Abstract
The circadian clock of cyanobacteria, which predicts daily environmental changes, typically includes a standard oscillator consisting of proteins KaiA, KaiB, and KaiC. However, several cyanobacteria have diverse Kai protein homologs of unclear function. In particular, Synechocystis sp. PCC 6803 harbours, in addition to a canonical kaiABC gene cluster (named kaiAB1C1), two further kaiB and kaiC homologs (kaiB2, kaiB3, kaiC2, kaiC3). Here, we identify a chimeric KaiA homolog, named KaiA3, encoded by a gene located upstream of kaiB3. At the N-terminus, KaiA3 is similar to response-regulator receiver domains, whereas its C-terminal domain resembles that of KaiA. Homology analysis shows that a KaiA3-KaiB3-KaiC3 system exists in several cyanobacteria and other bacteria. Using the Synechocystis sp. PCC 6803 homologs, we observe circadian oscillations in KaiC3 phosphorylation in vitro in the presence of KaiA3 and KaiB3. Mutations of kaiA3 affect KaiC3 phosphorylation, leading to growth defects under both mixotrophic and chemoheterotrophic conditions. KaiC1 and KaiC3 exhibit phase-locked free-running phosphorylation rhythms. Deletion of either system (∆kaiAB1C1 or ∆kaiA3B3C3) alters the period of the cellular backscattering rhythm. Furthermore, both oscillators are required to maintain high-amplitude, self-sustained backscatter oscillations with a period of approximately 24 h, indicating their interconnected nature.
Similar content being viewed by others
Introduction
Three genes, kaiA, kaiB, and kaiC, encode the core circadian oscillator in Cyanobacteria1. Over the last few decades, the biochemical interplay between these three proteins has been studied in great detail in Synechococcus elongatus PCC 7942 (hereafter Synechococcus). The KaiC protein forms a homohexamer and has autokinase, autophosphatase, and ATPase activities1,2,3,4. By associating with KaiC, KaiA stimulates the autokinase and ATPase activities of KaiC, and thus, the protein gets phosphorylated5,6,7. Upon phosphorylation of two neighboring residues (Ser431 and Thr432), KaiC undergoes structural rearrangements, exposing a binding site for KaiB8,9,10. After binding, KaiB sequesters KaiA from KaiC, promoting KaiC’s autophosphatase activity, and the protein reverts back to its unphosphorylated state8,9,11. The interplay between KaiA and KaiB is crucial for the KaiC phosphorylation cycle, which confers clock phase and rhythmicity to the cell12,13. For a more detailed review on the KaiABC oscillator and its regulatory network, see Cohen and Golden14, Swan et al.15 and Snijder and Axmann16.
Although most studies on prokaryotic circadian rhythms have focused on the cyanobacterium Synechococcus, the standard KaiABC system is functionally conserved in other cyanobacteria17. In addition to the standard KaiABC system, divergent homologs of KaiB and KaiC have been identified in cyanobacteria, other bacterial species, and archaea18. The structure, mechanism of function, and physiological roles of these homologs are often unclear. A few studies have demonstrated the role of KaiB and KaiC homologs in stress responses in e.g. Legionella pneumophila19 and Pseudomonas species20. Other Kai homologs are also involved in the regulation of diurnal rhythms outside the cyanobacterial lineage. These include e.g. KaiB and KaiC homologs from the phototrophic bacterium Rhodopseudomonas palustris21. Recently, a KaiA-independent hourglass timer was reconstituted using Rhodobacter sphaeroides (Rhodobacter) KaiC and KaiB homologs22. Rhodobacter KaiC exhibits a divergent extended C-terminus that is typically found in proteins belonging to the KaiC2 subgroup. This C-terminal extension mediates hexamer-hexamer interactions, allowing KaiA-independent phosphorylation. Rhodobacter KaiB controls the phosphorylation-dephosphorylation cycle of KaiC depending on the ATP-to-ADP ratio, suggesting that metabolic changes during the day and night cycles drive this KaiBC clock22 as it has been previously shown for Synechococcus KaiABC23.
The cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis) is a facultative heterotrophic cyanobacterium that, unlike Synechococcus, can utilize glucose as an energy and carbon source. In addition to the canonical kaiAB1C1 gene cluster, Synechocystis encodes two further kaiB homologs, kaiB2 and kaiB3, and two kaiC homologs, kaiC2 and kaiC324. For the Synechocystis KaiB3-KaiC3 timing system, Aoki and Onai suggested a function in the fine-tuning of the core oscillator KaiAB1C1 by modulating its amplitude and period25. This idea was supported by Wiegard et al., who investigated the characteristics of the KaiC3 protein and proposed an interplay between the KaiB3-KaiC3 system and the proteins of the standard clock system26. Furthermore, autophosphorylation and ATPase activities of Synechocystis KaiC3 have been verified, suggesting that enzymatic activities might be conserved across the KaiC protein family26,27,28. Recently, Zhao et al.17 used a luminescence gene reporter to study circadian gene expression in the Synechocystis wild type in comparison to mutant strains lacking each of the kai genes. They demonstrated that kaiAB1C1 and kaiB3C3 genes are both important for circadian rhythms in Synechocystis, whereas kaiC2 and kaiB2 deletion mutants still showed rhythmic gene expression, which is in agreement with previous suggestions by Aoki and Onai25. Phenotypic mutant analysis by our group revealed that two systems function in the autotrophy/heterotrophic switch, especially affecting heterotrophic growth. In contrast to the study by Zhao et al.17 that reported a small growth defect in Synechocystis in which kaiC3 has been deleted, our studies with the motile Synechocystis strain (PCC-M29) showed no such effect on growth under light/dark (LD) cycles. However, the mutant strain displayed a growth defect under chemoheterotrophic conditions in the dark compared with the wild type26,30. This impairment was less severe in comparison with the ΔkaiAB1C1-deficient strain, which completely lost its ability to grow in the dark. Notably, the complete deletion of kaiC2 was not possible in the wild-type strain used in our laboratory. Although Zhao et al.17 clearly showed that deletion of the kaiC3 and kaiB3 genes affects the circadian rhythm of Synechocystis, it remains unclear whether the KaiB3-KaiC3 system can function as an oscillator. How can such a minimal system maintain circadian rhythmicity without KaiA? Prochlorococcus MED4, which lacks a kaiA gene in its entire genome, lacks free-running circadian rhythmicity31,32. Moreover, Synechocystis KaiC3 lacks the extended C-terminus, which is crucial for the oscillation of the Rhodobacter KaiBC hourglass timer22.
In Synechococcus, the KaiA protein functions as a homodimer and harbors two distinct domains connected by a linker sequence33,34,35. The N-terminal domain is similar to bacterial response regulators, but lacks the aspartate residue crucial for phosphorylation; hence, it is designated as a pseudoreceiver domain (PsR domain)33. This domain was shown to bind the oxidized form of quinones and is therefore able to sense the onset of darkness and forward signals to the C-terminal domain33,36. The C-terminus has a four-helix bundle secondary structure and is highly conserved within Cyanobacteria. The domain harbors the KaiA dimer interface and the KaiC binding site, and is necessary to stimulate the autophosphorylation activity of KaiC33,35. Mutations in kaiA resulting in altered periodicity were mapped throughout both domains, indicating their importance in rhythmicity35,37.
To date, the regulatory network of the KaiB3-KaiC3 system in Synechocystis remains unclear, as it does not interact with KaiA and does not utilize the SasA-RpaA output pathway, suggesting alternative yet unidentified components for KaiB3-KaiC3-based signal transduction38. In a large-scale protein-protein interaction screen, a potential interaction partner of KaiC3 was identified39. This protein, Sll0485, was categorized as a NarL-type response regulator and could be a potential element in the KaiB3-KaiC3 signaling pathway40.
In this study, we computationally characterized Sll0485 and detected strong co-occurrences of the KaiB3-KaiC3 system with Sll0485 in the genomic context of Cyanobacteria and other bacteria. Because of this and the fact that its C-terminal domain shares similarities with KaiA homologs, we designated this protein KaiA3. Based on the in vitro analyses, we propose that Synechocystis KaiA3 forms an oscillator with KaiB3 and KaiC3. KaiA3 driven phosphorylation rhythms of KaiC3 are phase-locked with KaiC1 phosphorylation in the cell. Both systems, KaiA1B1C1 and KaiA3B3C3, appeared to control circadian rhythmicity and the phototrophy-to-heterotrophy switch in Synechocystis.
Results
KaiA3 is a chimeric protein harboring a NarL-type response regulator domain at the N-terminus and a conserved KaiA-like motif at the C-terminus
The canonical clock genes, kaiABC and kaiA1B1C1, form a cluster in Synechococcus and Synechocystis, respectively. In contrast, the kaiB3 and kaiC3 genes of Synechocystis are localized in different regions of the chromosome (Fig. S1a). Here, the kaiB3 gene forms a transcriptional unit with the upstream open reading frame sll0485 (kaiA3). KaiA3 has been annotated as a NarL-type response regulator40. Using reciprocal BLAST analyses, we detected orthologs of KaiA3 in 15 cyanobacterial species (16.5% of cyanobacterial species contained at least one KaiC homolog), mainly belonging to the order Chroococcales41 (Supplementary Data S1), and in five bacterial genera outside Cyanobacteria, namely, Roseiflexus, Chloroflexus, Chloroherpeton, Rhodospirillum, and Bradyrhizobium.
Owing to the genetic context, we aligned the cyanobacterial KaiA3 orthologs with both, a NarL-type response regulator (Fig. S2), and cyanobacterial KaiA proteins (Fig. 1a). The canonical NarL protein consists of an N-terminal receiver domain, a linker, and a C-terminal DNA-binding domain with a helix-turn-helix motif40,42. The N-terminus of the KaiA3 orthologs is conserved and indeed shows limited homology to NarL-type response regulators (Fig. S2). However, the similarities to the NarL protein decreased in the C-terminus (Fig. S2). Concurrently, the conservation between KaiA3 and the KaiA protein family increased (Fig. 1a). The conserved residues in the C-terminus correspond to structurally important features of the Synechococcus KaiA protein, such as α-helical secondary structures, the KaiA dimer interface, or residues critical for the KaiA-KaiC interaction33,34 (Fig. 1a). Additionally, the lack of conservation in the N-terminus compared to that observed in known KaiA orthologs is consistent with the results of Dvornyk and Mei, who proposed that different N-terminal domains exist for KaiA homologs for functional diversification43. Because of its similarity to KaiA and synteny with the kaiB3 gene, we named the hypothetical Sll0485 protein KaiA3. Furthermore, to facilitate the distinction of KaiA homologs, we will use the name KaiA1 for the Synechocystis KaiA core clock homolog Slr0756.
a Multiple sequence alignment and maximum likelihood-inferred phylogenetic reconstruction of KaiA3 and selected KaiA orthologs. The sequences were aligned with Mafft (L-INS-i default parameters, Jalview), trimmed to position 168 of the C-terminus of Synechococcus KaiA and are represented in the Clustalx color code with conservation visibility set to 25%. Marks above the alignment refer to Synechococcus KaiA as a reference. Light green bars and dots indicate residues critical for KaiC interaction, light pink bars and dots represent residues important for dimerization, and light gray blocks outline residues forming α-helices as secondary structures. Aligned sequences were used to infer a maximum likelihood protein tree. The scale bar indicates one substitution per position. Bootstrap values (n = 1000) are displayed on the branches. Bootstrap values of less than 50 are not shown. b Synteny analysis of kaiA1B1C1 compared to kaiA3, kaiB3, and kaiC3 genes for selected bacterial species. Analysis was performed with the online tool SyntTax, a prokaryotic synteny and taxonomy explorer (https://archaea.i2bc.paris-saclay.fr/synttax/; 2020-06-08). Default settings were used for analysis (best match, 10% norm. Blast). c Co-occurrence of KaiA3 with circadian clock proteins in cyanobacteria using pairwise right-sided Fisher’s exact test. Network of significant co-occurring circadian clock factors from Schmelling et al.27, including KaiA3 in Cyanobacteria. The line color corresponds to the level of significance resulting from pairwise Fisher’s exact test. Missing links were those with p-values higher than 0.01. The node size is proportional to the degree of that node.
A gene tree resulting from multiple sequence alignments (Fig. 1a) distinctly separated KaiA3 from the canonical KaiA orthologs. To further investigate the evolutionary relationship of KaiA3, multiple sequence alignments of the C-termini of orthologs of KaiA3, KaiA, and Slr1783 (Rre1) as a reference for NarL orthologs in Cyanobacteria44 were used to construct a phylogenetic tree (Fig. S3). Here, KaiA3 orthologs form a distinct clade at the basis of the KaiA orthologs when compared to both orthologous groups of Slr1783 (Rre1)/NarL (E. coli, UniProtKB - P0AF28) and KaiA simultaneously (Fig. S3). In summary, these findings strengthen the idea that the C-terminus of KaiA3 functions similarly to that of KaiA. We further constructed three-dimensional models of KaiA3 to gain a better understanding of its potential functions. To date, no structure is available for KaiA3, and it is difficult to generate a reliable three-dimensional model covering the full-length KaiA3 sequence because of the enigmatic structure of the linker region, for which no significant similarities could be detected. However, secondary structure prediction suggested that the N-terminus structurally aligns with NarL (Fig. S4a). Therefore, we modeled the N-terminus (residues 1-140) and the remaining part of the sequence separately (residues 141-299). For the N-terminus, numerous hits for response regulator domains were obtained, with E. coli NarL (PDB 1A04) showing the highest degree of sequence similarity. The 3D-model structures of KaiA3 are highly similar and display the canonical fold of response regulator domains: a central five-stranded parallel β-sheet flanked on both faces by five amphipathic α-helices and a phosphorylatable aspartate residue in the β3-strand (Fig. S4b). This aspartate residue (D65) plays a role in response regulator phosphorylation (Fig. S2, blue stars) and is conserved in all species, except Pleurocapsa and Microcystis. Thus, most KaiA3 homologs, including the Synechocystis protein, harbor a potential phosphorylation site. Furthermore, the structure superimposes well on the PsR domain of KaiA, even though the PsR domain lacks the phosphate-accepting aspartate residue and the α4-helix between the β4- and β5-strands (Fig. S4b). The amino acid sequence between the β4- and β5-strands shows the least conservation between KaiA and KaiA3; however, the level of sequence conservation in this region is generally low for KaiA and its homologs35. In contrast to the N-terminal response regulator domain, the C-terminal domain of KaiA3 revealed a unique fold, which has only been detected in KaiA thus far45, and the N-terminal domain of the phosphoserine phosphatase RsbU from Bacillus subtilis46, namely, a unique four-α-helix bundle constituting the KaiA-like motif (Fig. S4c). In conclusion, we propose that KaiA3 consists of two protein modules: (i) the N-terminal domain, resembling a NarL-type response regulator receiver domain, including its phosphorylation site, and (ii) the C-terminal domain displaying features of a KaiA-like motif. This is particularly intriguing because putative kaiA orthologs outside Cyanobacteria were not identified until recently43.
Conserved synteny and co-occurrence of KaiA3 and the KaiB3-KaiC3 system among prokaryotes
As in Synechocystis, we found the kaiA3 gene upstream of kaiB3 in all the analyzed cyanobacterial genomes. Furthermore, the kaiA3B3 cluster is usually extended by kaiC3, which resembles the structure of the canonical kaiABC gene cluster, with only two exceptions (Synechocystis and Microcystis aeruginosa NIES-843) (Fig. 1b). Interestingly, kaiA3B3C3 synteny was also found in other prokaryotic genomes that harbor orthologs of kaiA3, except for Chloroflexus aggregans DMS 9485 (Fig. 1b). Furthermore, we detected strong significant co-occurrences between KaiA3 and KaiB3 (p < 0.0001) as well as between KaiA3 and KaiC3 (p < 0.0001; Fig. 1c) in organisms encoding KaiC1. The co-occurrence of KaiB3 and KaiC3 has been previously shown27. Thus, KaiA3 forms a distinct set of proteins with KaiB3 and KaiC3, which showed no further significant co-occurrence with other clock components (Fig. 1c27). Altogether, both datasets suggest a functional relationship between KaiA3 and the KaiB3-KaiC3 system.
KaiA3 interacts with and promotes autokinase activity of KaiC3
Using yeast two-hybrid (YTH) experiments, we verified the interaction between the clock proteins KaiC3 and KaiA3 (Fig. 2a, Fig. S5), which is consistent with a previous large-scale protein-protein interaction analysis by Sato et al.39. Although KaiA3 clearly interacted with KaiC3, an interaction with KaiB3, the second element of the KaiB3-KaiC3 clock system, was not detected (Fig. S5b). This is not surprising, as it has been demonstrated that the interaction between the Synechococcus proteins KaiA and KaiB requires the presence of KaiC47. To further characterize the interaction of the proteins in vitro, we heterologously expressed different Kai proteins in E. coli and analyzed complex formation using clear-native PAGE (Fig. 2b and Fig. S6). The His-tagged KaiA3 protein (monomer: 35 kDa) migrated as a single band of approximately 100 kDa in size, indicating the formation of KaiA3 homo-oligomers, at least dimers. Synechococcus KaiA migrated at ~60 kDa, in line with previous results48, confirming the formation of KaiA dimers. The discrepancy in the migration pattern between KaiA3 (His-tagged) and KaiA (GST-tag removed) might be due to differences in their predicted charge (−19.17 for KaiA and −7.94 for KaiA3, respectively, at pH 7.0). Recombinant KaiB3 (monomer: 12 kDa) was shown to form monomers and tetramers after size exclusion chromatography26. KaiB3 displayed three distinct bands in the native gels (Fig. 2b). The two lower bands most likely represent the monomeric and tetrameric forms, whereas the uppermost band (~67 kDa) could be an impurity in the protein preparation. Recombinant KaiC3 was produced with an N-terminal Strep-tag26. Strep-tagged KaiC3 (monomer: 58 kDa) migrated as one band between 272 and 450 kDa and could represent a hexameric complex (348 kDa). Incubation of KaiC3 with KaiA3 alone led to protein accumulation in the wells in native PAGE, indicating precipitation of the KaiA3/KaiC3 complex in the absence of KaiB3 (Fig. 2b). However, the interaction between KaiA3 and KaiC3 was validated by immunoprecipitation-coupled liquid chromatography-mass spectrometry (LC-MS) analysis of FLAG-tagged KaiC3 (Fig. S7). Furthermore, the experiments did not reveal any interactions between KaiA3 and either KaiC1 or KaiC2 (Fig. S5, Fig. S7), indicating the specificity of the KaiA3-KaiC3 interaction. No complex formation was detected between KaiA3 and KaiB3 (Fig. 2b, Fig. S5, Fig. S6). In contrast, the formation of a large protein complex was observed when all three clock components, KaiA3, KaiB3, and KaiC3, were incubated together for 16 h at 30 °C (Fig. 2b; Fig. S6). The size matches that of a complex consisting of one KaiC3 hexamer, six KaiA3 dimers, and six KaiB3 monomers (840 kDa). The presence of KaiA3 in the complex was validated by western blot analysis using an anti-His antibody (Fig. 2b, Fig. S6). As expected, no such complex was formed when KaiA3 was replaced by Synechococcus KaiA (Fig. 2b). Moreover, no such complex was formed when KaiB3 was replaced by its isoform KaiB1, suggesting that KaiB3 is specific for KaiA3 as well and that KaiB3 might recruit KaiA3 to the KaiC3/KaiB3 complex (Fig. S6).
a YTH interaction analysis between KaiA3 and KaiC3. The KaiA1 dimer interaction was used as a positive control. YTH reporter strains carrying the respective bait and prey plasmids were selected by plating on complete supplement medium (CSM) lacking leucine and tryptophan (-Leu -Trp). AD, GAL4 activation domain; BD, GAL4 DNA-binding domain; empty, bait, and prey plasmids without protein sequence (only AD/BD domain). The physical interaction between bait and prey fusion proteins was determined by growth on complete medium lacking leucine, tryptophan, and histidine (-Leu -Trp -His) and the addition of 12.5 mM 3-amino-1,2,4-triazole (3-AT). The BD was fused to the N-terminus of KaiA3. For clarity, spots were assembled from several replicate assays (the original scans are shown in Fig. S5). KaiC3-KaiA3 interaction analysis was performed thrice. b Interaction analysis of recombinant Kai proteins on native polyacrylamide gel. Proteins were incubated for 16 h at 30 °C and subsequently subjected to 4–16% clear native PAGE. Gels were either stained with Coomassie blue (left side) or blotted and immunodecorated with a monoclonal anti-His antibody to detect recombinant KaiA3-His6 (right side). Representative images of three independent experiments. Recombinant Synechococcus KaiA was used for comparison. c KaiC3 phosphorylation depends on the presence of KaiA3 and KaiB3. KaiC3 was dephosphorylated by incubating for 18 h at 30 °C prior to the start of the assay. 0.2 µg/µl NP-KaiC3 was incubated at 30 °C in the presence or absence of 0.1 µg/µl Synechocystis KaiA3 (A3), KaiB3 (B3) and KaiB1 (B1) and Synechococcus KaiA (A), respectively. Aliquots were taken at 0 and 16 h, followed by separation on a high-resolution LowC SDS-PAGE gel in Tris-Tricine buffer and staining with Coomassie blue. A slow-migrating band representing the phosphorylated form of KaiC3 (P-KaiC3) was observed only in the presence of KaiA3. NP indicates dephosphorylated KaiC3. Phosphorylation analysis was performed at least three times, and a negative control for Synechococcus KaiA was performed twice. Source data are provided as a Source Data file.
Previous studies have shown that KaiC3 has autokinase activity that is independent of KaiA126,28. Since our studies revealed an interaction between KaiC3 and KaiA3, we were interested in probing the influence of KaiA3 on the phosphorylation of KaiC3. The recombinant Kai proteins described above were used for this purpose. KaiC3 was incubated for 16 h at 30 °C in the presence or absence of other Kai proteins, and its phosphorylation state was analyzed by LowC-SDS-PAGE (Fig. 2c), and LC-MS/MS (Fig. S8). Because KaiC3 was partially phosphorylated after purification from E. coli, the protein preparation was incubated for 18 h at 30 °C prior to the start of the assays. During this incubation period, KaiC3 was autodephosphorylated, as is typical for KaiC proteins (Fig. 2c, NP-KaiC3)45. The addition of KaiA3 led to the phosphorylation of KaiC3, while the presence of Synechococcus KaiA had no influence on the phosphorylation state of KaiC3. In contrast, KaiC3 dephosphorylation was enhanced by KaiB3 (Fig. 2c, upper panel). Replacing KaiB3 with its isoform KaiB1 in samples containing KaiA3 maintained KaiC3 in the phosphorylated state (Fig. 2c, lower panel). Analysis of KaiC3 phosphorylation by LC-MS/MS- identified the neighboring residues Ser423 and Thr424 as phosphorylation sites, which are conserved across KaiC1 and KaiC3 homologs (Fig. S8). Based on these analyses, we conclude that KaiA3 likely has a KaiA-like function in promoting the phosphorylation of KaiC3 and interaction with KaiB3, which in turn enhances dephosphorylation. Neither Synechococcus KaiA nor Synechocystis KaiB1 could substitute for KaiA3 or KaiB3, respectively, demonstrating that the Synechocystis KaiA3/KaiB3/KaiC3 proteins represent a functional complex.
KaiC3 phosphorylation cycles in vitro and in Synechocystis cells
The opposing effects of KaiA3 and KaiB3 on KaiC3 phosphorylation imply that these three Synechocystis proteins may form a functional in vitro oscillator. When Synechococcus KaiABC proteins were mixed in vitro, the KaiC phosphorylation rhythms were relatively insensitive to KaiB concentration but could occur only in a small window of KaiC:KaiA ratios. In the presence of KaiB, low KaiA concentrations were insufficient to increase KaiC phosphorylation, whereas excessive KaiA could not be counteracted by KaiB49. We observed the same effect for the Synechocystis proteins, when we incubated a constant KaiC3:KaiB3 ratio with various KaiA3 concentrations: 0.5 µM KaiA3 failed to change the phosphorylation of KaiC3 within 48 h. 8.4 µM KaiA3 stimulated hyperphosphorylation of KaiC3 within 6 h, and KaiC3 remained highly phosphorylated afterwards (Fig. 3, Fig. S9a). In the presence of 1.4–4.2 µM KaiA3, the KaiC3 protein was also phosphorylated within 6 h and then dephosphorylated until the 18 h time point. Thus, for the three intermediate KaiA3 concentrations, we observed one in vitro cycle of phosphorylation and dephosphorylation. The stimulating effect of KaiA3 on KaiC3 phosphorylation was saturated at a KaiA3 concentration of 4.2 µM, which corresponds to a KaiA3:KaiC3 stoichiometry of 1:0.8. Only in the presence of 1.4 µM and 2.8 µM KaiA3 (corresponding to a ~1:1.4 and 1:2.2 stoichiometry of KaiA3:KaiC3), we could reconstitute another weak phosphorylation cycle peaking at 30 h (Fig. 3c).
KaiC3 (3.4 µM) was incubated with KaiB3 (7.4 µM) and various concentrations of KaiA3 at 30 °C over a time course of 48 h. Aliquots incubated for the indicated time periods were applied to a high-resolution LowC SDS-PAGE gel and proteins were separated in Tris-glycine buffer. a Representative gels. b To assign the bands to the fully phosphorylated (PP-KaiC3), single-phosphorylated (P-KaiC3) and non-phosphorylated (NP-KaiC3) forms, KaiC3 was dephosphorylated with Lambda phosphatase (KaiC3/λ-PP) (n = 3) in the presence or absence of phosphatase inhibitors (PhosStop (n = 1) and vanadate (n = 2), KaiC3/λ-PP +Inh). c The ratio of PP-KaiC3 to total KaiC3 based on gel images (representative gels are shown in (a)). Each line/color represents a different KaiA3 concentration. The lines show the average from three assays (points). d Representative assay to test temperature compensation. KaiC3 (3.4 µM), KaiB3 (7.4 µM) and KaiA3 (2.8 µM) were mixed, aliquots were incubated at 25 (blue), 30 (green) or 35 °C(red) for the indicated times (n = 2 for all temperatures, except 35 °C (n = 1)) and the ratio of PP-KaiC3/total KaiC3 was determined as described above. Lines indicate the mean. Source data are provided as a Source Data file.
The KaiC3 phosphorylation cycles at intermediate KaiA3 concentrations fulfilled one of the defining criteria of a true circadian rhythm: they displayed a ~24 h period. They also continued for more than one cycle. However, the oscillation displayed a low amplitude and appeared dampened. Therefore, we tested another criterion of true circadian rhythm, which is temperature compensation. A mixture of KaiC3, KaiB3, and KaiA3 was incubated at different temperatures. We did not detect a period change between 30 °C and 35 °C. At 25 °C, the period was extended to approximately 30 h, indicating temperature compensation to a similar extent as that observed for the Synechococcus KaiABC oscillator1. In this experiment, we extended the analysis at 30 °C to 60 h and observed that the dampened oscillation continued.
These observations led us to investigate the potential persistence of KaiC3-phosphorylation rhythms in Synechocystis across successive free-running cycles. We entrained wild-type Synechocystis using two 12 h LD cycles. After release to constant light (LL), we collected samples over a course of three days and separated total proteins via SDS-PAGE for western blot analysis. Probing with a specific antibody (Fig. S9b) revealed stable, high-amplitude oscillations in KaiC3 phosphorylation (Fig. 4). The oscillations displayed a ~24 h period and persisted for (at least) three free-running cycles in the cellular context (Fig. 4). The timing of hypo- and hyperphosphorylation was comparable to that reported for Synechococcus KaiC6,50,51. KaiC3 was phosphorylated towards the end of the subjective day and dephosphorylated around the subjective dawn. In addition, the abundance of KaiC3 oscillated over a period of approximately 24 h (Fig. 4), as published for Synechococcus KaiC6,11,50,52.
a Western blots showing KaiC3 and KaiC1 phosphorylation over three days. The cells were cultured in liquid medium, entrained with two LD cycles, and then returned to LL. Cells were harvested every 4 h at LL. 8 μg of total protein were loaded in each lane and subjected to western blot analysis. We designated the upper bands as hyperphosphorylated (PP-KaiC) and the lower bands as hypophosphorylated (P- and NP-KaiC), analogous to our in vitro data and the Synechococcus KaiC, and based on Fig. S9b. The experiment was performed once over three LL cycles and confirmed once over one LL cycle. b Densitometric estimation of rhythms in the KaiC3 and KaiC1 fractions of PP-KaiC (left) and KaiC protein abundance (right) from the blots shown in (a). Source data are provided as a Source Data file.
KaiC1 and KaiC3 phosphorylation are phase-locked in Synechocystis cells
Our in vitro and in vivo phosphorylation data indicated that the Synechocystis KaiA3B3C3 system functions similarly to the well-studied KaiABC oscillator in Synechococcus. Initially, KaiA1B1C1 proteins were predicted to form this ortholog based on their sequences and interactions, as reported in previous studies26,28. However, it appears that the KaiA3B3C3 complex may serve as a functional equivalent to the Synechococcus KaiABC oscillator. Alternatively, it is possible that the two KaiABC systems coexist and are interconnected within Synechocystis. To answer these questions, we analyzed proteins from the above-described experiment, covering three free-running days after LD synchronization (Fig. 4), using an antibody specific for KaiC1 (Fig. S9b). KaiC1 phosphorylation displayed stable ~24 h rhythms which were phase-locked with the KaiC3 phosphorylation rhythm. However, the amplitude was reduced compared with that of KaiC3 phosphorylation, and KaiC1 abundance showed only weak changes.
The two KaiABC systems together drive circadian backscatter rhythms
The observed in vivo phosphorylation rhythms of KaiC1 (Fig. 4) implied that KaiA1B1C1 also forms a functional oscillator in Synechocystis. Oscillators can function as independent systems that drive separate rhythmic outputs, or only one system is a bona fide oscillator in the cell that controls the rhythmicity of the other. Alternatively, the oscillators may be dependent on each other, form only one complex, or are at least interconnected and integrated into one circadian output. Phase locking of the in vivo phosphorylation rhythms of KaiC1 and KaiC3 (Fig. 4), together with the observation that components of the two oscillators interact with each other in the cell26, support the latter hypothesis. Quantifying discrete outputs is challenging because of the yet-to-be-identified nature of the output components in the KaiA3B3C3 system. Therefore, we aimed to monitor a more general circadian rhythm that can be detected by backscatter measurements during growth in liquid cultures53. Even without knowing the distinct output of each oscillator, this enabled us to test the hypothesis that the two oscillators operate together.
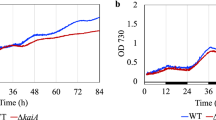
The backscatter properties of Synechocystis cells oscillate with a ~24 h period under LL, after cultures are synchronized by dilution with fresh medium. These circadian oscillations are temperature-compensated and driven by the kaiA1B1C1 gene cluster53. To understand whether this output can be used as a general readout of the circadian status of the cell, we investigated the effect of ∆kaiA3B3C3 deletion on backscatter oscillations in comparison with wild-type Synechocystis and ∆kaiA1B1C1 strains. We grew the three strains as two subsequent pre-cultures for ten days in LL, synchronized them by dilution to OD750nm of 0.9, and monitored the backscatter over time. The presence and loss of oscillations were already apparent in the raw backscatter data (ref. 53 and Fig. 5a–c). For a better presentation, we subtracted a polynomial regression fit from the raw backscatter signal to remove the influence of culture growth, and subtracted the average difference (Fig. 5d–h and Fig. S10). Deletion of the whole kaiA3B3C3 system led to an intermediate circadian output when compared to Synechocystis wild type and ∆kaiA1B1C1. Backscatter oscillations of the wild type could be described by a simple harmonic oscillation with a ~26 h period (26.46 ± 0.34 h, n = 3) (Fig. 5d, Fig S10a, b). In the kaiA1B1C1 deletion strain, the oscillation was almost abolished, as reported by Berwanger et al.53. Upon close inspection, however, we observed extremely low amplitude oscillations, which were best described by a simple harmonic oscillation with a ~33 h period (33.32 ± 3.57 h, n = 3) (Fig. 5e, Fig S10a, b). Deletion of the kaiA3B3C3 system resulted in reduced and potentially dampened oscillations, which were not well described by the simple harmonic cosine function (dotted line in Fig. 5f). The amplitude and period changed over the course of the experiment. To allow comparison with the wild type, we determined the amplitude and phase of the first backscattering peak as well as the period of the first backscattering cycle. We detected a phase shift of ~−7 h and a reduction in the amplitude to one-third in comparison to the first backscattering peak in the wild type (Fig. 5i). Furthermore, the period of the first backscattering cycle was significantly shortened by ~5 h in comparison to the wild type (Fig. 5j, Fig. S10c). Altogether, the backscattering data imply that the KaiA1B1C1 system mainly drives circadian rhythms, but requires KaiA3B3C3 to maintain the period and amplitude. On the other hand, KaiA3B3C3 may be able to drive low-amplitude oscillations but requires coupling to KaiA1B1C1 to maintain a circadian period and ensure a high amplitude.
a–c Growth of Synechocystis wild type (WT) (a), Synechocystis ∆kaiA1B1C1 mutant strain (b), and Synechocystis ∆kaiA3B3C3 mutant strain (c) in LL after initial synchronization by dilution. Graphs display the average backscatter signal as a rolling average (solid line) at 730 nm with SD (shaded) from a representative experiment with 4–5 wells per strain. Polynomial regression (dashed line) was fitted to the data. d, e For each replicate within the experiments with wild type (d) and Synechocystis ∆kaiA1B1C1 (e), the raw backscatter signal was subtracted from the polynomial regression fit to remove the contribution of the overall growth of the cultures. The average difference was subtracted for normalization. Displayed is the average of this normalized backscatter (solid lines) with SD (shaded). Curves are smoothed. The dotted line indicates a simple harmonic cosine fit. f Same as (d, e), but the two experiments investigating backscatter in the ∆kaiA3B3C3 mutant are shown. g, h Same as (d, e) for further Synechocystis kai gene mutants without the cosine fits. All data displayed in (d–h) were collected from the same representative experiment (except for the additional Synechocystis ∆kaiA3B3C3 in f). Data from all three independent experiments are shown along with the cosine fit in Fig. S10 (except for Synechocystis ∆kaiA3B3C3). i To determine the effect on the first cycle after synchronization, the phase shift and relative amplitude of each Synechocystis mutant were calculated for the first peak (approx. between 21–29 h) for three independent experiments (Fig. S10a). The circles, triangles, and squares represent the first, second, and third experiments, respectively. Synechocystis ∆kaiA3B3C3 was not included in the third experiment. Marker scales are proportional to the number of replicate wells within one experiment (Exp. 1: n = 5 for all strains except ∆kaiA1B1C1 (n = 4), Exp. 2: all strains n = 4, Exp 3: all strains n = 5). Error boundaries were calculated using formulas (3) and (5) (see Methods section). The X-axis is discontinuous. j For strains that displayed dampened backscatter oscillations, we derived the length of the first period from the distance of the first trough to the first peak (see materials and methods) for the three experiments in Fig. S10a (Synechocystis ∆kaiA3B3C3 only two experiments). The backscattering period of the wild type was determined in the same way for comparison. Boxes are based on all replicate wells (indicated as n) from all three experiments (discriminated by the same symbol) and range from the first to the third quartile. Whiskers extend to the furthest data point within 1.5x the interquartile distance. The red line indicates the median and the dashed line indicates the mean. Results of the pairwise statistical tests are shown in Fig. S10c. Source data are provided as a Source Data file.
To dissect the role of the single components of the new KaiA3B3C3 system, we monitored the effects of single and double mutants of kaiA3, kaiB3, and/or kaiC3 on backscatter oscillations. All strains in which kaiC3 was deleted, alone or in combination with other genes, displayed the same phenotype as Synechocystis ∆kaiA3B3C3 (Fig. 5g, i, j; Fig. S10a), confirming that KaiC3 was the central protein in this system. Deletion of either kaiA3 or kaiB3 had different effects (Fig. 5h–j; Fig. S10a). Knockout of kaiA3 led to a similar decrease in amplitude as observed in ∆kaiC3. However, the phase shift of the first peak of the dampened oscillation was less pronounced than that of ∆kaiC3, and the period of the first low-amplitude cycle was only slightly lower than that of the wild type (Fig. 5i, j, Fig. S10c). A double knockout of kaiA3 and kaiB3 resulted in a phenotype similar to that of the deletion of kaiA3 alone (Fig. 5h–j, Fig. S10). In contrast, the deletion of kaiB3 almost completely abolished these oscillations. In general, a high variance among the experiments was observed for this mutant (Fig. 5h, i; Fig. S10a). On average, single kaiB3 deletion drastically reduced the amplitude. Notably, when plotting the relative amplitude against the phase shift of the first peak, this mutant was grouped with the ∆kaiA1B1C1 mutant rather than with strains carrying mutations in the genes encoding the components of the KaiA3B3C3 system (Fig. 5i). Accordingly, the low-amplitude oscillations fitted well to harmonic oscillations without dampening (Fig. S10a) as was observed for the kaiA1B1C1 strain. The period of the simple harmonic oscillation fit to the backscattering oscillations in the ∆kaiB3 strain (39.04 ± 2.58 h) was extended in comparison to the wild type and ∆kaiA1B1C1 mutant (Fig. S10b).
Overall, this suggests that KaiC1 drives stable free-running oscillations, and is the core of the main system that drives the backscatter rhythms of the cell. However, backscatter rhythms persist only as self-sustained oscillations if a second post-translational putative oscillator, KaiA3B3C3, is also present, which implies that the two systems are directly or indirectly connected.
Mutation of kaiA3 impacts growth and viability during mixotrophic and chemoheterotrophic growth
Previously, we showed that deletion of the kaiA1B1C1 operon severely affects the viability of cells on agar plates30. When grown photoautotrophically in LL, the ΔkaiA1B1C1 mutant strain behaved like the wild-type strain. However, ΔkaiA1B1C1 was not able to grow under chemoheterotrophic conditions, and viability was reduced under mixotrophic conditions (in LL) as well as in LD cycles under both conditions26,30. Previously, we also revealed that deletion of kaiC3 had less detrimental effects: growth was reduced in chemoheterotrophic conditions, but not in LL or LD, independent of the presence or absence of glucose26. To further determine whether the two systems control similar physiological functions and, therefore, might be connected, we performed viability assays with mutants affecting the kaiA3 gene. Therefore, the ΔkaiA3B3C3 and ΔkaiA3 mutant strains, as well as the genomic ∆kaiA3/kaiA3 complementation strain, were analyzed under various growth conditions. These analyses were performed in the Synechocystis PCC-M wild-type background strain, which has been used in previous studies on the KaiA1B1C1 system and is known to grow in complete darkness28,30. Furthermore, this strain is motile and aggregates in liquid culture. Therefore, we performed spot assays and did not measure growth in liquid cultures. The cell suspensions were plated on agar at different dilutions and grown photoautotrophically (Fig. 6a) and photomixotrophically (Fig. 6b) in LL and 12 h LD cycles or chemoheterotrophically (Fig. 6c). Because the strains grew very slowly under chemoheterotrophic conditions, the cells were spotted at higher concentrations under these conditions. The mutant strain lacking kaiA3 and the triple-knockout strain showed a phenotype similar to that previously observed for a kaiC3 deletion mutant. There were almost no differences in the viability of the mutant strains compared to that of the wild type under photoautotrophic conditions under LL and LD cycles (Fig. 6a). However, they were unable to grow in the dark (Fig. 6c). This ability was fully restored when kaiA3 was reinserted into the kaiA3 deletion strain (Fig. 6c).
a–c Proliferation of the wild type (WT), the ΔkaiA3 and ΔkaiA3B3C3 deletion mutants, and the ΔkaiA3/kaiA3 complementation strain under different growth conditions. Strains were grown in liquid culture in LL, and different dilutions were spotted on agar plates and incubated under the indicated light conditions, with a light phase corresponding to 75 µmol photons m-2 s-1 white light. Representative result from three independent experiments are shown. a Cultures were diluted to an OD750nm value of 0.4, and tenfold dilution series were spotted on agar plates. Plates were analyzed after 6 or 8 d of LL and 12 h LD cycles, respectively (photoautotrophic growth). b Same as (a), but the cells were spotted on agar plates containing 0.2% glucose (photomixotrophic growth). c Cultures were diluted to OD750nm values of 1.2, 0.8, and 0.4, and spotted on agar plates supplemented with 0.2% glucose. The plates were analyzed after 26 d of continuous darkness (chemoheterotrophic growth). Because of the higher cell density, a second plate was prepared and incubated in LL for three days (photomixotrophic growth). Source data are provided as a Source Data file.
Under photomixotrophic conditions, the mutant strain lacking all three alternative kai genes (ΔkaiA3B3C3) exhibited a growth phenotype similar to that previously observed for ∆kaiC330. The strain proliferated well and, in LD cycles, seemed to have some advantages compared to the wild type (Fig. 6b). However, the ΔkaiA3 strain showed less viability under photomixotrophic conditions, a phenotype comparable to that of the ∆kaiA1B1C1 strain (Fig. 6b and ref. 30). Again, viability was partly restored by re-insertion of kaiA3 (Fig. 6b).
Surprisingly, overexpression of kaiA3 by insertion of a KaiA3-FLAG encoding plasmid in the wild-type background reduced viability to almost the same extent as kaiA3 deletion (Fig. S11). To exclude the possibility that this phenotype was caused by the FLAG-tag or expression from the plasmid, we inserted the same plasmid into the ∆kaiA3 deletion strain. The FLAG-tagged KaiA3 complemented the kaiA3 deletion in the same manner as genomic complementation with non-tagged KaiA3 (Fig. S11). These findings suggest that deregulation of the phosphorylation level of KaiC3 might affect the viability of Synechocystis under photomixotrophic and chemoheterotrophic conditions more than the deletion of kaiC3 or the whole KaiA3B3C3 system. In the absence of KaiA3 and the presence of excess KaiA3, when, according to the in vitro data, KaiC3 is constantly hypo- or hyperphosphorylated, respectively (Fig. 3c), the phenotypes are as detrimental as for the knockout of kaiA1B1C1. To confirm that KaiC3 phosphorylation is indeed dependent on KaiA3 levels in the respective mutants, we grew Synechocystis cells in an LD cycle, followed by constant illumination, separated whole-cell extracts on a Phos-tag gel, and identified KaiC3 phosphorylation forms by western blot analysis (Fig. 7a–c). Two or more bands were detected in vitro and in vivo, which partly overlapped with a non-specific band detected in the ∆kaiC3 strain (Fig. 7a). These bands might reflect single phosphorylated states of KaiC3, but were not included for densiometric quantification of KaiC3 phosphorylation to exclude the effects of potential cross-reactions of the antibody with KaiC1, KaiC2, or another protein (Fig. 7d). In the wild type, phosphorylation cycled with a period of about 24 h. In contrast, KaiC3 was mostly dephosphorylated in the ΔkaiA3 mutant strain. There is still some fully phosphorylated KaiC3 detectable in this mutant, which might originate from weak autophosphorylation in the absence of KaiA28. In the KaiA3 overexpression strain, KaiC3 was highly phosphorylated compared to the wild type. Phosphorylation after kaiA3 overexpression varied strongly between the experiments, most likely due to different KaiA3 levels when using the copper-dependent PpetJ promoter for overexpression. Thus, the phenotypes of the different mutants suggest that both clock systems are involved in regulating heterotrophic growth under light and darkness. The interconnection and (putative) role of the components in metabolic control and generation of backscatter oscillations are summarized in the model in Fig. 8.
Samples were collected every 6 h from cells grown in a 12 h LD cycle, followed by LL. Whole-cell extracts were separated using Phos-tag SDS-PAGE and immunodecorated with a KaiC3-specific antiserum. Representative blots are shown in (a). We detected 4-5 bands which partially overlapped or were slightly shifted compared to the bands detected in the ∆kaiC3 strain (12 h time point was loaded). The two indicated prominent bands were absent in the ∆kaiC3 strain. b Whole cell extracts of Synechocystis wild-type (WT), kaiA3 mutant (ΔkaiA3), and the overexpression (kaiA3-OE) strain grown for 6 or 24 h in a 12 h LD cycle were loaded together with in vitro phosphorylated recombinant KaiC3 (KaiA3-KaiC3/6 h), which was generated by 6 h incubation with 4.2 µM KaiA3 at 30 °C. c KaiC3 was dephosphorylated (KaiC3/λ-PP) using Lambda phosphatase and analyzed alongside with in vitro phosphorylated KaiC3 (KaiA3-KaiC3/6 h). Based on the band pattern of recombinant KaiC3, we assigned the indicated bands to fully phosphorylated (PP-KaiC3) and non-phosphorylated KaiC3 (NP-KaiC3). The control reactions in (b) and (c) were performed at least twice on different gels. d The ratio of PP-KaiC3 to NP-KaiC3 measured in wild-type and ΔkaiA3 mutant is plotted as average (line) with dots indicating biological replicates (n = 4). Data derived from two biological replicates of kaiA3-OE are presented as individual curves. The white and dark gray boxes represent the light and dark periods, respectively, and the light gray box represents the subjective night. Source data are provided as a Source Data file.
KaiC1 and KaiC3 display phase-locked phosphorylation rhythms. KaiA3 and KaiB3 regulate auto-phosphorylation and dephosphorylation of KaiC3 and form a second oscillator. The KaiA1B1C1 oscillator appears to be the main driver of backscatter rhythms, whereas KaiC3 and KaiA3 are required to maintain the amplitude and period. KaiB3 deletion abolished the rhythms to a similar extent as kaiA1B1C1 deletion. Because KaiB proteins were shown to also interact with the KaiC proteins of the other system26, we assume that the absence of competing KaiB3 leads to enhanced KaiB1 binding to phosphorylated KaiC3 (dashed line), thereby disturbing KaiA1B1C1-based oscillations. Altogether, this implies that phosphorylated KaiC3 stabilizes backscatter oscillations. The interconnection between the KaiC1 and KaiC3 systems is also metabolically relevant. Deletion of kaiA1B1C1 and both up- and downregulation of kaiA3 reduced mixotrophic growth, whereas deletion of the entire KaiA3B3C3 system and kaiC3 had no effect. This implies that KaiA1B1C1 mainly contributes to the switch from autotrophic to heterotrophic growth, but the phosphorylation rhythms of KaiC3 can interfere with it. Whether this interference occurs directly via the KaiC1-KaiC3 interaction26 or indirectly via output pathways needs to be clarified. This figure was created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Discussion
Our knowledge of the function, composition, and network of clock systems in prokaryotes, including cyanobacteria, is steadily increasing. Even though multiple copies of the core clock proteins KaiB and KaiC are encoded in bacterial genomes, the canonical KaiA was found only as a single copy in Cyanobacteria yet27,28,43,54. By identifying a chimeric KaiA3 and verifying its interaction with the KaiB3-KaiC3 complex, we added another component to the diversity of bacterial clock systems.
New putative KaiA orthologs have been bioinformatically identified in prokaryotes other than Cyanobacteria43,55. Therefore, we suggest that such proteins may play a previously overlooked role in KaiB-KaiC-based systems. Exploring this possibility could provide valuable insights into unanswered research questions, such as the mechanism responsible for the rhythmic processes observed in Rhodospirillum rubrum. Indeed, this purple bacterium lacks KaiB1 and KaiC1 orthologs, but possesses KaiA3, KaiB3, and KaiC3 (ref. 56 and Fig. 1). Notably, the recently described primordial oscillator from Rhodobacter, which consists of homologs of KaiC2 and KaiB2, can form an hourglass timer without KaiA22. A similar primordial clock has been suggested to be present in Rhodopseudomonas palustris21 and the cyanobacterium Prochlorococcus MED431,32, while other bacterial KaiB and KaiC homologs, including the KaiC2-KaiB2 system from Synechocystis, are believed to have clock-independent functions17,57,58.
It has been proposed that kaiC is the oldest evolutionary member of circadian clock genes54. KaiC homologs can be found even in Archaea where it was found to control e. g. motility of Sulfolobus acidocaldarius by protein interaction59. The later addition of KaiB was enough to form a primordial timekeeper which needs a signal for daily resetting of the clock21,22,31,32. In Rhodobacter KaiC2, dephosphorylation is regulated by the stability of coiled-coil interactions between two connected hexamers as well as by KaiB22. Whether autophosphorylation or dephosphorylation dominates depends primarily on the ATP/ADP ratio. Hence, the KaiC2-KaiB2 timer cannot oscillate autonomously but responds to changing ATP/ADP levels. Therefore, it was suggested that the Rhodobacter clock represents an ancient timer that depends on changes in photosynthetic activity during the day-night switch22.
With the evolution of KaiA, a self-sustained oscillator was developed that allowed for true circadian oscillations in gene expression, which can be observed in cyanobacteria. Why does KaiC require KaiA to drive persistent oscillations? By default, the A-loops of Synechococcus KaiC hexamers adopt a buried conformation, which inhibits autophosphorylation. Only the binding of KaiA favors phosphorylation by stabilizing A-loop exposure5. In contrast, Rhodobacter KaiC2 constantly exposes its A-loops, sterically allowing high intrinsic phosphorylation22. Furthermore, introducing KaiA as a factor stimulating autophosphorylation of KaiC allows coupling between different KaiC molecules, e.g. by KaiA sequestration, which is needed for synchrony and thus high-amplitude oscillation13,48,60,61,62.
The interacting residues between KaiA and KaiC are less conserved in both Synechocystis KaiA3 and KaiC328,63 (Fig. 1). Since we demonstrated an interaction between KaiC3 and KaiA3, it is likely that co-evolution of the two proteins occurred. KaiC3 does not display the extended C-terminus that contributes to homododecamer formation in Rhodobacter KaiC222,28, and we only observed the formation of hexamers or smaller oligomers26 (Fig. 2).
KaiA3 formed a distinct clade at the basis of the KaiA clade. Apart from its presence in the N-terminal domain of the phosphatase RsbU of Bacillus subtilis, a distinctive structure of the KaiA C-terminus has rarely been observed46. RsbU acts as a positive regulator of the alternative sigma factor B, which is involved in the general stress response64. The N-terminal domain of RsbU forms dimers similar to KaiA, and the proposed binding site for its corresponding activator, RsbT, is in an equivalent location to the KaiC-binding site on KaiA46. These findings may reflect how protein domains change during evolution, while their original functions are conserved. However, a link between RsbU and the recently proposed circadian clock in Bacillus subtilis has not yet been identified65. Moreover, circadian rhythms have been observed in several prokaryotes that do not encode Kai orthologs, suggesting convergent evolution of circadian rhythms in prokaryotes65,66. Further in-depth analyses are needed to elucidate whether KaiA3, together with KaiB3 and KaiC3, or the well-studied Synechococcus circadian clock present a more ancestral system, because analysis of a larger dataset recently suggested that the canonical kaiA gene evolved at the same time as cyanobacteria43.
In this work, we broadly define an oscillator to include systems that may be dampened but nevertheless have a natural frequency. Taken together, our data are consistent with a model in which KaiA3 fulfills the functions of a true KaiA homolog, such as dimerization, binding to KaiC3, and enhancing KaiC3 autophosphorylation. Other mechanistic processes, such as sequestration to the CI ring by binding to KaiB3, remain to be investigated but are clearly possible. By mixing KaiA3, KaiB3, and KaiC3, we reconstituted a dampened in vitro oscillator (Fig. 3), suggesting that the observed in vivo rhythm of KaiC3 phosphorylation is driven by KaiA3 and KaiB3, and that the amount of KaiA3 is critical for the phosphorylation rhythm.
In Synechococcus KaiC, ATPase activity directly correlates with the clock period and mediates temperature compensation4. We observed temperature compensation of dampened KaiC3 phosphorylation in the presence of KaiA3 and KaiB3, although the ATPase activity of KaiC3 alone is temperature dependent26. Future work might reveal how the presence of KaiA3 and KaiB3 contributes to temperature compensation.
The in vivo phosphorylation of KaiC3 displayed a higher amplitude than the in vitro oscillation, implying that rhythms might be stabilized by other mechanisms in the cell. Whether direct crosstalk between the KaiA1B1C1 and KaiA3B3C3 systems contributes to stabilization remains to be investigated. We can only speculate on the nature of a direct interconnection. KaiC3 is the central protein of the newly identified in vitro oscillator; however, its absence has less severe consequences for backscatter oscillations than the absence of KaiB3. Removing KaiA3 together with KaiB3 restored the dampened oscillation. This suggests that hyperphosphorylated KaiC3 interferes with KaiA1B1C1 driven backscatter oscillations, which could occur directly via sequestration of KaiB1 (see model in Fig. 8).
The Rhodobacter hourglass-like timer required environmental cues for daily resetting. However, entrainment by metabolites has also been described for more elaborate and true circadian oscillators. In addition to entrainment by the input kinase CikA67, the Synechococcus clock can be entrained directly by the ATP/ADP ratio and oxidized quinones36,68. Moreover, CikA does not sense light directly, but perceives the redox state of the plastoquinone pool69,70. Furthermore, glucose feeding can entrain Synechococcus when engineered to take up glucose71. In plants, it has been demonstrated that both exogenous sugars and internal sugar rhythms resulting from cyclic photosynthetic activity entrain the clock72. Synechocystis can naturally utilize glucose, which may make it even more susceptible to metabolic entrainment by sugars. In addition, the need for metabolic compensation73 may be particularly pronounced. Notably, the Synechocystis PCC-M wild-type strain could grow in complete darkness when supplemented with glucose. This is different from an earlier study that showed that Synechocystis requires a 5 min blue-light pulse at least once a day to grow heterotrophically in the dark74. The authors described this behavior as light-activated heterotrophic growth. There are no studies that explain why cells require this short light pulse, but it is also clear that the PCC-M strain grows fully chemoheterotrophically30.
In contrast to Synechococcus, CikA from Synechocystis is a true photoreceptor that binds a chromophore75. Thus, it remains unclear whether CikA has a similar function in both cyanobacteria, and whether it interacts with both circadian clock systems in Synechocystis. The high structural similarity of the N-terminal domain of KaiA3 to response regulator domains from other organisms indicates that the core structure and activity are maintained, while adaptivity and variation provide specificity for distinct pathways25. Within KaiA3, the aspartate residue crucial for phosphorylation is conserved. Theoretically, the protein can receive an input signal from a cognate histidine kinase that has not yet been identified. Thus, there are potentially important differences related to input and output factors, and possibly entrainment of different cyanobacterial circadian clock systems.
The physiological function of the KaiA3B3C3 clock system seems to be related to the different metabolic modes of Synechocystis. Mutants deficient in kaiA3 lose the ability to grow chemoheterotrophically on glucose, which is an aggravated effect compared to kaiC3-deficient mutants that merely show reduced growth rates during heterotrophy26. Similarly, in Synechococcus, disruption of kaiA led to one of the most severe effects on activity loss and was traced back to the unbalanced output signaling of the circadian clock76. Overaccumulation of KaiA3 also appeared to disturb the Synechocystis system (Fig. 6). Such an effect was also shown for the Synechococcus clock system, in which increased KaiA levels promote the hyperphosphorylation of KaiC6,77, thereby deactivating rhythmic gene expression78. Surprisingly, inactivation of the complete KaiA3B3C3 system resulted in a different phenotype. Although growth in darkness on glucose was strongly affected, similar to the single mutants, photomixotrophic growth was slightly better in the ∆kaiA3B3C3 strain than in the wild type.
It is possible that, in the absence of KaiA3, an altered interaction of the KaiC3 system with the KaiC1 system leads to the aggravated growth defects of ΔkaiA3. Constant dephosphorylation of KaiC3 may also change its interactions with the KaiA1B1C1 system. Therefore, when the complete KaiA3B3C3 system is missing, KaiA1B1C1 may be able to compensate for this under certain growth conditions. In Synechocystis, ΔkaiA3-like phenotypes, such as impaired viability during LD cycles or complete loss of chemoheterotrophic growth on glucose, were also observed for ΔkaiA1B1C1, ΔsasA, and ΔrpaA mutants30,79. For ΔsasA, it was shown that the mutant strain was able to accumulate glycogen but was unable to utilize the storage compound to grow heterotrophically, probably because of its inability to catabolize glucose79, whereas Synechocystis ΔkaiA1B1C1 displays a highly reduced glycogen level53. A recent metabolomics study suggested that the growth inhibition of ΔkaiA1B1C1 and ΔrpaA mutants in an LD cycle might be at least partly related to a defect in the inhibition of the RuBisCO enzyme in the dark and increased photorespiration, leading to the accumulation of the potentially toxic product, 2-phosphoglycolate80. This previous study also revealed an enhanced growth defect in ΔkaiA1B1C1 and ΔrpaA mutants under photomixotrophic conditions in LD cycles, similar to the ΔkaiA3 strain in the current study. This further supports the idea that the KaiA3B3C3 system is interconnected with the core clock system KaiA1B1C1.
Clearly, there is a difference in the phenotypes between our study and the results demonstrated by Zhao et al.17, who analyzed single and double kaiB3 and kaiC3 knockout strains. In LD cycles, the kaiB3C3 knockout strain showed a reduced growth rate compared to the wild-type control under photoautotrophic conditions. Even in LL, this mutant showed a reduced growth rate and was outcompeted by the wild-type cells in mixed cultures. Photoheterotrophic and heterotrophic conditions were not tested in this study. Synechocystis strains used in different laboratories can vary in their genome and phenotypic characteristics, including glucose sensitivity (see for example29,81). As the input and output pathways of the newly discovered KaiA3B3C3 system are unknown, it is possible that mutations in different wild-type variants lead to variations in the expression of phenotypic effects in clock mutants. However, oscillations were observed not only in one particular laboratory strain, but also in different Synechocystis variants using different equipment (Fig. 4 and Fig.7). Another reason why we used different Synechocystis variants in this study was to allow comparison with previous studies. In addition, different laboratory strains were better suited for specific analyses (e.g., recording backscatter signals required non-aggregating strains). In addition, mutations in kai genes have similar effects on the backscatter rhythm of the strain investigated here (originating from the University of Uppsala) and on bioluminescence rhythms in the strain reported by Zhao et al.17 (Vanderbuilt University). In both background strains, kaiA1B1C1 deletion abolished the recorded oscillations. Furthermore, kaiC3 deletion dampened backscatter rhythms with a reduced amplitude in the first cycle (Fig. 5). Interestingly, Zhao et al. also revealed a reduced amplitude peak in luminescence rhythms in their kaiC3 mutant strain17.
Both studies (ref. 17, this work) suggest that the KaiA1B1C1 system is the master clock in Synechocystis, and that the KaiA3B3C3 system provides some redundancy and might stabilize oscillations. The nature of backscattering rhythms is not clearly resolved yet, but was suggested to be related to glycogen metabolism, because glycogen is known to display circadian synthesis and degradation rhythms in Synechococcus68,82,83. Consistent with this, no backscattering oscillations were detected in a glgC mutant, which is defective in glycogen synthesis53,84. Alternatively, the oscillations might reflect cell division, which is known to be regulated in a circadian fashion in Synechococcus85,86,87. Circadian clock systems that are composed of multiple oscillators are widespread in multicellular eukaryotic organisms88. Coupling of oscillators between cells has also been observed in filamentous cyanobacteria, but not between unicellular Synechococcus cells89,90. Early studies using the dinoflagellate Gonyaulax polyedra (renamed Lingulodinium polyedra) have suggested that at least two oscillators can exist in a unicellular organism91,92. Two detectable circadian rhythms (bioluminescence and aggregation) were relatively independent, with different periods and phase responses under certain conditions, and hence, able to decouple. Therefore, the authors concluded that separate oscillators individually control each rhythm91,92,93. The molecular oscillators have not been identified yet94. In Synechocystis, the deletion of each single Kai system had different effects on the period of backscatter oscillation (Fig. 5), which implies that two distinct oscillators exist. The KaiA3B3C3 oscillator may drive the long-period oscillation with an extremely low amplitude, which we detected in the kaiA1B1C1 mutant. The KaiA1B1C1 system might generate a dampened rhythm in the kaiA3B3C3 deletion strain, which started with a reduced period compared to the wild type. In the wild type, the phosphorylation cycles of KaiC1 and KaiC3 were synchronized, and a 24 h backscatter rhythm was detected, indicating that both systems operated jointly to regulate the same functions within a coupled system. However, we cannot rule out the possibility that the two systems function separately or have different phases under untested conditions. In eukaryotic organisms, circadian clock systems have been described that are composed of multiple oscillators88. This allows oscillators to respond to different environmental signals and to control rhythms in different output paths. The coupling strength of multiple oscillators balances the stability and precision of the timing mechanism with the flexibility of entrainment95. For Lingulodinium polyedra, it has been suggested that the coupling of the two oscillators provides higher adaptivity to the availability of resources93. In particular, the connection between circadian rhythms and metabolism opens up new perspectives in the field96.
In summary, we demonstrated that KaiA3 is a novel KaiA homolog and an element of the KaiC3-based signaling pathway with canonical KaiA functions. The N-terminal half of KaiA3 may still have a response regulatory function and may connect the whole system to other regulatory elements. KaiA3 must be located within the regulatory and metabolic networks of Synechocystis. Finally, our findings demonstrate that Synechocystis encodes two KaiABC-based protein oscillators. Both systems are required to drive rhythmicity and ensure the growth of Synechocystis with exogenously supplied glucose. Compared to the well-understood Synechococcus circadian clock system, the more versatile lifestyle of Synechocystis may require a more complex and redundant regulatory network.
Methods
Reciprocal BLAST of Sll0485 (KaiA3) and Slr1783 (Rre1)
Reciprocal BLAST analysis was performed as described at https://doi.org/10.17504/protocols.io.q3rdym6 and by Schmelling et al.27 The 2017 database was used for comparison with existing data on other circadian clock proteins. The protein sequences of Sll0485 (KaiA3) and Slr1783 (Rre1), as references for NarL response regulators44 from Synechocystis, were used as query sequences for this reciprocal BLAST search.
Co-occurrence analysis
The co-occurrence of KaiA3 with other circadian clock proteins in cyanobacteria containing KaiC1 was examined according to Schmelling et al.27. A right-sided Fisher’s exact test was used97. P-values were corrected for multiple testing after Benjamini-Hochberg98, with an excepted false discovery rate of 10−2. All proteins were clustered according to their corrected p-values.
Synteny analyses using SyntTax
The conservation of gene order was analyzed using the web tool ‘SyntTax’99. If not mentioned otherwise, default settings (Best match, 10 % norm. BLAST) were applied. Chromosomes were selected manually according to the results of Schmelling et al.27.
Multiple sequence alignments with Mafft and Jalview
Sequence alignments, visualization, and analysis were performed with ‘Jalview’100. The sequences were aligned with Mafft, and if not mentioned otherwise, default settings (L-INS-i, pairwise alignment computation method - localpair using Smith-Waterman algorithm, gap opening penalty: 1.53, gap opening penalty at local pairwise alignment: -2.00, group-to-group gap extension penalty: 0.123, matrix: BLOSUM62) were applied101. For analyses of the C-terminus, alignments were trimmed to position 168 in the KaiA reference sequence of Synechococcus. After trimming, the alignment was recalculated with Mafft, using the default parameters mentioned above.
2D and 3D structure predictions
The alignments generated in Jalview were then used with ‘Ali2D’ for secondary structure prediction102 (https://toolkit.tuebingen.mpg.de). The identity cut-off to invoke a new PSIPRED run was set to 30%. Three-dimensional protein structures were modeled using either Phyre2 or SWISS-MODEL103,104 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index; https://swissmodel.expasy.org/). The resulting structures were analyzed and illustrated using UCSF Chimera105 (https://www.cgl.ucsf.edu/chimera/).
Phylogenetic reconstruction of protein trees
Phylogenetic reconstruction of the protein trees of Sll0485 (KaiA3), Slr1783 (Rre1)/NarL (E. coli, UniProtKB - P0AF28), and KaiA was achieved with MEGA X106,107 using the above constructed alignments. For all alignments, a neighbor-joining tree and maximum likelihood tree were constructed and compared. To construct neighbor-joining trees, 1000 bootstrap iterations with a p-distance substitution model and a gamma distribution with three gamma parameters were used. To construct maximum likelihood trees, an initial tree was constructed using the maximum parsimony algorithm. Further trees were constructed using 1000 bootstrap iterations with an LG-G substitution model, a gamma distribution with three gamma parameters, and nearest-neighbor-interchange (NNI) as the heuristic method.
Yeast two-hybrid assay
AH109 yeast cells (Clontech) were used for YTH experiments. Transformation of yeast cells was performed according to the manufacturer’s guidelines using the Frozen-EZ Yeast Transformation Kit (Zymo Research). Genes of interest were amplified from wild-type genomic DNA using Phusion Polymerase (NEB), according to the manufacturer’s guidelines. The indicated restriction sites were introduced using oligonucleotides listed in Table S1A. Vectors and PCR fragments were cut with the respective restriction enzymes, and the gene of interest was ligated into the vector, leading to a fusion protein with a GAL4 activation domain (AD) or GAL4 DNA-binding domain (BD) either at the N- or C-terminus. All constructed plasmids are listed in Table S1B. The detailed protocol for the growth assay can be found at https://doi.org/10.17504/protocols.io.wcnfave26. Successfully transformed cells were selected on a complete supplement mixture (CSM) lacking leucine and tryptophan (-Leu -Trp) dropout medium (MP Biochemicals) at 30 °C for 3–4 days. Cells containing bait and prey plasmids were streaked on CSM lacking leucine, tryptophan, and histidine (-Leu -Trp -His) dropout medium (MP Biochemicals) with the addition of 12.5 mM 3-amino-1,2,4-triazole (3-AT, Roth) and incubated for 6 days at 30 °C to screen for interactions.
Expression and purification of recombinant Kai proteins
Synechocystis KaiB3, KaiB1 and Synechococcus KaiA (plasmids kindly provided by T. Kondo, Nagoya University, Japan) were produced as GST-fusion proteins in E. coli BL21(DE3) as described at https://doi.org/10.17504/protocols.io.48ggztw26. Briefly, proteins were purified by affinity chromatography using glutathione-agarose 4 B (Macherey and Nagel), and the N-terminal GST-tag was removed using PreScission Protease (Cytiva) prior to elution of the untagged proteins from the glutathione resin. Synechocystis KaiC3 was produced with an N-terminal- Strep-tag (Strep-KaiC3) in E. coli Rosetta-gami B (DE3) cells and purified via affinity chromatography using Strep-Tactin XT superflow (IBA-Lifesciences) (see https://doi.org/10.17504/protocols.io.meac3ae26). The Synechocystis ORF sll0485, encoding KaiA3, was inserted into the vector pET22b to create a C-terminal His6-fusion. KaiA3-His6 was expressed in E. coli Tuner (DE3) cells and purified by immobilized metal affinity chromatography (IMAC) using PureProteome™ Nickel Magnetic Beads (Millipore). For a detailed protocol, see https://doi.org/10.17504/protocols.io.bu5bny2n. Recombinant proteins were stored at −80 °C in buffer containing 20 mM Tris, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, and 1 mM ATP.
KaiC3 phosphorylation in in vitro assays and liquid chromatography-mass spectrometry (LC-MS/MS)
Recombinant Strep-KaiC3 purified from E. coli exists mainly in its phosphorylated form (KaiC3-P). Fully dephosphorylated Strep-KaiC3 (KaiC3-NP) was generated by incubating the protein for 18 h at 30 °C in assay buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, and 1 mM ATP). The autokinase activity of KaiC3-NP was investigated by incubating 0.2 µg/µl KaiC3 for 16 h at 30 °C in 20 µl assay buffer in the presence or absence of 0.1 µg/µl KaiA3-His6, KaiB3, KaiB1 and Synechococcus KaiA, respectively. Aliquots of 10 µl were taken before and after incubation at 30 °C, and the reaction was stopped with SDS sample buffer. Samples were stored at −20°C prior to application to a high resolution LowC SDS gel (10% T, 0.67% C)108 using the Hoefer Mighty Small II gel electrophoresis system and Tris-Tricine running buffer (cathode buffer: 100 mM Tris, 100 mM Tricine, 0.1 % SDS, pH 8.25; anode buffer: 100 mM Tris, pH 8.9, according to Schägger and von Jagow109). Gels were stained with Coomassie Blue R.
For the 48–60 h assay, pools containing 0.2 µg/µl (3.4 µM) KaiC3-NP, 0.1 µg/µl KaiB3 (7.4 µM) and various concentrations of KaiA3-His6 (corresponding to 0.5–8.4 µM) were prepared in assay buffer supplemented with 5 mM ATP, split in 10 µl aliquots for the desired timepoints and stored at −80 °C. Samples were thawed on ice for 10 min prior to incubation at 25 °C, 30 °C, or 35°C for different time periods. The reaction was stopped at specific time points by adding SDS sample buffer. Samples were stored at −80 °C prior to application to a LowC SDS gel (10% T, 0.67% C)27 using the Biorad Mini PROTEAN gel electrophoresis system and Tris-glycine running buffer (25 mM Tris, 192 mM glycine, 0.1 % SDS, according to Laemmli110). The gels were stained with ROTI®Blue quick stain. In Tris-glycine buffer, three KaiC3 bands were separated, whereas two KaiC3 bands were separated in Tris-tricine buffer. Gels were imaged using a Bio-Rad’s ChemiDoc XRS+ Imaging System, and densitometric analysis was performed in ImageLab 6.1. (Bio-Rad). The ratio of PP-KaiC3 to total KaiC3 was calculated in Excel and plotted as the average using GraphPad Prism 10.2.3. To evaluate the band patterns of PP-KaiC3 and NP-KaiC3, KaiC3 was incubated with 10U/µl Lambda phosphatase (NEB) and 1 mM MnCl2 for 14–18 h at 30 °C. As a control, Lambda phosphatase activity was blocked by the addition of PhosSTOP (Roche), or PhosSTOP (Roche) and 10 mM vanadate.
For LC-MS/MS analysis of KaiC3 phosphorylation sites, Strep-KaiC3 and KaiA3 were co-incubated in vitro as described above. Samples were taken directly after mixing and after 2 and 6 h of incubation, and separated by SDS-PAGE. For each sample, gel regions containing proteins of the size of Strep-KaiC were cut using a scalpel. For the 6 h time point, a gel region at the potential size of the Strep-KaiC3/A3 complex was also extracted. In-gel protein digestion with trypsin was performed according to the protocol described by Shevchenko et al.111. In brief, gel bands were treated with dithiothreitol and subsequently with iodoacetamide to reduce disulfide bonds and irreversibly alkylate the resulting cysteine thiol groups. Proteomics grade trypsin (Promega) was added to digest proteins overnight at 37 °C. The generated peptides were extracted and purified using the stage tip protocol112. Of the resulting peptide solution, 20% was used for nanoLC-MS/MS analysis. Therefore, peptides were separated in a 37 min reverse-phase linear gradient and directly ionized in an online coupled ESI source upon elution for analysis on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) operated in data-dependent acquisition mode. The 12 most abundant multiply charged ions in each full scan were separately fragmented by HCD, and the generated fragment ions were analyzed in consecutive MS/MS scans. Raw data files were processed using MaxQuant software (version 1.5.2.8) and default settings. Phosphorylation of Ser, Thr, and Tyr was defined as a variable modification. The acquired m/z spectra were searched against the proteome databases of Synechocystis and E. coli (downloaded from Cyanobase and Uniprot, respectively). Annotated MS/MS spectra were visualized using the MaxQuant integrated viewer.
Clear native protein PAGE and immunodetection
Kai proteins (containing 0.2 µg/µl dephosphorylated Strep-KaiC3, 0.1 µg/µl KaiA3-His6, KaiB3, KaiB1 or Synechococcus KaiA, respectively) were incubated for 16 h at 30 °C in phosphorylation assay buffer, followed by separation of the native proteins in 4–16% native PAGE at 4 °C using a clear native buffer system (Serva) without anionic dye. Thus, only proteins with a pI<7 at physiological pH were separated. Protein bands were visualized with Coomassie staining (ROTIBlue Quick, Carl Roth) or immunodetected with a monoclonal anti-6x-His Tag antibody conjugated to HRP (MA1-21315-HRP, Thermo Fisher, LOT number YH374751, 1:2000 diluted) and imaged using a ChemiDoc XRS+ Imaging System (BioRad). A detailed protocol can be found at https://doi.org/10.17504/protocols.io.bu67nzhn.
Strains and growth conditions
Three laboratory strains of Synechocystis PCC 6803 were used in this study: PCC-M (resequenced29), ‘Uppsala’ (kindly provided by Pia Lindberg, Uppsala University), and’Chicago’ (kindly provided by Dr. Amin Omairi-Nasser, University of Chicago).
Wild-type Synechocystis (PCC-M), the derived deletion strains ΔrpaA38, ΔkaiC326, ΔkaiA3, and ΔkaiA3B3C3 (Fig. S1), and complementation strain ΔkaiA3/kaiA3 (Fig. S1) were cultured photoautotrophically in BG11 medium113 supplemented with 10 mM TES buffer (pH 8) under constant illumination with 75 μmol photons m-2 s-1 white light (Philips TLD Super 80/840) at 30 °C. The cells were grown in Erlenmeyer flasks with constant shaking (140 rpm) or on plates (0.75% Bacto-Agar; Difco) supplemented with 0.3% thiosulfate. For photomixotrophic experiments, 0.2% glucose was added to the plates. For chemoheterotrophic growth experiments in complete darkness, Synechocystis cells were spotted at different dilutions on BG11 agar plates containing 0.2% glucose and incubated either mixotrophically for three days with continuous illumination or chemoheterotrophically in the dark for 26 days.
Wild-type Synechocystis (Chicago) and its derived kaiC mutant strains were grown in BG-11 M liquid medium supplemented with 20 mM HEPES (pH 8.0) at 30 °C with shaking (165 rpm) in a Percival incubator under a light intensity of ~50 µmol m−2s−1, provided by cool white fluorescent light bulbs (Philips Alto II, USA). The kaiC mutant strains were grown in the presence of the appropriate antibiotics.
Wild-type Synechocystis (Uppsala) and it’s derived deletions strains ∆kaiA1B1C153, ∆kaiA3B3C3, ∆kaiA3, ∆kaiB3, ∆kaiC3, ∆kaiA3B3, ∆kaiA3C3, ∆kaiB3C3 were grown in BG11 medium supplemented with 10 mM TES buffer (pH 8) in a Multitron Infors HT® Incubator at 30 °C, 0.5% CO2, and 75% humidity under constant illumination with 80 μmol photons m−2 s−1 of white light. Growth occurred on plates (0.75% Bacto-Agar; Difco) or in Erlenmeyer flasks with shaking at 150 rpm. For backscatter measurements, cultures were incubated in a biolector equipped with a light array module (Beckman Coulter) (see Detection of in vivo oscillations via backscatter analysis).
Construction of mutants of the KaiC3 based clock system
Mutants constructed in the Synechocystis (PCC-M) background strain
To construct the kaiA3 (sll0485) deletion strain, Synechocystis wild-type cells were transformed with the plasmid pUC19-Δsll0485. For plasmid construction, PCR products were generated using the oligonucleotides P13/P14 and pUC19 as template, P15/16 and P19/20 with genomic Synechocystis wild-type DNA as template and P17/25 with pUC4K as template. Homologous recombination led to the replacement of the kaiA3 gene with a kanamycin resistance cassette (Fig. S1). For genomic complementation of the ΔkaiA3 strain, cells were transformed with the plasmid pUC19-Δsll0485-compl. Overlapping fragments were generated using oligonucleotides P15/28 and P24/32 with genomic Synechocystis wild-type DNA as template, P13/26 and pUC19 as template, and P22/23 and the vector pACYC184 as template. In the resulting complementation strain ΔkaiA3/kaiA3, the kanamycin resistance cassette was replaced with kaiA3, and a chloramphenicol resistance cassette was introduced downstream of the kaiB3 gene (Fig. S1). For the triple-knockout mutant ΔkaiA3B3C3, ΔkaiC3 cells were used as the background strain for transformation with the pUC19-ΔkaiA3B3 plasmid. PCR products were generated using the oligonucleotides P13/26 and pUC19 as template, P17/27 and pUC4K as template, P15/16 and P25/28 with genomic Synechocystis wild-type DNA as template. The operon kaiA3kaiB3 was replaced with a kanamycin resistance cassette (Fig. S1). Complete segregation of the mutant alleles was confirmed using PCR. For the ΔkaiA3 strain, oligonucleotides P15/29 were used. Segregation of the complementation strain was confirmed by PCR with P15/29, P30/31, and P19/32. For the triple knockout mutant ΔkaiA3B3C3, deletion of the kaiA3B3 operon was confirmed by PCR using the primer pairs P15/33 and P19/30. The kaiA3B3 chromosomal region of the mutants is shown in Fig. S1. Ectopic expression of kaiA3 was achieved in wild-type and ΔkaiA3 cells after transformation with the plasmid pUR-NFLAG-sll0485. The plasmid was constructed via restriction digestion of the vector pUR-N-Flag-xyz, and the PCR product was amplified with the oligonucleotide pair P29-P34 using genomic Synechocystis wild-type DNA as a template. Restriction digestion using EcoRI and BamHI was followed by ligation. Successful transformation was confirmed by PCR using P35/36. The oligonucleotides and plasmids used are listed in Table S1.
Mutants constructed in the Synechocystis (Chicago) background strain
The kaiC1 gene was amplified from Synechocystis genomic DNA using oligonucleotide primers P41/42. The kaiC1 DNA fragment (1566 bp) amplified by P41/42 was cloned into the pGEMT-Easy plasmid (pGEMT-Easy+ kaiC1 plasmid). A kanamycin gene cassette was amplified using primers P43/44) and PsasA-Nina plasmid (kindly provided by Carl H. Johnson) as a template. The kaiC1 ko plasmid was constructed by Gibson assembly using the following DNA fragments: a KpnI-digested pGEMT-Easy+ kaiC1 plasmid backbone and a 1340 bp sequence fragment of the kanamycin cassette, resulting in pGEMT-∆kaiC1. The pUC19_ΔkaiC3 plasmid (table S1) was modified and the chloramphenicol cassette was removed and replaced with a kanamycin cassette. The knockout plasmids were incorporated into the cyanobacterial genome by natural transformation30. Fully segregated mutants were generated by streaking the transformants multiple times with appropriate antibiotics.
Mutants constructed in the Synechocystis (Uppsala) background strain
To generate the plasmid pUC19-ΔkaiB3, PCR products containing overlapping fragments were produced using P37/38 and P25/28 and wild-type DNA as a template. The kanamycin resistance cassette, substituting kaiB3, was amplified from pUC4K using oligonucleotides P27/39. The pUC19 backbone was amplified using P26/40. The plasmid was assembled by ligating overlapping fragments. Wild-type Synechocystis (Uppsala) was transformed with plasmids pUC19_Δsll0485, pUC19_ΔkaiB3, pUC19_ΔkaiC3, or pUC19_ΔkaiA3B3 to generate deletion strains ∆kaiA3, ∆kaiB3, ∆kaiC3, ∆kaiA3B3, respectively (see table S1). Transformation of Synechocystis ∆kaiC3 with plasmids pUC19_ΔkaiA3B3, pUC19_Δsll0485 or pUC19_ΔkaiB3 was performed to obtain ∆kaiA3B3C3, ∆kaiA3C3, and ∆kaiB3C3, respectively. Segregation was achieved by selection on plates with appropriate antibiotics.
Detection of in vivo oscillations via SDS-PAGE and immunodetection
Wild-type Synechocystis sp. PCC 6803 strain (Chicago)and derived kaiC mutant strains were exposed to two LD cycles (12 L:12D) to synchronize the circadian clock, then released to constant light conditions (LL) (∼50 µmol m−2 s−1) and cultures were harvested (centrifuged at 2800 × g for 10 min at room temperature) every 4 h. Cell pellets were flash frozen in liquid nitrogen and stored at −80 °C until samples were processed for western blot analysis. Total protein extraction and western blot analysis were performed as previously described23. In brief, each frozen cell pellet was resuspended in 200 μl of lysis buffer (8 M urea, 20 mM HEPES pH 8.0) and cells were broken by vigorous vortexing (30 s vortex and 1 min cooling in ice for 7 cycles) with glass beads (0.1 mm, acid-washed). The supernatant fraction containing total protein was collected after centrifugation (1000 × g for 3 min at 4 °C) the cell suspension. The protein concentration was measured using the Bradford assay. Equal amounts of total protein (8 μg) were mixed with 4X DTT-containing SDS polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The samples were heated at 95 °C for 4 min and were loaded onto big-format SDS-PAGE gels (10%). The gel was run for 4.5 h at a constant current of 35 mA per gel and 12 °C for protein separation. The protein samples were transferred onto PVDF membranes, blocked with 4% w/v nonfat dry milk/ Tris-buffered saline with 0.1% Tween-20 (TBS-T) for 2 h at room temperature. Membranes were then incubated overnight at 4 °C with anti-KaiC1 (1:3750 dilutions in TBST) and anti-KaiC3 antibody (1:7500 dilutions in TBST). Membranes were washed 3 times (20 minutes each at room temperature) in TBST and finally probed with Goat anti-rabbit IgG (H + L) Secondary Antibody, HRP (Thermo Fisher Scientific, 1:10000 dilution). Chemiluminescent detection was performed using Pierce SuperSignal West Pico detection reagent (Thermo Scientific). Blots were photographed using a ChemiDoc MP Imaging System (Bio-Rad). Total protein abundance and the ratio of PP-KaiC to total KaiC protein were estimated by densitometry of the blot images and plotted using GraphPad Prism 9.5.1.
Detection of in vivo oscillations via Phos-tag SDS-PAGE and immunodetection
To analyze the in vivo phosphorylation of KaiC3, Synechocystis (PCC-M) wild type, ΔkaiA3, and ΔkaiC3 cells were cultivated in BG11 or copper-depleted medium for kaiA3 overexpression. The cells were grown in two consecutive 12 h LD cycles followed by 24 h of LL conditions. After the initial 12 h LD cycle, 10 ml of cells were collected every 6 h for analysis. The cells were cooled in liquid nitrogen for 5 s and harvested by centrifugation (3220 × g, 2 min, 4 °C). The pellet was frozen in liquid nitrogen and stored at −20 °C until further processing. To lyse the cells, the pellets were resuspended to an OD750 of 25 in phosphorylation buffer (50 mM NaOH-HEPES pH 7.5, 300 mM NaCl, 0.5 mM Tris-(2-carboxyethyl)-phosphine, 10 mM MgCl2). The cells were disrupted twice in a cell mill at 30 Hz for 1 min at 4 °C, using glass beads. The crude cell extract was obtained by centrifugation (500 × g, 1 min, 4 °C). For mobility shift detection of phosphorylated and dephosphorylated KaiC3, a Zn2+-Phos-tag® SDS-PAGE assay (Wako Chemicals) was used. A 9% SDS-PAGE gel containing 25 µM Phos-tag acrylamide was prepared and 12 µL of the cell extract was run at 150 V for 3 h at 4 °C. Proteins were blotted onto a nitrocellulose membrane (Amersham Protran®) using wet blotting. The membranes were blocked with 5% dry milk in TBS-T for 1 h at room temperature. Immunodetection was performed using the αKaiC328 antibody(1:7500 in TBS-T) at 4 °C overnight, and subsequently with anti-rabbit αHRP (Thermo Fisher Scientific Inc., USA) antibody (1:20000 in TBS-T) for 2 h at room temperature. Between the steps, the membrane was washed with TBS-T for 10 min at room temperature. Protein detection was performed using Pierce ECL western blotting Substrate (Thermo Scientific). The blots were visualized using a Fusion SL chemiluminescence detector (Vilber Lourmat). The ratios of phosphorylated to non-phosphorylated KaiC3 were quantified via mean gray value measurements (ImageJ114) and plotted as average with GraphPad Prism 10.2.3.
Detection of in vivo oscillations via backscatter analysis
Prior to monitoring the backscatter properties of wild-type Synechocystis (Uppsala variant) and the indicated kai deletion mutants, cells were grown for 10 days under LL as two subsequent pre-cultures in BG11 medium without antibiotics. For pre-culture 1, cells from agar plates were inoculated in 20 ml medium and grown in a 100 ml Erlenmeyer flask under constant illumination with 80 µmol photons m−2 s−1 in a Multitron Infors HT® incubator set to 30 °C, 75% humidity, 150 rpm and 0.5% CO2 supply. After 7 days, cultures reached an OD750nm of ~4-6. For pre-culture 2, the cells were diluted to OD750 nm of approx. 0.6 (Fig. 5a–f) or 0.7 (Fig. S10) in a total volume of 100 ml medium and incubated for 3 further days in 250 ml Erlenmeyer flasks under the same conditions. The cells reached an OD750nm of ~1.8-2.4. At the start of the experiment, cells from pre-culture 2 were diluted to an OD750nm of 0.9 with BG11 without antibiotics. Per strain, 4-5 wells of a 48-well FlowerPlate MTP (Beckman Coulter) were filled with 1 ml cells each (originating from the same pre-culture), the plate was sealed with gas-permeable foils and transferred to a BioLector XT microbioreactor equipped with a Light Array Module (LAM) (Beckmann Coulter). This process took approx. 1-2 hours. Cells were incubated at 30 °C and 600 rpm, with a flow of 20 ml min-1 of humidity-controlled air with 1% CO2. The LAM module was programmed to roughly mimic the light spectrum of LEDs in the Infors HT® incubator and was set to a constant illumination of ~80 µmol m−2 s−1. Backscatter at 730 nm (gain 6) was measured every 5 min and analyzed for 84 h. The BioLection and LUA protocols can be found together with the raw data as Supplementary Data S4. All optical densities mentioned above were measured in a Specord 200 plus (Analytic Jena) using dilutions that allowed measurements in the range of OD750nm 0.2-0.7.
Processing of the data has been done using Python (version 3.9.16), with packages: pandas (version 1.5.3), matplotlib (version 3.7.1), regex (version 2022.7.9), numpy (version 1.24.3), scipy (version 1.10.1), scikit-learn (version 1.2.2). The script53 was modified and is available at https://github.com/flo-sti/cyano-backscatter115. The backscatter data were imported from.xlsx files and raw data were plotted. To isolate the oscillation signal, we removed growth effects using sklearn.LinearRegression. A fourth degree polynomial was fitted to the observed raw backscatter values, excluding data from the first 3 h. Regression values were predicted using sklearn.predict, and subtracted from the observed raw backscatter values. The resulting signal was normalized by subtracting the arithmetic mean and smoothed with the numpy.convolve function with a kernel size of 40 and the mode ‘same’. The smoothened signal was used for further graphical and statistical analysis.
To determine the period of the oscillation for each strain, we first attempted to fit an equation for a simple harmonic oscillation of the following form to the data (A: amplitude, ω: angular frequency, φ:phase angle):
The fit was performed with the scipy.optimize.fit_curve function in Python, and the resulting parameters were used to generate a curve that could be plotted along with the signal. The boundaries for the fit_curve function were chosen as follows: amplitude: 0 ≤ A ≤ max(data) – mean(data); angular frequency: 2*π/(maximum estimated period) ≤ ω ≤ 2*π/(minimum estimated period) with the minimum estimated period between 18 h and 35 h and the maximum estimated period between 26 h and 45 h depending on the strain; phase angle: 2*π/12 < = φ < = 2*π/1. The formula and amplitude estimates were based on the study by Santos et al.116. This fit performed well for the wild type, ΔkaiA1B1C1, and ΔkaiB3 mutants and was used to derive the period of these strains, but the fit failed to accurately capture the phenotype of the kaiA3 and kaiC3 knockouts.
To calculate the length of the first period for strains that could not be fitted with a simple harmonic oscillation, the scipy.find_peaks function (distance=150; Height=50) was used to identify the first trough and peak. Since the time difference between these values is only half of a period, the result was multiplied by two and the absolute value was taken. This method was chosen to measure the period length because it was sometimes impossible to determine the later peaks using the same parameters for each strain. However, the first trough and peak were detected reliably. The values of the length of the first period of all strains for all three experiments were collected, and the arithmetic mean, median, and standard deviation were calculated. Pairwise statistical tests were performed for each strain combination. First, a Levene’s test was performed to determine whether the compared strains had the same variance. If the variance was the same, a student’s t-test was performed. If the variances differed significantly (p < 0.05), a Welch’s t-test was performed. Differences between two strains were considered significant if the p-value of the statistical test was <0.05.
To determine the phase shift and relative amplitude after synchronization in comparison to the wild type for all strains, independent of whether they displayed an undampened or dampened oscillation, peaks were identified using the scipy.find_peaks function with a distance of 150, width of 65, and a minimum height of the mean of the signal + 20 % of the maximum value. The time point and height of the first peak were determined, and for each strain, the arithmetic mean and standard deviation (SD) of the 4-5 replicates within one experiment were calculated. Peak detection in the normalized backscatter data of the kaiA1B1C1 mutant was difficult due to the low amplitude. In the first two experiments, we observed a broad peak with a slightly higher bump in the front compared with the harmonic fit (Fig. 5e) and used the first detectable peak to calculate the phase shift and amplitude. For each experiment, the results were correlated with the wild type as follows:
Screening of KaiC3 and KaiC1 binding partners by immunoprecipitation-coupled liquid chromatography mass spectrometry (LC-MS/MS)
Synechocystis (PCC-M) WT/FLAG-kaiC3, WT/FLAG-kaiC1, and WT/FLAG-sfGFP (control) strains were cultivated in BG11 medium (100 ml, copper depleted), harvested by centrifugation at 6000 × g for 10 min at 4 °C and resuspended in purification buffer (50 mM HEPES/NaOH (pH 7.5), 5 mM MgCl2, 25 mM CaCl2, 10 % (v/v) glycerol, 150 mM NaCl, 5 mg ml−1 6-aminohexaonic acid, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 4 mM p-aminobenzamidine, 1 mM ATP). Cells were disrupted in a mixer mill, followed by solubilization with n-dodecyl-β-maltoside (detergent-to chlorophyll ratio 20:1) for 1 h at 4 °C. The supernatant was used for FLAG purification in pull-down assays with Anti-Flag® M2 Magnetic Beads (Sigma-Aldrich), following the manufacturer’s protocol. The resulting elution fractions were loaded onto a NuPAGE™ Bis-Tris Gel and run following the manufacturer’s protocol (Invitrogen). Protein bands were allowed to migrate for only a short distance of approximately 10 mm. After staining the gel for 60 min with InstantBlue™ (Expedeon), the protein-containing gel regions were excised. Two independent replicates were produced for each condition (KaiC3, KaiC1, and control pull-down). In-gel protein digestion with trypsin was performed as described above, and the resulting peptide solutions were purified using stage tips. Approximately 20% of the sample was subjected to nanoLC-MS/MS analysis as described above on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) operated in data-dependent acquisition mode. Raw data of KaiC3 or KaiC1 pull-downs were separately processed using the MaxQuant software (version 1.5.2.8) embedded MaxLFQ algorithm as described by Cox et al.117. Raw spectra were searched against the proteome databases of Synechocystis and E. coli (downloaded from Cyanobase and Uniprot, respectively) and bait protein sequences. Significantly enriched proteins were identified using Perseus software (version 1.6.5.0) significance B analysis with a p-value of 0.01.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw and processed data generated in this study are available on figshare: Raw data: https://doi.org/10.6084/m9.figshare.25218143. Processed Data: https://doi.org/10.6084/m9.figshare.25218137, Alignments: https://doi.org/10.6084/m9.figshare.25218122, Phylogeny: https://doi.org/10.6084/m9.figshare.25218134), processed KaiA3 hits are also available as Supplementary Data S1. The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository118, with the dataset identifier PXD042846 (analysis of KaiC3 phosphorylation) https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/07/PXD042846, PXD042845 (screening of KaiC3 and KaiC1 binding partners) https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/07/PXD042845, and summarized data are available as Supplementary Data S2 and S3. Datasets S1 to S4 are available as Supplementary Data. Source data are provided with this paper.
Code availability
The script for reciprocal BLAST analysis is available at https://github.com/schmelling/reciprocal_BLAST/blob/master/notebooks/. The script for backscatter data analysis is available at https://github.com/flo-sti/cyano-backscatter115. Raw data, Biolection and LUA protocols are available as Supplementary Data S4.
References
Nakajima, M. et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 (2005).
Egli, M. et al. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry 51, 1547–1558 (2012).
Pattanayek, R. et al. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol. Cell 15, 375–388 (2004).
Terauchi, K. et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl Acad. Sci. USA 104, 16377–16381 (2007).
Kim, Y. I., Dong, G., Carruthers, C. W., Golden, S. S. & LiWang, A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc. Natl Acad. Sci. USA 105, 12825–12830 (2008).
Iwasaki, H., Nishiwaki, T., Kitayama, Y., Nakajima, M. & Kondo, T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl Acad. Sci. USA 99, 15788–15793 (2002).
Pattanayek, R. & Egli, M. Protein–protein interactions in the cyanobacterial circadian clock: structure of KaiA dimer in complex with C-terminal KaiC peptides at 2.8 Å resolution. Biochemistry 54, 4575–4578 (2015).
Chang, Y.-G., Kuo, N.-W., Tseng, R. & LiWang, A. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc. Natl Acad. Sci. USA 108, 14431–14436 (2011).
Chang, Y.-G., Tseng, R., Kuo, N.-W. & LiWang, A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl Acad. Sci. USA 109, 16847–16851 (2012).
Nishiwaki, T. et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl Acad. Sci. USA 101, 13927–13932 (2004).
Kitayama, Y., Iwasaki, H., Nishiwaki, T. & Kondo, T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22, 2127–2134 (2003).
Tseng, R. et al. Cooperative KaiA–KaiB–KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J. Mol. Biol. 426, 389–402 (2014).
Rust, M. J., Markson, J. S., Lane, W. S., Fisher, D. S. & O’Shea, E. K. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318, 809–812 (2007).
Cohen, S. E. & Golden, S. S. Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 79, 373–385 (2015).
Swan, J. A., Golden, S. S., LiWang, A. & Partch, C. L. Structure, function, and mechanism of the core circadian clock in cyanobacteria. J. Biol. Chem. 293, 5026–5034 (2018).
Snijder, J. & Axmann, I. M. In Macromolecular Protein Complexes II: Structure and Function (eds Harris, J. R. & Marles-Wright, J.) (Springer, 2019).
Zhao, C., Xu, Y., Wang, B. & Johnson, C. H. Synechocystis: a model system for expanding the study of cyanobacterial circadian rhythms. Front. Physiol. 13, 1085959 (2022).
Schmelling, N. M., Scheurer, N., Köbler, C., Wilde, A. & Axmann I. M. in Circadian Rhythms in Bacteria and Microbiomes (eds Johnson, C. H. & Rust, M. J.) (Springer International Publishing, 2021).
Loza-Correa, M., Gomez-Valero, L. & Buchrieser, C. Circadian clock proteins in prokaryotes: hidden rhythms? Front. Microbiol. 1, 1–11 (2010).
Terrettaz, C., Cabete, B., Geiser, J., Valentini, M. & Gonzalez, D. KaiC-like proteins contribute to stress resistance and biofilm formation in environmental Pseudomonas species. Environ. Microbiol 25, 894–913 (2023).
Ma, P., Mori, T., Zhao, C., Thiel, T. & Johnson, C. H. Evolution of KaiC-dependent timekeepers: a proto-circadian timing mechanism confers adaptive fitness in the purple bacterium Rhodopseudomonas palustris. PLoS Genet. 12, e1005922 (2016).
Pitsawong, W. et al. From primordial clocks to circadian oscillators. Nature 616, 183–189 (2023).
Rust, M. J., Golden, S. S. & O’Shea, E. K. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 331, 220–223 (2011).
Kanesaki, Y. et al. Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 19, 67–79 (2012).
Aoki, S. & Onai, K. in Bacterial Circadian Programs (eds Ditty, J. L., Mackey, S. R. & Johnson C. H.) (Springer-Verlag, 2009).
Wiegard, A. et al. Synechocystis KaiC3 displays temperature- And KaiB-dependent ATPase activity and is important for growth in darkness. J. Bacteriol. 202, 1–36 (2020).
Schmelling, N. M. et al. Minimal tool set for a prokaryotic circadian clock. BMC Evolut. Biol. 17, 169 (2017).
Wiegard, A. et al. Biochemical analysis of three putative KaiC clock proteins from Synechocystis sp. PCC 6803 suggests their functional divergence. Microbiology 159, 948–958 (2013).
Trautmann, D., Voß, B., Wilde, A., Al-Babili, S. & Hess, W. R. Microevolution in cyanobacteria: Re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 19, 435–448 (2012).
Dörrich, A. K., Mitschke, J., Siadat, O. & Wilde, A. Deletion of the Synechocystis sp. PCC 6803 kaiAB1C1 gene cluster causes impaired cell growth under light-dark conditions. Microbiology 160, 2538–2550 (2014).
Axmann, I. M. et al. Biochemical evidence for a timing mechanism in Prochlorococcus. J. Bacteriol. 191, 5342–5347 (2009).
Holtzendorff, J. et al. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythms 23, 187–199 (2008).
Williams, S. B., Vakonakis, I., Golden, S. S. & LiWang, A. C. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc. Natl Acad. Sci. USA 99, 15357–15362 (2002).
Vakonakis, I. et al. NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction. Proc. Natl Acad. Sci. USA 101, 1479–1484 (2004).
Ye, S., Vakonakis, I., Ioerger, T. R., LiWang, A. C. & Sacchettini, J. C. Crystal structure of circadian clock protein KaiA from Synechococcus elongatus. J. Biol. Chem. 279, 20511–20518 (2004).
Kim, Y. I., Vinyard, D. J., Ananyev, G. M., Dismukes, G. C. & Golden, S. S. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc. Natl Acad. Sci. USA 109, 17765–17769 (2012).
Nishimura, H. et al. Mutations in KaiA, a clock protein, extend the period of circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 148, 2903–2909 (2002).
Köbler, C., Schultz, S. J., Kopp, D., Voigt, K. & Wilde, A. The role of the Synechocystis sp. PCC 6803 homolog of the circadian clock output regulator RpaA in day–night transitions. Mol. Microbiol. 110, 847–861 (2018).
Sato, S. S. et al. A large-scale protein protein interaction analysis in Synechocystis sp. PCC 6803. DNA Res. 14, 207–216 (2007).
Baikalov, I. et al. Structure of the Escherichia coli response regulator NarL. Biochemistry 35, 11053–11061 (1996).
Komarek, J., Kaštovský, J., Mares, J. & Johansen, J. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 86, 295–335 (2014).
Galperin, M. Y. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 (2006).
Dvornyk, V. & Mei, Q. Evolution of kaiA, a key circadian gene of cyanobacteria. Sci. Rep. 11, 9995 (2021).
Ashby, M. K. & Houmard, J. Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol. Mol. Biol. Rev. 70, 472–509 (2006).
Vakonakis, I. & LiWang, A. C. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc. Natl Acad. Sci. USA 101, 10925–10930 (2004).
Delumeau, O. et al. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J. Biol. Chem. 279, 40927–40937 (2004).
Iwasaki, H., Taniguchi, Y., Ishiura, M. & Kondo, T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 18, 1137–1145 (1999).
Clodong, S. et al. Functioning and robustness of a bacterial circadian clock. Mol. Syst. Biol. 3, 90 (2007).
Nakajima, M., Ito, H. & Kondo, T. In vitro regulation of circadian phosphorylation rhythm of cyanobacterial clock protein KaiC by KaiA and KaiB. FEBS Lett. 584, 898–902 (2010).
Tomita, J., Nakajima, M., Kondo, T. & Iwasaki, H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254 (2005).
Qin, X., Byrne, M., Xu, Y., Mori, T. & Johnson, C. H. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 8, e1000394 (2010).
Imai, K., Nishiwaki, T., Kondo, T. & Iwasaki, H. Circadian rhythms in the synthesis and degradation of a master clock protein KaiC in cyanobacteria. J. Biol. Chem. 279, 36534–36539 (2004).
Berwanger, L. C. et al. Self-sustained rhythmic behavior of Synechocystis PCC 6803 under continuous light conditions in the absence of light-dark entrainment. bioRxiv https://doi.org/10.1101/2023.09.26.559469 (2023).
Dvornyk, V., Vinogradova, O. & Nevo, E. Origin and evolution of circadian clock genes in prokaryotes. Proc. Natl Acad. Sci. USA 100, 2495–2500 (2003).
Géron, A., Werner, J., Wattiez, R. & Matallana-Surget, S. Towards the discovery of novel molecular clocks in Prokaryotes. Crit. Rev. Microbiol. 50, 491–503 (2024).
Van Praag, E., Degli Agosti, R. & Bachofen, R. Rhythmic activity of uptake hydrogenase in the prokaryote Rhodospirillum rubrum. J. Biol. Rhythms 15, 218–224 (2000).
Loza-Correa, M. et al. The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments. Environ. Microbiol. 16, 359–381 (2014).
Min, H., Guo, H. & Xiong, J. Rhythmic gene expression in a purple photosynthetic bacterium, Rhodobacter sphaeroides. FEBS Lett. 579, 808–812 (2005).
de Sousa Machado, J. N. et al. Autophosphorylation of the KaiC-like protein ArlH inhibits oligomerization and interaction with ArlI, the motor ATPase of the archaellum. Mol. Microbiol. 116, 943–956 (2021).
Brettschneider, C. et al. A sequestration feedback determines dynamics and temperature entrainment of the KaiABC circadian clock. Mol. Syst. Biol. 6, 389 (2010).
Qin, X. et al. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc. Natl Acad. Sci. USA 107, 14805–14810 (2010).
van Zon, J. S., Lubensky, D. K., Altena, P. R. & ten Wolde, P. R. An allosteric model of circadian KaiC phosphorylation. Proc. Natl Acad. Sci. USA 104, 7420–7425 (2007).
Dong, P. et al. A dynamic interaction process between KaiA and KaiC is critical to the cyanobacterial circadian oscillator. Sci. Rep. 6, 25129 (2016).
Yang, X., Kang, C. M., Brody, M. S. & Price, C. W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10, 2265–2275 (1996).
Eelderink-Chen, Z. et al. A circadian clock in a nonphotosynthetic prokaryote. Sci. Adv. 7, eabe2086 (2021).
Paulose, J. K., Wright, J. M., Patel, A. G. & Cassone, V. M. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS ONE 11, e0146643 (2016).
Schmitz, O., Katayama, M., Williams, S. B., Kondo, T. & Golden, S. S. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289, 765–768 (2000).
Pattanayak, GopalK., Phong, C. & Rust, MichaelJ. Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr. Biol. 24, 1934–1938 (2014).
Ivleva, N. B., Gao, T., Liwang, A. C. & Golden, S. S. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc. Natl Acad. Sci. USA 103, 17468–17473 (2006).
Kim, P. et al. CikA, an input pathway component, senses the oxidized quinone signal to generate phase delays in the cyanobacterial circadian clock. J. Biol. Rhythms 35, 227–234 (2020).
Pattanayak, GopalK., Lambert, G., Bernat, K. & Rust, MichaelJ. Controlling the cyanobacterial clock by synthetically rewiring metabolism. Cell Rep. 13, 2362–2367 (2015).
Haydon, M. J., Mielczarek, O., Robertson, F. C., Hubbard, K. E. & Webb, A. A. R. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692 (2013).
Johnson, C. H. & Egli, M. Metabolic compensation and circadian resilience in prokaryotic cyanobacteria. Annu. Rev. Biochem. 83, 221–247 (2014).
Anderson, S. L. & McIntosh, L. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J. Bacteriol. 173, 2761–2767 (1991).
Narikawa, R., Kohchi, T. & Ikeuchi, M. Characterization of the photoactive GAF domain of the CikA homolog (SyCikA, Slr1969) of the cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 7, 1253–1259 (2008).
Welkie, D. G. et al. Genome-wide fitness assessment during diurnal growth reveals an expanded role of the cyanobacterial circadian clock protein KaiA. Proc. Natl Acad. Sci. USA 115, E7174–E7183 (2018).
Xu, Y., Mori, T. & Johnson, C. H. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 22, 2117–2126 (2003).
Xu, Y. et al. Circadian Yin-Yang regulation and its manipulation to globally reprogram gene expression. Curr. Biol. 23, 2365–2374 (2013).
Singh, A. K. & Sherman, L. A. Pleiotropic effect of a histidine kinase on carbohydrate metabolism in Synechocystis sp. strain PCC 6803 and its requirement for heterotrophic growth. J. Bacteriol. 187, 2368–2376 (2005).
Scheurer, N. M. et al. Homologs of circadian clock proteins impact the metabolic switch between light and dark growth in the Cyanobacterium Synechocystis sp. PCC 6803. Front. Plant Sci. 12, 675227 (2021).
Tichý, M. et al. Strain of Synechocystis PCC 6803 with aberrant assembly of photosystem II contains tandem duplication of a large chromosomal region. Front Plant Sci. 7, 648 (2016).
Diamond, S., Jun, D., Rubin, B. E. & Golden, S. S. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc. Natl Acad. Sci. 112, E1916–E1925 (2015).
Puszynska, A. M. & O’Shea, E. K. Switching of metabolic programs in response to light availability is an essential function of the cyanobacterial circadian output pathway. eLife 6, e23210 (2017).
Gründel, M., Scheunemann, R., Lockau, W. & Zilliges, Y. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology (Reading) 158, 3032–3043 (2012).
Mori, T., Binder, B. & Johnson, C. H. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl Acad. Sci. USA 93, 10183–10188 (1996).
Kondo, T. et al. Circadian rhythms in rapidly dividing cyanobacteria. Science 275, 224–227 (1997).
Dong, G. et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140, 529–539 (2010).
Bell-Pedersen, D. et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556 (2005).
Mihalcescu, I., Hsing, W. & Leibler, S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature 430, 81–85 (2004).
Arbel-Goren, R. et al. Robust, coherent, and synchronized circadian clock-controlled oscillations along Anabaena filaments. Elife 10, e64348 (2021).
Roenneberg, T. & Morse, D. Two circadian oscillators in one cell. Nature 362, 362–364 (1993).
Morse, D., Hastings, J. W. & Roenneberg, T. Different phase responses of the two circadian oscillators in Gonyaulax. J. Biol. Rhythms 9, 263–274 (1994).
Roenneberg, T. The complex circadian system of Gonyaulax polyedra. Physiologia Plant. 96, 733–737 (1996).
Dagenais-Bellefeuille, S., Beauchemin, M. & Morse, D. miRNAs do not regulate circadian protein synthesis in the Dinoflagellate Lingulodinium polyedrum. PLoS ONE 12, e0168817 (2017).
Micklem, C. N. & Locke, J. C. W. Cut the noise or couple up: coordinating circadian and synthetic clocks. iScience 24, 103051 (2021).
Laothamatas, I., Rasmussen, E. S., Green, C. B. & Takahashi, J. S. Metabolic and chemical architecture of the mammalian circadian clock. Cell Chem. Biol. 30, 1033–1052 (2023).
Fisher, R. A. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 85, 87–94 (1922).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B: (Methodol.) 57, 289–300 (1995).
Oberto, J. SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinform. 14, 4 (2013).
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Gabler, F. et al. Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinform. 72, e108 (2020).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Stecher, G., Tamura, K. & Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239 (2020).
Nishiwaki, T. et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 26, 4029–4037 (2007).
Schägger, H. & von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 (1987).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Shevchenko, A., Tomas, H., Havliš, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2007).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Rippka, R., Deruelles, J., Herdman, M., Waterbury, J. B. & Stanier, R. Y. Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. Microbiology 111, 1–61 (1979).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Stirba, F. P. Two KaiABC systems control circadian oscillations in one cyanobacterium. Dataset and Code on Zenodo (2024).
De los Santos, H., Collins, E. J., Hurley, J. M. & Bennett, K. P. Circadian rhythms in Neurospora exhibit biologically relevant driven and damped harmonic oscillations. In: Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, 455–463 (Association for Computing Machinery, 2017).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 13, 2513–2526 (2014).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Acknowledgements
The authors thank all members of the Research Unit FOR2816 ‘SCyCode’ (The Autotrophy-Heterotrophy Switch in Cyanobacteria: Coherent Decision-Making at Multiple Regulatory Layers) for fruitful discussions and the German Research Foundation (DFG) for financial support (project number 397695561, A.W., B.M.). This work was further supported by grants from the DFG (WI2014/5-3 (A.W.); WI2014/10-1 and 10-2 (A.W.); AX 84/1-3 (I.M.A.); MA 4918/4-1 (B.M.)), and SFB1535 - Project ID 458090666 (I.M.A.). I.M.A. and A.Wie were further supported by the DFG under Germany’s Excellence Strategy – EXC-2048/1, project ID 390686111 (CEPLAS). NIH R01 GM135382 provided funding to M.J.R. We thank Pauline Morys, Katerine Cheronis, Annika Klopp, Isabell Bleile, and Werner Bigott for technical assistance. Figure 8 was created using BioRender.com.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
C.K., I.M.A., A. Wie, and A.W. designed the study. C.K., N.M.S., A.P., P.S., N.M.Sche., K.N.S., A. Wie, G.K.P., F.P.S., P.K., and L.B. performed and analyzed the experiments. All authors interpreted and discussed the data. C.K., N.M.S., A.P., P.S., G.K.P., M.J.R., B.M., A. Wie, I. M.A., and A.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interest.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Köbler, C., Schmelling, N.M., Wiegard, A. et al. Two KaiABC systems control circadian oscillations in one cyanobacterium. Nat Commun 15, 7674 (2024). https://doi.org/10.1038/s41467-024-51914-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51914-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.