investigation.
To further delineate relevant molecular pathways at play, we employed a proximity-labeling proteomics technique to describe the interacting partners of PRMT1 in ccRCC cells. Consistent with previous reports, the bulk of interactors identified were RNA binding proteins (RBPs) that regulate key RNA metabolic activities including mRNA transcription, splicing, transport, translation, and turnover18. These included BCLAF1 and THRAP3, which are known to directly influence the splicing and transport of important DDR protein transcripts, including key members of the homologous recombination and Fanconi anemia DDR pathways47,48. Many of these same proteins were also downregulated in response to type I PRMT inhibition or PRMT1 knockdown. Our data demonstrates decreased expression and nucleocytoplasmic transport following prolonged exposure to MS023 for the DDR genes analyzed in our study. While we were able to confirm differential asymmetric dimethylation of BCLAF1 in the presence of MS023, the exact influence of this PTM on the ability of BCLAF1 and THRAP3 to act as splice regulators is unknown. Conceivably, PRMT1 inhibition and the resulting loss of aDMA PTMs on BCLAF1 may impact its ability to effectively regulate splicing, resulting in compromised transcript quality and contributing to disruptions of the DDR targets described in this study.
In addition to BCLAF1 and THRAP3, many additional RNA-binding protein interactors were identified in our proximity labeling experiment. Given the role for RBP-arginine methylation in modulating protein-protein interactions, nucleic acid binding and splicing functions more generally18,52, we sought to investigate RBP function in the context of R-loop regulation and alternative splicing events. The significant accumulation of R-loops detected in our cell line models with MS023 treatment and PRMT1-specific knockdown is of particular interest for two main reasons: First, it suggests that PRMT1 inhibition leads to a compromise in the protective function of RBPs, which normally bind and stabilize nascent ssRNA to initiate mRNA maturation, thus preventing R-loop formation by sterically inhibiting the interaction between nascent RNA and the exposed ssDNA template50,51. Second, abnormal R-loops are a widely recognized source of cellular stress and genomic instability51,53, and may account for the accumulation of DSBs noted in this study. Stabilized, co-transcriptional R-loops are believed to disrupt replication fork progression during S-phase of the cell cycle resulting in stalled and collapsed replication forks, double-strand breaks and incomplete DNA synthesis50,53. Intriguingly, several of the RBPs identified as PRMT1 interactors in our study have also been shown to play a role in R-loop resolution (e.g. DDX1, THRAP3, TAF15, RNF20)54,55,56,57, suggesting that PRMT1 inhibition not only induces R-loop formation but may also impair the cells’ ability to resolve them. In addition, research has shown that R-loop homeostasis is intimately linked with DDR pathways and specifically the Fanconi Anemia proteins FANCM and FANCD258,59, as well as BRCA259,60 and RAD51AP161. The link between downregulation of these proteins in PRMT1-inhibited ccRCC models and the accumulation of unresolved R-loops warrants further investigation. Finally, the significant perturbations in alternative splicing events highlighted from our in silico analysis corroborates a breakdown in RBP-mediated splicing functionality and indicates widespread splicing changes relative to control conditions.
Although a collapse in RBP function and splicing is one plausible explanation for the DNA damaged/growth arrested phenotypes seen with MS023 treatment, it should be noted that PRMT1 has been reported to directly methylate an array of specific DNA damage proteins, including BRCA1, hnRNPK, hnRNPUL1, MRE11 and 53BP162,63,64,65. These proteins are key players in DSB repair and their PRMT1-mediated aDMA PTMs are known to affect their abilities in this regard. MS023 treatment could plausibly lead to unresolved DNA damage by impairing the function of these proteins leading to cell cycle arrest and eventual cell death. However, we note that key proteins from the Fanconi anemia pathway, a related yet mechanistically separate DDR pathway specialized for the removal of DNA interstrand crosslinks, are also critically downregulated by MS023 treatment. The functional compromises we described in this pathway are not readily explained by changes in aDMA PTMs for MRE11, 53BP1, hnRNPK, hnRNPUL1 or BRCA1.
A relative black box in our mechanistic exploration of PRMT1 in ccRCC revolves around the influence of its histone substrate, H4R3me2a on the overall cellular transcriptome and resulting proteome. Despite recent advancements in high throughput sequencing technologies, the epigenomic profiling of arginine histone substrates has remained an outstanding challenge owing in part to a lack of high quality, validated antibodies against arginine-methylated histones66. We know that H4R3me2a is associated with transcriptional activation, but to our knowledge no successful genome wide mapping of this PRMT1 mediated mark has yet been accomplished. Since our data suggests that MS023 treatment diminishes this histone mark in our disease models, we must consider the possibility that H4R3me2a-mediated transcriptional changes also contribute to the observed growth arrested phenotypes.
Finally, we note that type I PRMT inhibition resulted in varying levels of growth inhibition across our ccRCC cell line panel, a result that is consistent with previously reported type I PRMT efficacy studies in other cancer types67. This spectrum of sensitivity likely results from the heterogenous genomic and epigenomic character of our cell lines. Previous reports have linked PRMT1 sensitivity to factors like MTAP loss, SRSF mutations and gene expression signatures enriched for interferon response and antiviral signaling21,23,67. While these genetic factors are uncommon in ccRCC, the conserved evolutionary driver subtypes seen clinically most likely play a role in the cells’ sensitivity to PRMT1 inhibition. For example, BAP1 driven ccRCCs are known to have inherently compromised BRCA-mediated DDR pathways and may exhibit enhanced sensitivity to further DDR perturbations, while SETD2 driver mutations already display splicing defects and may have enhanced responses11. Further investigation of PRMT1 sensitivity in ccRCC subtypes is warranted and would help facilitate a potential molecular-based stratification for drug responses.
To summarize, our findings reveal the central role of PRMT1 as a regulator of RNA metabolism, including splicing, and add to the growing body of knowledge linking splicing disruptions to the DNA damage response. We propose a model in which RNA metabolic processes, such as transcription, splicing, and R-loop resolution, are dependent upon asymmetric dimethylation of RBPs by PRMT1; inhibition of PRMT1 leads to accumulation of R-loops, and the decreased expression of essential DNA repair proteins such as FANCM, FANCD2 and BRCA2 impedes the cells’ ability to repair the accumulated damage, leading to growth arrest followed by cell death (Fig. 9). We demonstrate that PRMT1 plays a critical role in the maintenance and growth of ccRCC and may represent a key therapeutic vulnerability. The potential also exists for this target to synergize with current DNA-damaging therapies including radiation and traditional chemotherapeutics. In addition, the inability of MS023 treated cells to repair DSBs suggests the possibility of synergy with other DNA repair targeted drugs such as PARP inhibitors. Finally, the induction of DNA damage may lead to increased neoantigen load and/or stimulate the cGAS/STING pathway68, leading to increased sensitivity to immune checkpoint blockade. Further studies are needed to investigate these possibilities.
PRMT1 inhibition leads to loss of aDMA post-translational modifications on RBPs involved in RNA processing, resulting in the accumulation of R-loops. Some RBPs in the PRMT1 interactome are reported to be involved in R-loop resolution (e.g. DDX1, RNF20, THRAP3, TAF15), thus in addition to promoting R-loop formation, PRMT1 inhibition may also prevent R-loop resolution. In addition, DDR genes such as FANCM, FANCD2 and BRCA2 are decreased at both the transcriptional and protein level, thus DSBs that result from the presence of unresolved R-loops are not repaired. It remains unclear whether this decreased expression is occurring via effects on RBPs, or other mechanisms such as transcriptional regulation via the H4R3me2a histone mark. FANCM loss also plays a role in R-loop resolution, thus contributing to the phenotype at multiple levels. Unrepaired DSBs lead to growth arrest and ultimately cell death. RBP: RNA-binding protein; aDMA: asymmetric dimethylation; Me: methyl group. Figure 9 created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Methods
Cell lines and cell culture
The human kidney cancer cell line 786-0 was obtained from American Type Culture Collection (ATCC). Cell lines RCC22, RCC162, RCC222, RCC243, RCC323, RCC364 and RCC407 were generated as previously described in Lobo et al.14. The continued use of these patient-derived lines is approved under UHN Research Ethics Board approval, protocol #15-9559. All cell lines in this study were routinely cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Wisent #319-105-CL) supplemented with 10% FBS (Thermo #12483020), and 1% penicillin/streptomycin (Wisent #450-201-EL) in a humidified incubator at 37 oC with 5% CO2, 2%O2. Cells were plated in culture flasks coated with rat tail collagen type I (5ug/cm2; Thermo #A1048301) and passaged no more than 20 times. Cell cultures were monitored for mycoplasma infection (Universal Mycoplasma Detection Kit, Cedarlane #30-1012 K) and cell culture identity verification was done by short tandem repeat profiling (GenePrint® 10 System, Promega #B9510). FASTA files underwent genome alignment using Burrows-Wheeler Aligner (BWA) followed by BAM file conversion. SNVs and indels were called using Mutect2 in the GATK (version 3.5) software package and true calls were annotated with Oncotator.
Targeted DNA sequencing
DNA was extracted using a QiaAMP DNA mini kit (Qiagen). Libraries were constructed using a KAPA Hyper Prep kit using custom unique molecular identifier BIOO Scientific NextFlex adapters to barcode the samples, as instructed by the manufacturer. Target capture for the genes of interest was carried out using custom xGen® Lockdown® probes purchased from IDT. Following end repair, A-tailing and adapter ligation, Agencourt AMPure XP beads were used for library clean up and ligated fragments were amplified 4 cycles using 0.5 μM custom unique xGen Predesigned Hybrid Panel index primers (IDT). Post amplification cleanup was performed using Agencourt AMPure XP beads. Final library quality control was performed using a Bioanalyzer 2100. Sequencing was done on an Illumina HiSeq2000 using a 100-cycle paired-end protocol.
Epigenetic chemical probe screen
All ‘epiprobe’ compounds were purchased from Cayman Chemical, Millipore-Sigma or MedChemExpress. The detailed resources for each compound are listed in Supplementary Table 1 the additional file. Chemical purity was validated at the SGC at more than 99%. Cell lines were plated at the density of 2500 cells/well (except 786-0 which was plated at 150 cells/well) in 96-well plates and allowed to adhere. Epiprobes were dissolved in DMSO and added to achieve a final concentration as indicated in Fig. 1a. Each plate contained three replicates per compound and three replicates of a 0.1% DMSO control condition. After seven days exposure to each probe, the cell permeable, far-red DNA fluorescent dye DRAQ5TM was added to the wells and fluorescence was quantified using the LI-COR imaging system. Data was normalized to the DMSO control wells and the average log2(probe/DMSO) readings are presented in the heatmap.
Western blot analysis
Cells were lysed and proteins were extracted in ice cold RIPA buffer (150 mM NaCl, 1.0%NP40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) supplemented with HaltTM Protease Inhibitor Cocktail (Thermo # 78425). Protein concentrations were determined using the PierceTM BCA Protein Assay Kit according to the manufacturer’s instructions. Protein concentrations were normalized to 15 μg each, and samples were denatured in Laemmli sample buffer (BioRad #1610737) supplemented with 5% β-mercaptoethanol and boiled at 95 °C for 5 min. Proteins were separated on 4–15% Mini-PROTEAN® TGXTM Precast Protein Gels (BioRad, #4568084) using the Mini-PROTEAN® Electrophoresis System (BioRad). Separated proteins were then transferred to Immobilon®-P PVDF Membranes (Milipore Sigma #ISEQ00010) using the BioRad Transblot SD Semi-dry Transfer Cell as per manufacturer’s recommendations.
Membranes were blocked in 5% milk Tris-buffered saline with 0.1% Tween® 20 Detergent (TBST) and incubated overnight with primary antibody. Antibodies used: asymmetric dimethylarginine ASYM25 (Sigma, #09-814, 1:1000), Tubulin (Santa Cruz #sc-5286, 1:1000), Histone H4 (Abcam, #ab174628, 1:1000), Histone H4R3me2a (Active Motif, #39705, 1:1000), PRMT1 (Thermo, PA5-17299, 1:1000), GAPDH (Santa Cruz, #sc-69778, 1:1000), BUB1B (Cell Signalling Technology (CST), #4116S, 1:1000), CENP-I (CST, #49426S, 1:1000), CENP-A (CST, #2186 T, 1:1000), Phospho-Histone H3 (Ser10) (Millipore, #06-570, 1:1000), Actin (Abcam #ab7817, 1:1000), BRCA2 (CST, #107415, 1:1000), RAD51AP1 (Thermo/Proteintech, #11255-1-AP, 1:1000), FANCM (Bethyl Labs, #A302-637A, 1:1000), FANCD2 (CST, #16323, 1:1000), BCLAF1 (Thermo/Bethyl Labs, #A300-608A, 1:1000), CAS9 (Abcam, #ab191468, 1:1000), FLAG (Sigma, F3165, 1:1000), Streptavidin-HRP (BD Pharmingen, #554066, 1:1000).
Membranes were washed 5x in TBST and probed with species specific HRP-conjugated secondary antibody (anti-rabbit-HRP (CST, #7074S, 1:2500), or anti-mouse-HRP (CST, #7076S, 1:1000)) for 1 h. Membranes were then rewashed 5x in TBST and developed with the chemiluminescence AmershamTM ECLTM start Western blotting detection reagent and visualized on Hyblot CL film (Cedar Lane, #DV-E3012). Uncropped scans of Western blots are provided in the attached Source Data file.
Western Blotting of tumor tissue
50 mg fragments of snap frozen tissue were homogenized using a Qia-TissueRuptor at medium speed for 30 s or until completely homogenous, on ice in RIPA buffer containing protease and phosphatase inhibitors. Samples were sonicated twice for 10 s at 50% amplitude. Western blots were carried out as described above.
Plasmids, transfections, transductions and engineered cell line development
All lentiviral preparations were made via co-transfection of target and packaging plasmids in HEK293T cells using the XtremeGENE HP DNA transfection reagent (Sigma, #6366236001). Packaging plasmids: pMD.G and CMVdR8.74 (gift from the Naldini lab). Viral supernatant was harvested at 48 and 72 h, filtered through a 0.45 μm syringe and frozen at −80 °C or used fresh. Cell transductions were performed using a 1:4 dilution of lentiviral supernatant to media for 24 h and culture media was subsequently changed.
ccRCC cell lines were transduced with lentivirus lentiCas9-Blast (Addgene Plasmid #52962) and selected with 4 μg/mL blasticidin for 10 days. Monoclonal lines were subsequently selected following limiting dilutions. Functional testing (Supplementary Fig. 3C) was performed after transduction with lentivirus pXPR_011 (Supplementary Fig. 3B, Addgene Plasmid #59702) and 7 days in culture.
Guide RNA sequences targeting PRMT1, PRMT3, PRMT4/CARM1, PRMT6, RPA3 and the human ROSA26 locus were selected from the Broad Insitutute’s ‘Brunello Library’ (https://portals.broadinstitute.org/gppx/crispick/public, Supplementary Table 2). Respective oligonucleotides were ordered from IDT and then cloned into the sgRNA delivery LRG plasmid (Addgene Plasmid #65656) using a BsmBI digestion.
Dox-inducible shRNA constructs targeting PRMT1 (shRNA target sequence: GTGTTCCAGTATCTCTGATTA) and Luciferase (non-targeting control, shRNA target sequence: CAAATCACAGAATCGTCGTAT) were obtained as a gift from the Structural Genomics Consortium. Protein coding sequences for PRMT1v1 (ENST00000391851.8) and PRMT1v2 (ENST00000454376.7) were ordered from GenScript (https://www.genscript.com) and PCR- cloned into pHIV-Luc-ZsGreen (Addgene plasmid #39196) using and EcoRI and XbaI digestion.
Mini-Turbo plasmid (pSTV6-N-miniTurbo-BirA) was obtained as a gift from Dr. Brian Raught’s lab. Open reading frame for PRMT1v1 (ENST00000391851.8) was cloned into the Mini-Turbo plasmid via gateway cloning using pDONR223 (gift from Dr. Brian Raught’s lab) and BP/LR combined cloning reagents (Gateway BP clonase II enzyme mix, Thermo #11789020 and LR Clonase II Enzyme Mix, Thermo, # 11791019).
All cloned plasmids were amplified in OneShotTM Stbl3TM E. Coli (ThermoFisher, #C737303) and prepared using the PureLinkTM Quick Plasmid Miniprep Kit (Invitrogen, #K210010). Plasmid identification was verified via Sanger sequencing.
CRISPR/Cas9 competition assay and GFP drop out screening
Cas9 expressing cell lines were plated at ~60% confluency in 6 well plates and transduced with their respective cloned-LRG plasmid. Fresh media was replaced on cells 48 h later (day 0), the culture was passaged at 1:4 and a baseline GFP+ percentage was measured using a BD LSR2 analytical flow cytometer. Cells were subsequently passaged every 4 days at a ratio of 1:4 and the percentage of GFP+ cells was measured at each split. Dropout values represent the fold decrease in GFP+ cells at each passage, relative to the GFP+ percentage on day 0.
RNA Isolation,Sequencing, Pathway and Splicing Analysis
RNA isolation was performed using the Qiashredder kit (Qiagen, #79654) and the RNeasy mini kit (Qiagen, #74104). RNA quality was assessed on an RNA 6000 Pico chip (Agilent Technologies) using the Agilent Bioanalyzer to determine sample RNA integrity number (RIN) and quantified by the Qubit RNA HS assay kit (Life Technologies). For MS023 vs DMSO treated cells, all samples had RIN values > 9.7. RNA was sent to Genome Quebec at McGill University for sequencing. NEB stranded mRNA libraries were constructed and paired-end (100 bp) next generation sequencing was performed on a Illumina NovaSeq 6000 S4 system. For PRMT1 knock down lines, all samples had RIN values = 10.0 and libraries were prepared with the Illumina mRNA kit. Paired-end (150 bp) next generation sequencing was performed at the Princess Margaret Genomics Centre on the NovaSeq XTM platform.
Raw reads were processed using TrimGalore v0.6.6 to remove adaptor sequences (via cutadapt v3.0) and to assess read quality (via fastqc v0.11.5). Reads then were mapped to the Genome Reference Consortium Human Build 38 patch release 12 (GRCh38.p12) reference genome using STAR v2.7.9a. Read counts per gene were subsequently obtained using the htseq-count command in the HTSeq v0.11.0 package alongside Gencode’s human genome annotation release 44. Differential gene expression analysis based on the negative binomial distribution was performed on treated vs untreated cells using the DESeq2 R package v1.36.0. Genes were considered differentially expressed if they were found with a minimum |log2(fold-change)|≥ 1 and FDR adjusted p-value ≤ 0.01.
Gene Ontology (GO) overrepresentation analysis was performed on differentially expressed lists using the PANTHER17.0 analysis tool69, which uses Fisher’s exact test and references the GO Ontology database for biological processes (https://doi.org/10.5281/zenodo.6399963, released 2022-03-22)70,71. Pathways were considered significant if they were found overrepresented at a minimum level of 2-fold above expectation and with a FDR adjusted p-value ≤ 0.05.
Splicing analysis was done on aligned data using rMATS v 4.3.0 to identify differential alternative splicing events between conditions as indicated in the figures72. Analysis was conducted using rMATS modified version of the generalized linear mixed model to detect differential alternative splicing events from RNA-Seq data. The junction count exon count (JCEC) methodology was used and events were filtered based on absolute Includsion Level Difference (ΔPSI) > 0.1 between indicated conditions with FDR < 0.05.
Cell cycle/DNA content analysis
RCC243 cells were treated with 5 μM MS023 for the indicated time periods and then cells were trypsinized and stained using the Propidium Iodide Flow Cytometry Kit (Abcam, #ab139418, Abcam) based on the manufacturer’s instructions. A BD LSR2 analytical flow cytometer was used to acquire fluorescence data. All the flow cytometric data were analyzed using FlowJo software version 10.7.1 to quantify DNA content of individual cells and assess cell cycle status.
Immunofluorescence and Foci Staining
Cells were grown on chamber slides and treated as indicated with MS023, 0.1% DMSO, 1μg/mL dox or no-dox. Following treatment, cells were washed three times in PBS and fixed in 4% paraformaldehyde for 15 min on ice. Cells were again washed 3 times with ice cold PBS and then permeabilized with two different blocking buffers: for Anti-phospho-Histone H2A.X (Ser139)—5% BSA, 10% FBS, 0.25% Triton X-100, 1% fish skin gelatin in PBS; and for FANCD2—5% Normal Goat Serum, 0.3% Triton X-100, 1% BSA in PBS. Primary antibody was added in blocking buffer and incubated at 4 °C overnight. Primary antibodies used: Anti-phospho-Histone H2A.X (Ser139) (Millipore, #05-636, 1:1000) and FANCD2 (CST, #16323, 1:100). Cells were washed 3 times in 0.25% Triton X-100 in PBS (PBS-T) and secondary antibodies: Goat anti-Rabbit IgG H + L -Alexa FluorTM 594 (Invitrogen #A11037, 1:400) or Goat anti-Mouse IgG (H + L) -Alexa FluorTM 488 (Invitrogen #A10667, 1:1000) was added in blocking buffer for 1 h at room temperature. Cells were again washed 3 times in 0.25% PBS-T and 1:1000 Hoechst was added in PBS for 1 min. A final wash in PBS-T was performed and coverslips were subsequently mounted using Mowiol with PPD antifade. Images were collected on the Zeiss Axioimager Z1 wide-field fluorescence microscope. Image processing was performed using ZEN3.6 (blue edition) software and foci quantification was performed using Image J.
R-loop staining
Cells were grown on coverslips in 24 well TC-treated plates and treated as indicated with MS023, 0.1% DMSO, 1μg/mL dox or no-dox. Following treatment, cells were washed 3 times for 5 min each on ice with PBS and fixed with methanol kept at −20 °C for 20 min. Cells were again washed 3 times on ice with PBS and then permeabilized with 0.2% Triton X-100 in PBS for 5 mins. Cells were then blocked in 5% non-fat dried milk in PBS for 1 h at RT. Coverslips were placed in a humidity chamber and primary antibody was added in blocking buffer and incubated at 4 °C overnight. Primary antibody used: S9.6 (Fischer Scientific, #MABE1095MI, 1:100). All subsequent steps were performed at RT and in the dark. The following day, cells were washed once with PBS and incubated with secondary antibody in blocking buffer for 2 h. Secondary antibody used: Goat anti-Mouse IgG (H + L)-Alexa FluorTM 488 (Invitrogen #A10667, 1:500). Cells were washed once with PBS then stained with DAPI (Fisher Scientific, # 15757405, 1:1000) in PBS for 10 mins. Cells were washed 3 times with PBS for 5 mins each with rocking then mounted onto glass slides using Mowiol with PPD antifade. Images were collected on the Zeiss Axioimager Z1 wide field fluorescence microscope. Nuclear intensity quantification was performed using Image J and image processing was performed using ZEN 3.6 (blue edition) software.
BioID sample processing
Cells were transduced with Flag-miniTurbo-PRMT1v1 lentiviral supernatant for 48 h before media was changed and puromycin selection applied. Cells were cultured for 1 week in the presence of puromycin and expression of mini-Turbo fusion protein was induced by the addition of doxycyline (1 μg/mL) for 24 h in surviving cells and confirmed via Western blotting (Supplementary Fig. 5). A total of 250 mg of Biotin (BioBasic ref# BB0078) was dissolved in 2.04 ml of NH4OH 28-30% (SIGMA ref# 221228) to produce a 500 mM solution. The solution was neutralized by gently adding 18 ml of 1 N HCl to get a final stock solution at 50 mM. Biotin was then added to mini-Turbo expressing cells for 90 min to allow biotin tagging to proceed. Cells were harvested and frozen at −80 °C. Pellets were lysed in modified RIPA (1% Triton X-100, 0.1% SDS, 50 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 0.5% sodium deoxycholate) supplemented with protease inhibitors and benzonase. Samples were incubated 1 h at 4 °C and sonicated prior to centrifugation at 15,000 × g for 30 min. Cleared lysates were incubated for 3 h at 4 °C and rinsed with 50 mM NH4HCO3 for 6 cycles. Trypsin (1 μg/sample; diluted in 50 mM NH4HCO3) was added to the beads and samples were incubated for 16 h at 37 °C. Additional trypsin (0.5 μg/sample) was added and samples were incubated for 2 h at 37 °C. The supernatant was collected, beads were rinsed with 50 mM NH4HCO3 eluates were pooled prior to the peptides being lyophilized in a vacuum centrifuge.
Liquid chromatography-mass spectrometry (LC-MS)
Dried peptides were reconstituted in 0.1% HCOOH and injected on a loading column (C18 Acclaim PepMapTM 468 100, 75 μM × 2 cm, 3μm, 100Å) prior to separation on an analytical column (C18 Acclaim PepMapTM 469 RSLC, 75 μm × 50 cm, 2μm, 100Å) by HPLC over a reversed-phase gradient (120-min gradient, 5–30% CH3CN in 0.1% HCOOH) at 225nL/min on an EASY-nLC1200 pump in-line with a Q-Exactive HF (Thermo Scientific) mass spectrometer operated in positive ESI mode. An MS1 ion scan was performed at 60,000 FWHM followed by MS/MS scans (HCD, 15,000 fwhm) of up to 20 parent ions (minimum activation of 1000). Fragmented ions were added to a dynamic exclusion list (10 ppm) for 5 s. For peptide and protein identification, raw files (.raw) were converted to.mzML format with Proteowizard (v3.0.19311) and searched using X!Tandem (v2013.06.15.1) and Comet (v2014.02.rev.2) search engines against the human proteome RefSeqV104 database (36,113 entries). Parent ion mass tolerance was set at 15 ppm and an MS/MS fragment ion mass tolerance at 0.4 Da. Up to two missed cleavages was allowed. No fixed modification was set. Deamidation (NQ), oxidation (M), acetylation (protein N-term) were set as variable modifications. Search results were further processed using the trans-proteomic pipeline (TPP v4.7) using iProphet. Proteins were identified with an iProphet cut-off of 0.9 and at least two unique peptides. Putative proximity interactors were identified using Significance Analysis of INTeractome (SAINT)73 comparing Flag- miniTurbo-only samples to Flag-miniTurbo-PRMT1 samples using a Bayesian false discovery rate (BFDR) cut-off of ≤0.01 (1%). For visualization representation, the protein names of interactors identified in both RCC243 and 786-0 cells (n = 41) were imported into Cytoscape 3.9.1 and enrichment analysis was carried out using the STRING Enrichment app using the categories GO biological process, GO molecular function and COMPARTMENTS.
Immunoprecipitation
RCC243 cells were seeded in 10 cm plates and allowed to reach ~60% confluence. Cells were treated with 5 μM MS023 or 0.1% DMSO for three days. Cells were collected and lysed in 1 mL ice cold RIPA buffer supplemented with HaltTM Protease Inhibitor Cocktail (Thermo # 78425) and sonicated 2 times for 10 s @ 50% output followed by a 10 min incubation on ice. After cell lysis and sonication, tubes were centrifuged, and supernatant was transferred into a new Eppendorf tube.
Protein A Sepharose beads (Abcam, #ab193256) were washed with 1 mL of RIPA buffer and added to each sample tube to facilitate pre-clearing of the samples. Beads and samples were shaken for 1 h at 4 °C on a nutator and the supernatant was transferred to a new Eppendorf tube. BCLAF1 antibody (Bethyl A300-608A) or mouse IgG was added and incubated at 4 °C overnight with continuous shaking on a nutator. New protein A beads were washed in RIPA and added to each sample the next day and incubated for 3 h at 4 °C on a nutator. Samples were then centrifuged for 1 min at 1000 × g and the beads collected and washed 3 times in RIPA. Beads were resuspended in 50 μL SDS loading buffer, boiled at 95 °C and run on a Western blot (as described in section 2.5.4) probed with BCLAF1 antibody (Bethyl A300-608A, 1:1000) and Asymmetric Di-Methyl Arginine ASYM25 (Sigma, #09-814, 1:1000).
Cytoplasmic/nuclear RNA fragmentation
Cells were typsinized and pelleted by centrifugation at 300 × g for 5 min. The plasma membrane was then lysed with 175 µl of cooled RLN buffer (50 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% (v/v) Nonidet P-40 (1.06 g/ml) for 5 min on ice. Lysate was then centrifuged at 4 °C for 2 min at 300 × g. The supernatant, containing the cytoplasmic RNA, was transferred to a new tube and the remaining pellet contained the nuclear fraction. RNA was then extracted from both the cytoplasmic and nuclear fractions using the Qiagen RNeasy mini kit. The purity of cytoplasmic and nuclear fractions were assessed for the expression of the nuclear enriched MALAT1 RNA via quantitative reverse transcriptase (qRT)-PCR.
qRT-PCR
Synthesis of cDNA was performed on 1 µg of DNAse-free RNA using the SuperScript VILO cDNA Synthesis Kit (Invitrogen, Thermo Fisher). Power SYBR Green PCR Master Mix (Invitrogen, Thermo Fisher) was used to perform qPCR on 10 ng of cDNA for each sample (in triplicate) using primers specific to each transcript. Samples were loaded into a Bio-Rad CFX96 real-time PCR detection system following the manufacturer’s protocol. Post-spliced transcripts (exon-exon spanning primers) and pre-spliced transcripts (exon-intron spanning primers) were normalized to ACTB expression levels within the same samples. The expression of each gene of interest was normalized to ACTB and then the relative amounts of expression were calculated by the delta delta Ct method. The primers used were as follows:
MALAT1 Forward: 5′- GACGGAGGTTGAGATGAAGC-3′, MALAT1 Reverse: 5′-ATTCGGGGCTCTGTAGTCCT- 3′, ACTB Forward: 5′-AGACCTGTACGCCAACACAG-3′, ACTB Reverse: 5′- GGAGCAATGATCTTGATCTTCA-3′ FANCD2_Exon36_Forward: 5′- CCCAGAACTGATCAACTCTCCT-3′, FANCD2_Exon37_Reverse: 5′- CCATCATCACACGGAAGAAA-3′, FANCD2_Intron37_Reverse: 5′- ACAGGTGTGTGCCACCGTG-3′, BRCA2_Exon18_Forward: 5′- CCTGATGCCTGTACACCTCTT-3′, BRCA2_Exon19_Reverse: 5′- GCAGGCCGAGTCACTGTTAGC-3′, BRCA2_Intron18_Reverse: 5′- TACATCTAAGAAATTGAGCATCCT-3′, Rad51_Exon6_Forward: 5′- TGAGGGTACCTTTAGGCCAGA-3′, Rad51_Exon7_Reverse: 5′- CACTGCCAGAGAGACCATACC-3′, Rad51_Intron6_Reverse: 5′- AGAGACATTCTTCGGCCAAACT-3′ and FANCA_Exon2_Forward: 5′- TCCTGAAAGGGCACAGAAATTA-3′, FANCA_Exon2/3_Reverse: 5′- GGGCCTTCTACCTCAAGCAAA-3′, FANCA_Intron2_Reverse: 5′-CCAGCTTCCTCTTACCTCAAG-3’, RAD51 AP1_Exon2_Forward: 5′- CCAGTCAATTACTCACAGTTTGAC-3′, RAD51 AP1_Exon3_Reverse: 5′-TAACTCCTTTGGTGCTGTTCT-3′, RAD51 AP1_Intron2_Reverse: 5′-TCCGAGGAAA TGAGTTTCCAA-3′.
In vivo tumor studies and drug treatments
ccRCC samples were obtained from the University Health Network (UHN) and the Cooperative Health Tissue Network from patients providing written consent under UHN Research Ethics Board approval, protocol #09-0828-T. PDX models were generated by implanting patient tissue under the renal capsule of 6-8 week old male NOD/SCID/IL2Rγ−/− (NSG) mice and allowed to expand until humane endpoint was reached. All animal studies were approved by the University Health Network Animal Care Committee under protocol #4896. Once tumors were established in the renal capsule, tissue was harvested and finely minced using a scalpel, then incubated in 1× Collagenase/Hyaluronidase and 125 Units/mL DNase (Stem Cell Technologies) with frequent pipetting at 37 °C for 2 h. Red blood cells were lysed with ammonium-chloride/potassium (ACK) lysing buffer (Gibco) and cell clumps were filtered using 70 μm nylon mesh. Dissociated cells were stained with trypan blue, viable cells were counted for re-implantation into mice. 1-5 million viable cells were injected in 100 μL of phosphate-buffered saline (PBS):Matrigel (1:1) in the subcutaneous left flank of 6–8-week-old NSG mice. Tumors were allowed to grow to 100–200 mm3 then randomized into each treatment group of 5 mice per arm. MS023 was administered 3 days sequentially at 80 mg/kg intraperitoneally, with 4 days off for a period of three weeks or until vehicle control reached humane endpoint. Tumor measurements were collected twice a week for the duration of the experiment. Cell lines were injected as described above. For inducible cell line studies, dox treatment was initiated once tumors reached between 100-200 mm3 and was delivered into the drinking water at 1 mg/ml. Water was changed twice a week until endpoint. Tumors were excised at endpoint and tumor fragments were snap frozen in liquid nitrogen and stored at −80 °C for further analysis.
Comet assay
Cells were plated in 10 cm culture dishes at ~60% confluence and incubated for 24 h with 5 μM MS023, 2 μM NU7441 (DNA-PK inhibitor, gift from Dr. Shane Harding) or 0.1% DMSO. Cells were then exposed to 10 Gy ionizing radiation (IR) using a Cs137 irradiator and were collected at indicated time points post (IR) exposure. Single-cell electrophoresis was carried out in the CometAssay® Electrophoresis System II (Trevigen, #4250-050-ES) in accordance with the manufacturer’s protocol for neutral conditions (double-stranded break quantification). Microscopy was carried out on the Zeiss Axioimager Z1 wide-field fluorescence microscope and comet tail moments were calculated in ImageJ with the OpenComet plugin v1.3.
Statistics and reproducibility
Statistical analyses and representations were performed using GraphPad Prism 9.3.1 or R/R-Studio. Complete information regarding entity and exact n values, test statistics and displayed error bars are reported in the figure legends along with descriptions of statistical tests used to determine the significance between group comparisons. Experiments were independently repeated a minimum of two times with consistent results unless otherwise stated in figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw and processed RNAseq data generated in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE229357. The proximity labeling proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the MassIVE repository with the dataset identifier PXD051931. All other data supporting the findings of this study are provided in the accompanying Supplementary Information and Source data files. Source data are provided with this paper.
References
Hsieh, J. J. et al. Renal cell carcinoma. Nat. Rev. Dis. Prim. 3, 17009 (2017).
Motzer, R. J. et al. Kidney Cancer, Version 3.2015. J. Natl Compr. Cancer Netw. 13, 151–159 (2015).
Gupta, K., Miller, J. D., Li, J. Z., Russell, M. W. & Charbonneau, C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat. Rev. 34, 193–205 (2008).
Speed, J. M., Trinh, Q.-D. D., Choueiri, T. K. & Sun, M. Recurrence in localized renal cell carcinoma: a systematic review of contemporary data. Curr. Urol. Rep. 18, 15 (2017).
Siegel, R. L., Miller, K. D., Jemal, A. & Society, A. C. Cancer statistics, 2020. Ca. Cancer J. Clin. 70, 7–30 (2020).
Gossage, L., Eisen, T. & Maher, E. R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer 15, 55–64 (2015).
Young, A. P. et al. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat. Cell Biol. 10, 361–369 (2008).
Rankin, E. B., Tomaszewski, J. E. & Haase, V. H. Renal cyst development in mice with conditional inactivation of the von hippel-lindau tumor suppressor. Cancer Res 66, 2576 LP–2572583 (2006).
Turajlic, S. et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 173, 595–610.e11 (2018).
de Cubas, A. A. & Rathmell, W. K. Epigenetic modifiers: activities in renal cell carcinoma. Nat. Rev. Urol. 15, 599–614 (2018).
Walton, J. et al. PBRM1, SETD2 and BAP1—the trinity of 3p in clear cell renal cell carcinoma. Nat. Rev. Urol. 20, 96–115 (2023).
Nargund, A. M. et al. The SWI/SNF protein PBRM1 restrains VHL-loss-driven clear cell renal cell carcinoma. Cell Rep. 18, 2893–2906 (2017).
Gu, Y.-F. F. et al. Modeling renal cell carcinoma in mice: Bap1 and Pbrm1 inactivation drive tumor grade. Cancer Discov. 7, 900–917 (2017).
Lobo, N. C. et al. Efficient generation of patient-matched malignant and normal primary cell cultures from clear cell renal cell carcinoma patients: clinically relevant models for research and personalized medicine. BMC Cancer 16, 485 (2016).
Scheer, S. et al. A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat. Commun. 10, 19 (2019).
Wu, Q. et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat. Commun. 10, 1915 (2019).
Blanc, R. S. & Richard, S. Arginine methylation: the coming of age. Mol. Cell 65, 8–24 (2017).
Bedford, M. T. & Clarke, S. G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 (2009).
Jarrold, J. & Davies, C. C. PRMTs and arginine methylation: cancer’s best-kept secret? Trends Mol. Med. 25, 993–1009 (2019).
Li, X., Wang, C., Jiang, H. & Luo, C. A patent review of arginine methyltransferase inhibitors (2010–2018). Expert Opin. Ther. Pat. 29, 97–114 (2019).
Fedoriw, A. et al. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell 36, 100–114.e25 (2019).
Giuliani, V. et al. PRMT1-dependent regulation of RNA metabolism and DNA damage response sustains pancreatic ductal adenocarcinoma. Nat. Commun. 12, 4626 (2021).
Fong, J. Y. et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209.e9 (2019).
Huang, K. et al. A novel EZH2 inhibitor induces synthetic lethality and apoptosis in PBRM1-deficient cancer cells. Cell Cycle 19, 758–771 (2020).
Sun, C. et al. EZH2 Expression is increased in BAP1-mutant renal clear cell carcinoma and is related to poor prognosis. J. Cancer 9, 3787–3796 (2018).
Kim, K. H. et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 21, 1491–1496 (2015).
Lewin, J. et al. Phase Ib trial with birabresib, a small-molecule inhibitor of bromodomain and extraterminal proteins, in patients with selected advanced solid tumors. J. Clin. Oncol. 36, 3007–3014 (2018).
Ameratunga, M. et al. First-in-human Phase 1 open label study of the BET inhibitor ODM-207 in patients with selected solid tumours. Br. J. Cancer 123, 1730–1736 (2020).
Eram, M. S. et al. A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem. Biol. 11, 772–781 (2016).
Yang, Y. & Bedford, M. T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 (2013).
Wu, Q., Schapira, M., Arrowsmith, C. H. & Barsyte-Lovejoy, D. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 20, 509–530 (2021).
Wang, J. et al. PRMT1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Theranostics 11, 5387–5403 (2021).
Lee, J., Sayegh, J., Daniel, J., Clarke, S. & Bedford, M. T. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280, 32890–32896 (2005).
Lin, A., Giuliano, C. J., Sayles, N. M. & Sheltzer, J. M. CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. Elife 6, e24179 (2017).
Sanson, K. R. et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184 (2016).
Irion, S. et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 25, 1477–1482 (2007).
Thiebaut, C., Eve, L., Poulard, C. & Le Romancer, M. Structure, activity, and function of prmt1. Life 11, 1–23 (2021).
Prigent, C. & Dimitrov, S. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 116, 3677–3685 (2003).
Garcia-Higuera, I. et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 (2001).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801 LP–801810 (2012).
Bao, X. et al. CSNK1a1 regulates PRMT1 to maintain the progenitor state in self-renewing somatic tissue. Dev. Cell 43, 227–239.e5 (2017).
Guo, A. et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteom. 13, 372–387 (2014).
Li, W.-J. et al. Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 12, 1946 (2021).
Guccione, E. & Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 20, 642–657 (2019).
Savage, K. I. et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell 54, 445–459 (2014).
Vohhodina, J. et al. The RNA processing factors THRAP3 and BCLAF1 promote the DNA damage response through selective mRNA splicing and nuclear export. Nucleic Acids Res. 45, 12816–12833 (2017).
Brickner, J. R., Garzon, J. L. & Cimprich, K. A. Walking a tightrope: the complex balancing act of R-loops in genome stability. Mol. Cell 82, 2267–2297 (2022).
Chakraborty, P. New insight into the biology of R-loops. Mutat. Res.—Fundam. Mol. Mech. Mutagen. 821, 111711 (2020).
Allison, D. F. & Wang, G. G. R-loops: formation, function, and relevance to cell stress. Cell Stress 3, 38–46 (2019).
Thandapani, P., O’Connor, T. R., Bailey, T. L. & Richard, S. Defining the RGG/RG Motif. Mol. Cell 50, 613–623 (2013).
Chan, Y. A., Hieter, P. & Stirling, P. C. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 30, 245–253 (2014).
Cargill, M., Venkataraman, R. & Lee, S. Dead-box rna helicases and genome stability. Genes (Basel). 12, 1471 (2021).
Kang, H. J. et al. Thrap3 promotes R-loop resolution via interaction with methylated DDX5. Exp. Mol. Med. 53, 1602–1611 (2021).
Britton, S. et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 42, 9047–9062 (2014).
Li, J. et al. The RPA–RNF20–SNF2H cascade promotes proper chromosome segregation and homologous recombination repair. Proc. Natl Acad. Sci. 120, e2303479120 (2023).
García-Rubio, M. L. et al. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 11, 1–17 (2015).
Stirling, P. C. & Hieter, P. Canonical DNA repair pathways influence R-loop-driven genome instability. J. Mol. Biol. 429, 3132–3138 (2017).
Groelly, F. J. et al. Mitotic DNA synthesis is caused by transcription-replication conflicts in BRCA2-deficient cells. Mol. Cell 82, 3382–3397.e7 (2022).
Ouyang, J. et al. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 594, 283–288 (2021).
Guendel, I. et al. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PLoS ONE 5, e11379 (2010).
Gurunathan, G., Yu, Z., Coulombe, Y., Masson, J.-Y. & Richard, S. Arginine methylation of hnRNPUL1 regulates interaction with NBS1 and recruitment to sites of DNA damage. Sci. Rep. 5, 10475 (2015).
Auclair, Y. & Richard, S. The role of arginine methylation in the DNA damage response. DNA Repair (Amst.) 12, 459–465 (2013).
Yang, J.-H. et al. Arginine methylation of hnRNPK negatively modulates apoptosis upon DNA damage through local regulation of phosphorylation. Nucleic Acids Res. 42, 9908–9924 (2014).
Rakow, S., Pullamsetti, S. S., Bauer, U.-M. & Bouchard, C. Assaying epigenome functions of PRMTs and their substrates. Methods 175, 53–65 (2020).
Wu, Q. et al. PRMT inhibition induces a viral mimicry response in triple-negative breast cancer. Nat. Chem. Biol. 18, 821–830 (2022).
Concannon, K., Morris, B. B., Gay, C. M. & Byers, L. A. Combining targeted DNA repair inhibition and immune-oncology approaches for enhanced tumor control. Mol. Cell. https://doi.org/10.1016/j.molcel.2022.12.031 (2023).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019).
The Gene Ontology Consortium, The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Wang, Y. et al. rMATS-turbo: an efficient and flexible computational tool for alternative splicing analysis of large-scale RNA-seq data. Nat. Protoc. 19, 1083–1104 (2024).
Choi, H. et al. SAINT: probabilistic scoring of affinity purification–mass spectrometry data. Nat. Methods 8, 70–73 (2011).
Acknowledgements
The authors thank the patients who graciously donated their samples for the study. We would like to thank Dr. Shane Harding for advice and reagents relating to DNA damage assays. We would also like to thank Dr. Jian Jin and Yudao Shen for the synthesis of MS023 used for in vivo studies. This research is supported by the Cancer Research Society (grant #23145), the Ontario Institute for Cancer Research (IA-016) through funding provided by the Government of Ontario, and the Princess Margaret Cancer Foundation (to LA). JW was supported by an Ontario Graduate Scholarship provided by the Government of Ontario and by the Canadian Urological Association in collaboration with Kidney Cancer Research Network of Canada (KCRNC). The Structural Genomics Consortium is a registered charity (no: 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and US), Pfizer and Takeda.
Author information
Authors and Affiliations
Contributions
J.W., C.H.A., B.R., and L.A. designed research studies. J.W., A.S.N.N., K.A., A.A., J.M., C.K., J.S.G., J.D., and E.C. conducted experiments and acquired data. J.W., A.A., J.S.G. analyzed data. P.P., C.H.A. provided reagents. J.W. and L.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mehdi Mollapour and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Walton, J., Ng, A.S.N., Arevalo, K. et al. PRMT1 inhibition perturbs RNA metabolism and induces DNA damage in clear cell renal cell carcinoma. Nat Commun 15, 8232 (2024). https://doi.org/10.1038/s41467-024-52507-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52507-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.