Abstract
Cholera is a life-threatening gastrointestinal infection caused by a toxigenic bacterium, Vibrio cholerae. After a lull of almost 30 years, a first case of cholera was detected in Lebanon in October 2022. The outbreak lasted three months, with 8007 suspected cases (671 laboratory-confirmed) and 23 deaths. In this study, we use phenotypic methods and microbial genomics to study 34 clinical and environmental Vibrio cholerae isolates collected throughout this outbreak. All isolates are identified as V. cholerae O1, serotype Ogawa strains from wave 3 of the seventh pandemic El Tor (7PET) lineage. Phylogenomic analysis unexpectedly reveals the presence of two different strains of the seventh pandemic El Tor (7PET) lineage. The dominant strain has a narrow antibiotic resistance profile and is phylogenetically related to South Asian V. cholerae isolates and derived African isolates from the AFR15 sublineage. The second strain is geographically restricted and extensively drug-resistant. It belongs to the AFR13 sublineage and clusters with V. cholerae isolates collected in Yemen. In conclusion, the 2022-2023 Lebanese cholera outbreak is caused by the simultaneous introduction of two different 7PET strains. Genomic surveillance with cross-border collaboration is therefore crucial for the identification of new introductions and routes of circulation of cholera, improving our understanding of cholera epidemiology.
Similar content being viewed by others
Introduction
Cholera is an acute life-threatening diarrhoeal disease caused by two cholera toxin-producing serogroups—O1 and, less frequently, O139—of a Gram-negative bacterium, Vibrio cholerae; it occurs following the ingestion of contaminated water and food in endemic and epidemic settings1. Cholera continues to be a major public health problem, with 1.3–4.0 million cases and 21,000–143,000 deaths annually, according to the World Health Organisation (WHO)2. Cholera outbreaks are still raging globally, particularly in countries already bearing the brunt of natural disasters, human turmoil, and weak economic systems. Indeed, the two countries hardest hit by outbreaks in modern history are Yemen (2016–present) and Haiti (2010–2019; September 2022–present)) with record numbers of cases, at 2.5 million (as recorded in April 2021) and 820,000, respectively3,4.
Lebanon has witnessed several cholera outbreaks throughout its history, the most recent of these past outbreaks occurring between July and December 1993, with a total of 344 cases and 29 deaths5. On October 6, 2022, the Lebanese Ministry of Public Health (MoPH) notified the WHO of two laboratory-confirmed cases of cholera meeting all diagnostic criteria in the absence of an epidemiological link with a confirmed cholera outbreak (i.e. culture, including seroagglutination with V. cholerae O1-specific antisera, and confirmation of the presence of the cholera toxin genes by PCR) reported by the North and Akkar governorates in northern Lebanon. The index case, a 51-year-old Syrian man living in an informal settlement in Minieh-Donniyeh district (North governorate), was reported on October 5, 2022. This patient was admitted to the hospital on October 1 with rice–water diarrhoea and severe dehydration. The second case occurred in a 47-year-old health worker, probably through healthcare-associated transmission during care of the index case, resulting in the first nosocomial infection of this outbreak. Shortly after these two cases were identified, the epidemiological surveillance unit began detecting active cases in the informal settlement inhabited by the index case. In total, about 10 additional cases were diagnosed by laboratory testing. V. cholerae was also found in sources of drinking water, irrigation, and sewage (October 9, 2022). In parallel, two culture-confirmed cases were identified in Halba (the capital of Akkar Governorate). On October 10, an additional four cases were confirmed by culture in Syrian nationals living in an informal settlement in Aarsal, a town in the Baalbek district.
Within three months, the cholera outbreak spread across all eight governorates and 20 of the 25 districts in Lebanon (Fig. 1). The last positive case was recorded on January 5, 2023. As of June 2023, the cumulative number of reported cholera cases was 8007, with 671 cumulative culture-confirmed cases and 23 deaths. The outbreak was concentrated in the northern governorates of Akkar and North Lebanon and in the Beqaa Valley (Baalbek-Hermel and Beqaa governorates) (Fig. 1). About 29% of the cases concerned children aged 0–4 years, and 16% of all cases required hospitalisation. Daily hospitalisation rates peaked one week into the outbreak, with more than 220 patients hospitalised per day. During this period, the case fatality rate (CFR) for cholera reached 11% and the attack rate was highest in the northern districts of Akkar and Minieh-Donniyeh. The intensity of the outbreak necessitated the activation of a multisectoral response to increase cholera preparedness, including greater laboratory capacity for cases of acute watery diarrhoea (AWD) (testing of stools and water) and the training of surveillance and rapid response teams in the early identification of AWD cases. The response to cholera was also strengthened by introducing the oral cholera vaccine (OCV) and ensuring the maintenance of adequate support for intensive care units to prepare for emergencies and the provision of training in the management of cholera and other forms of AWD. The massive cholera vaccination campaign (more than 1 million people) conducted up to January 2023 helped to contain the disease but did not entirely eliminate the risk, particularly as only a single dose of vaccine was administered, rather than the recommended two doses, thus providing protection against cholera for about six months, rather than two years. There is, therefore, a risk of a resurgence of the disease.
a Number of suspected cholera cases per day vs. the confirmed number of cases per day in Lebanon until January 5, 2023. The dates of first isolation for the antimicrobial-resistant (AMR) and extensively drug-resistant XDR isolates are shown above the epidemic curve. The dates on which the isolates sequenced in this study were obtained are shown under the epidemic curve. b Cumulative attack rates (per 100,000) per governorate and district (within the governorate) of Lebanon are colour coded with shades of blue, where darker shades correspond progressively to a higher cumulative attack rate. The different governorates are labelled, and their limits are indicated by bold lines. Coloured circles indicate the number and type of sequenced isolates per governorate (Yellow: antimicrobial drug-resistant (AMR) and light purple: extensively drug-resistant (XDR)). These circles are randomly placed within each governorate. The five main cities are indicated in red (the city of Beirut belongs to the Beirut governorate). The small inlay map—which shows the geographic location of Lebanon (in red) in the Middle East—was created with MapChart (https://www.mapchart.net/). The map of Lebanon is a modified version of the image named Lebanon divisions (https://fr.wikipedia.org/wiki/Fichier:Lebanon_divisions.svg), created by Crates under the license CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0/deed.en).
Two different antimicrobial drug susceptibility profiles with different prevalences were observed in the V. cholerae O1 isolates obtained during the 2022–2023 Lebanese outbreak: a major profile with a narrow antimicrobial drug-resistant (AMR) spectrum and a minor profile with an extensively drug-resistant (XDR) spectrum. The first isolate with an AMR profile was recovered at the beginning of the outbreak (October 3), and the first XDR isolate was recovered 19 days later (October 22). Two scenarios could potentially account for this distribution: the first, considered the most likely a priori, involves the acquisition of a mobile genetic element, such as an MDR plasmid, by the strain with a narrower AMR profile initially responsible for the outbreak, following some kind of selection pressure, resulting in an XDR profile (as in Yemen)6. The second scenario, which is uncommon outside the Bay of Bengal, involves the circulation of two different strains introduced separately into Lebanon. We used whole-genome sequencing (WGS) and genomic epidemiology approaches to determine which of these two scenarios had occurred in Lebanon. We also delved into the genetic basis of antibiotic resistance and virulence in the circulating isolates.
Results
Serotyping and antimicrobial drug susceptibility testing
Serotyping and antimicrobial drug susceptibility testing in the Lebanese laboratories (LMSE and AUB) identified two different profiles—one AMR and one XDR (Table 1 and Supplementary Data 1)—in the 671 Vibrio cholerae O1 isolates recovered during the outbreak. The AMR profile predominated, accounting for 94.7% (636/671) of the isolates collected across Lebanon, including the 19 isolates tested at Institut Pasteur. It displayed resistance to nitrofurantoin, the vibriostatic agent O/129, and nalidixic acid only, and decreased susceptibility to ciprofloxacin. The second profile, XDR, was a minor profile accounting for only 5.6% (35/671) of the isolates. It was characterised by resistance to third-generation cephalosporins (cefotaxime and ceftazidime), macrolides (erythromycin and azithromycin), sulfonamides, the vibriostatic agent O/129, cotrimoxazole, nitrofurantoin and nalidixic acid, and decreased susceptibility to ciprofloxacin. This XDR profile was found exclusively in isolates originating from the South of Lebanon (Tyr) and the Beqaa Valley.
Phylogenetic and genomic features of V. cholerae O1 isolates
We selected 34 V. cholerae O1 isolates for further analysis on the basis of their AMR profiles (Fig. 1); 31 presented the predominant AMR profile (limited resistance) and three presented the minor XDR profile (Table 1). Genome sequencing confirmed that all 34 isolates belonged to serogroup O1, serotype Ogawa and biotype El Tor (sequence type ST69). All the V. cholerae O1 isolates (Table 1) displayed the following genomic features: (i) the ctxB7 variant of the cholera toxin subunit B gene, (ii) the toxin-coregulated pilus gene subunit A gene variant tcpACIRS101, and (iii) a deletion (ΔVC0495–0512) in the Vibrio seventh pandemic island II (VSP-II) (Table 1).
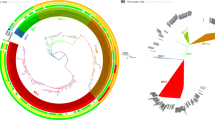
All 34 V. cholerae O1 isolates had (i) deletion of about 10 kb in the chromosomal ICEVchInd5 integrative and conjugative element, resulting in the loss of four genes encoding resistance to streptomycin (strA and strB), sulfonamides (sul2), and chloramphenicol (floR), but not the fifth gene encoding resistance to the vibriostatic agent O/129 (dfrA1), (ii) mutations of the chromosomal VC0715 (resulting in the R169C substitution) and VCA0637 (resulting in a premature codon stop at Q5) nitroreductase genes, leading to nitrofurantoin resistance, (ii) a mutation of the chromosomal VC1320 (vprA) (D89N) gene re-establishing susceptibility to polymyxin B, and (iv) mutations of the chromosomal DNA gyrase gyrA (S83I) and topoisomerase IV parC (S85L) genes, leading to nalidixic acid resistance and decreased susceptibility to ciprofloxacin. The three XDR isolates also carried an IncC of about 139 kb (formerly IncA/C2). This plasmid displayed 100% nucleotide sequence identity to the MDR plasmid, pCNRVC190243 (GenBank accession number OW443149.1)6 found in Vibrio cholerae O1 isolates from Yemen in 2018-2019 (Fig. 2)6. The backbone of pCNRVC190243 harboured a 20 kb pseudo-compound transposon, YemVchMDRI, flanked by IS26 insertion sequences and encompassing genes encoding aminoglycoside resistance (aadA2), a quaternary ammonium compound efflux pump (qac), sulfonamide resistance (two copies of the sul1 gene), an extended-spectrum beta-lactamase (ESBL; blaPER-7), and macrolide resistance (mph(A), mph(E), and msr(E)). The resistance to cotrimoxazole of the XDR isolates probably resulted from the simultaneous presence of the plasmid-borne sul1 gene and the chromosomal dfrA1 gene. The pCNRVC190243 plasmid had a nucleotide sequence 99.98% identical to that of pYA00120881 (GenBank accession number MT151380) identified in Zimbabwean V. cholerae O1 isolates collected in 2015 and 2018, but it carried a different multidrug-resistance region, containing, in particular, the ESBL gene blaCTX-M-157.
Circles from innermost to outermost indicate (1) the nucleotide position of the plasmid of the VIC_202210_72 isolate (alternative name pVIC-11A), (2) the alignment throughout the plasmids between pVIC-11A (Lebanon, 2022) in the dark green, pCNRVC190243 (Yemen, 2019) (GenBank accession number OW443149.1) in medium green, and pYA00120881 (Zimbabwe, 2018) (GenBank accession number MT151380) in light green, (3) the G + C content map of pVIC-11A, and (4) the coding sequences (CDS) map of pVIC-11A, in which green arrows indicate antimicrobial drug resistance CDS, dark blue arrows transposase and integrase CDS, red arrows the CDS involved in conjugative transfer, yellow arrows those involved in the structure and cellular processes, and light blue CDS with other functions. The names of resistance genes within the YemVchMDRI are indicated above the corresponding CDS.
One isolate (VIC_202211_60) also harboured a putative small Col3M colicin plasmid encoding a plasmid-mediated quinolone resistance protein, QnrD1, a member of the Qnr family protecting DNA-gyrase and topoisomerase IV against quinolones. However, this had no effect on the quinolone resistance profile of this isolate.
We then placed these 34 V. cholerae O1 isolates from the Lebanon 2022–2023 outbreak in a global context by constructing a maximum-likelihood phylogeny of 1471 7PET genomic sequences using 10,647 SNVs evenly distributed over the non-repetitive, non-recombinant core genome. All 34 isolates clustered in the genomic wave 3 clade of the 7PET lineage, and more particularly in the subclade containing isolates with the ctxB7 allele (Fig. 3). These 34 isolates also differed from other isolates previously recovered in Lebanon, including those isolated in 1970 and 1993, which clustered within genomic waves 1 and 2 of the 7PET lineage, respectively (Fig. 3).
A6 was used as the outgroup. Blue arrows represent the three genomic waves and the black arrow indicates the acquisition of the ctxB7 allele. The colour coding in the first column shows the geographic origins of the isolates, and African sublineages (AFR1, AFR3–AFR15) are shown on the left. The red colour in the second column indicates the Lebanese origin of the isolates. A magnification of the clades containing the two strains from Lebanon (red square corresponding to the predominant strain and blue square corresponding to the multidrug-resistant minor strain) is shown on the left with red text indicating the Lebanese isolates. For each genome, its name (or accession number), the country in which contamination occurred and the year of sample collection are indicated at the tip of the branch. Scale bars indicate the number of nucleotide substitutions per variable site. Blue dots correspond to bootstrap values ≥ 90%.
Our phylogenomic analysis revealed that the 34 V. cholerae O1 isolates were distributed between two different clusters according to their AMR profiles (Fig. 3). Indeed, the 31 isolates with the predominant AMR profile clustered together (median pairwise distance of 1.5 [range 0–9] core-genome SNVs) and with many other isolates originating from the Pakistan 2022 outbreak, from India (2019–2022), and one isolate from the Iraq 2022 outbreak. The three remaining isolates with the XDR profile clustered together (no SNV between them) and the Yemeni isolates recovered between 2016 and 2019.
Discussion
After approximately three decades without cholera, Lebanon recently suffered an outbreak extending from October 2022 to January 2023 (Fig. 1)8. The two different AMR profiles observed in the Lebanese isolates initially suggested the possibility of a mother strain relatively susceptible to antibiotics acquiring an XDR plasmid early in the outbreak, or of two different strains circulating simultaneously, this second possibility being considered less likely given that the first XDR isolate was recovered 19 days after the first AMR isolate. However, the high discriminatory power of WGS made it possible to distinguish two different strains of V. cholerae O1 serotype Ogawa harbouring the ctxB7 allele from genomic wave 3 of the 7PET lineage, and, thus, to conclude that the 2022-2023 cholera outbreak in Lebanon was caused by two phylogenetically distant strains rather than a single strain that subsequently acquired an MDR plasmid. The two-strain outbreak scenario was initially considered unlikely because, in countries in which cholera is not endemic, outbreaks generally occur following a single introduction of a single 7PET strain, contrasting with the situation in the Bay of Bengal, where many lineages circulate simultaneously9,10. This two-strain outbreak in Lebanon is, thus, unusual, as it stems from two different introductions outside the endemic setting. The two 7PET strains concerned had different AMR profiles and different patterns of circulation in Lebanon. The strain with the narrower AMR profile predominated in all affected regions of Lebanon, including North Lebanon in particular, whereas the XDR strain was found only in South Lebanon and the Beqaa Valley. This more limited distribution of the XDR strain may be due to the movements of people between these parts of Lebanon and Yemen in the current geopolitical context.
Globally, the 7PET lineage has been responsible for the repeated spread of the seventh pandemic from the Bay of Bengal in South Asia to the rest of the world through three epidemic waves11,12 The wave 3 clade carrying the ctxB7 allele first emerged in Kolkata (India) in 200613, subsequently spreading to other parts of the world, including Haiti and Yemen, and across Africa6,7,14,15.
In the global phylogenetic tree, the predominant Lebanese strain clustered with isolates from South Asia, including isolates from the 2022 Pakistani outbreak collected locally (GenBank bioproject PRJNA916827, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA916827) or from travellers from the US and Australia with a history of travel to Pakistan16 These isolates were considered the direct ancestors of the Lebanese strain (Fig. 3). Two published studies revealed that the strain circulating in the 2022 Pakistani outbreak was also the ancestor of a strain circulating in South Africa and Malawi considered to belong to the AFR15 sublineage16,17 We show here that the predominant Lebanese strain belongs to a clade containing African isolates from the AFR15 sublineage. This sublineage may, therefore, have been imported into the Middle East region directly from South Asia or indirectly via Africa. Interestingly, one isolate (PNUSAV00294, SRR20325463)16 collected in the US in June 2022 from a traveller with a history of recent travel to Iraq clustered with the Lebanese isolates, suggesting that the predominant strain in this Lebanese outbreak was the same strain that swept the region (Iraq and Syria) shortly before the Lebanese outbreak, causing outbreaks beginning on June 20, 2022, in Iraq18 and September 10, 2022, in Syria19 One month after the declaration of the Syrian outbreak, Lebanon declared its first index case in a Syrian refugee residing in North Lebanon, providing additional support for the theory that the predominant strain in Lebanon was imported from Syria. According to the United Nations High Commission for Refugees, Lebanon has the largest refugee population per capita in the world, with an estimated 1.5 million Syrian refugees living on its soil. Nevertheless, our ability to infer precise transmission routes through genomic analysis is hampered by the lack of availability of isolates from the various affected countries in the region, including Syria and Iraq.
The minor strain with an XDR profile grouped with isolates from the 2019 outbreak in Yemen, suggesting probable direct introduction of this strain from Yemen into South Lebanon and Beqaa, the only areas of Lebanon in which it was isolated, or indirect introduction via other unsampled countries in the region. Like the Yemeni isolates, this strain carried an IncC plasmid (pCNRVC190243) bearing determinants of resistance to cotrimoxazole, macrolides, third-generation cephalosporins, and aminoglycosides. The Yemeni epidemic comprised several waves but was seeded by a single introduction linked to the 7PET sublineage AFR13, which was recently transmitted from South Asia into East Africa and from there to Yemen6,20 Before 2018–2019, AFR13 isolates had features resembling those of the predominant Lebanese strain, including a narrow AMR profile, due partly to an ICEVchInd5 deletion and the subsequent loss of four of five AMR genes. However, a plasmid-carrying AFR13 clone, the pCNRVC190243-carrying AFR13 clone, began to emerge in late 2018 and displayed resistance to many therapeutically relevant drugs. The spread of this XDR clone was driven by the therapeutic overuse of macrolides6. Our XDR minor strain is, therefore, a Yemeni AFR13 clone carrying a self-transmissible MDR plasmid. The spread of this strain would greatly decrease treatment options and jeopardise cholera case management. This bleak scenario might occur if the XDR Yemeni clone manages to expand beyond its current geographic location and spread throughout Lebanon or if it passes its plasmid to the predominant strain (sublineage closely related to AFR15).
Similar IncC plasmids were previously observed in other V. cholerae strains, including the pYAM00120881 plasmid identified in Zimbabwe in 2018, which has a backbone almost identical to that of pCNRVC1902437. Intriguingly, the pYAM00120881-carrying AFR13 clone caused a 6-month-long outbreak in Zimbabwe, with over 10,000 suspected cases7 demonstrating a certain degree of plasmid stability in a 7PET V. cholerae strain sustaining an outbreak and even providing evidence against the presumed plasmid instability in this species6. It has been suggested that the 10 kb deletion in SXT/R391 ICE (ICEVchInd5) seen in our isolates might render these isolates fit to host MDR IncC plasmids stably, with deleterious consequences for antibiotic susceptibility in the future6.
Our genomic analysis revealed general consistency with the phenotypic AMR profile. All the isolates also harboured the catB9 gene, which does not confer chloramphenicol resistance11,21. Resistance to polymyxin B has been used as a marker of V. cholerae O1 biotype El Tor since the start of the seventh cholera pandemic, as a means of differentiating this biotype from the classical biotype, which was susceptible to polymyxin B. The susceptibility to polymyxins of our isolates may be due to the VprA (VC1320) D89N substitution, as previously reported in the AFR13 and AFR14 sublineages15,20
One limitation of this study is that only a small proportion (5%, 34/671) of the outbreak isolates were sequenced. We cannot, therefore, rule out the possibility that some of the 637 non-sequenced V. cholerae O1 isolates do not belong to the 7PET lineage, the only lineage currently causing epidemic cholera. The intensive use of culture media for the isolation of Vibrio species (such as TCBS) during cholera outbreaks can lead to the isolation of a non-pathogenic Vibrio spp. from a patient with diarrhoea, not due to cholera. Lassalle and coworkers6 sequenced 5.7% (250/4,375) of the V. cholerae O1 isolates recovered from suspected cholera patients in Yemen between 2018 and 2019 and found that 8% (n = 21) of these isolates were not toxigenic and did not belong to the 7PET lineage. The proportion of non-7PET isolates reached 30% (3/10) for their environmental isolates. However, only two of these 24 non-7PET isolates—from clades VcK and ST170—were assigned to serogroup O1; the others belonged to other O groups (O7, O34, O146, O167). Here, we include only V. cholerae isolates clearly demonstrated to belong to the O1 serogroup. Furthermore, a random selection of 60 non-sequenced isolates was shown to contain the ctx gene. However, the presence of the ctx gene in V. cholerae O1 is not a 100% reliable indicator of membership of the 7PET lineage because this feature is also observed in other V. cholerae O1 lineages, such as L3, which includes the US Gulf Coast clone14,16. These non-7PET lineages are associated with sporadic seafood-borne infections but not epidemic cholera14. Based on our data and the epidemiological context of this explosive but short-lived cholera outbreak in Lebanon, we are confident that only a very small proportion of the 637 non-sequenced isolates were not the true toxigenic agent of cholera.
Lebanon has been struggling with an unprecedented, multifaceted crisis since 2019, including severe economic collapse, the COVID-19 pandemic, the explosion in the Port of Beirut in August 2020, and a high burden of refugees. This major crisis and the fragile infrastructure of Lebanon favoured this outbreak. The economic crisis, with all its implications, affected all aspects of the outbreak. Access to safe water was hampered by the lack of electricity and the inadequacy of the sewage system. The country was already suffering from a shortage of medical and diagnostic supplies in addition to the global shortage of oral cholera vaccines, and laboratory supplies for cholera diagnosis. Nevertheless, tremendous collaborative efforts were initiated, under the auspices of the Ministry of Public Health in Lebanon and in collaboration with several national and international organisations including, but not limited to, the WHO, UNICEF, UNHCR, and ICRC, making it possible to halt the spread of the disease within three months of the declaration of the index case. However, we are well aware of the possibility of disease resurgence and of another outbreak, particularly as the neighbouring countries have not yet brought their own outbreaks under control.
In conclusion, the outbreak in Lebanon was caused by two different 7PET strains: one with a narrower AMR profile related to South Asian isolates and the other with an XDR profile similar to the Yemeni AFR13 V. cholerae strain. However, as isolates from the neighbouring countries are missing from the phylogenetic analysis, it may be difficult to establish a comprehensive history for this outbreak. Regional surveillance of the causal agent of cholera by microbial genomics methods is, thus, paramount for the reliable inference of transmission routes and for tracking and monitoring the emergence of any AMR, particularly after the worrying switch of several AFR13 strains from a limited AMR to an XDR phenotype following the acquisition of IncC-type plasmids6.
Methods
Ethics statement
In this study, only deidentified data was used as it was based exclusively on bacterial isolates and the corresponding metadata collected for nationwide surveillance of the cholera outbreak by the Lebanese Ministry of Public Health (MoPH) in collaboration with the American University of Beirut (AUB), Rafic Hariri University Hospital, and the Laboratoire Microbiologie Santé et Environnement (LMSE).
Vibrio cholerae isolates
Stools, sewage, water and plant samples were collected by the MoPH and delivered to the bacteriology and molecular microbiology research laboratory at AUB, a WHO collaborating centre for reference and research on bacterial pathogens. In total, 671 clinical isolates of V. cholerae were identified, with 144 isolates from North Lebanon collected and stored in CMUL (la Collection Microbiologique de l’Université Libanaise) at LMSE at the Lebanese University. The remaining 527 isolates were stored at AUB. We included 18 isolates from AUB and 16 from the LMSE in this study (Supplementary Data 1). These isolates were recovered between October and December 2022, for continuous surveillance and prevention, the last positive case being reported on January 5th, 2023.
Bacterial culture and identification
The approach to bacterial culture and identification differed between the LMSE and AUB laboratories. At the LMSE, part of each stool sample was plated directly on two different media, a non-selective nutrient-rich agar medium (pH = 8.5, 10 g/l NaCl), and a selective agar medium, thiosulfate-citrate-bile salts-sucrose (TCBS) agar (BioMérieux, Marcy-l’Etoile, France). Another portion of each sample was incubated in alkaline peptone water (Bio-Rad, Marnes-la-Coquette, France; #12013259 with 10 g/l NaCl) for 6–8 h at 35–37 °C and was then plated on the same solid media. By contrast, at AUB, the stool sample was incubated in alkaline peptone water for 6 h at 35 °C and was then plated on the surface of TCBS agar, MacConkey agar (Bio-Rad #3564154), and Vibrio Chromagar (CHROMagar, Paris, France, VB912). After standard microbiological identification by microscopy (a comma-shaped Gram-negative bacterium) and oxidase tests, V. cholerae isolates were identified with API 20E test strips (BioMérieux, 20160) and by matrix-assisted laser desorption ionisation–time of flight mass spectrometry (MALDI-TOF), with Vitek® MS (BioMérieux) at LMSE or MALDI Biotyper (Bruker Daltonics, Germany) at AUB. Agglutination was performed with specific antisera (V. cholerae O1 Inaba Monovalent Antiserum (M11003), V. cholerae O1 Ogawa Monovalent Antiserum (M11004), V. cholerae O1 Polyvalent (Inaba, Ogawa) Antiserum (M11002) and V. cholerae O139 Antiserum (M15001) (Mast Assure, Liverpool, UK)), for all 671 isolates.
DNA preparation
The presence of cholera toxin genes was confirmed with a multiplex PCR method developed by Hoshino and colleagues22 for the isolates from the first five cases and 60 randomly selected non-sequenced isolates. Briefly, the bacterial isolates were cultured overnight in LB broth at 37 °C. The culture was then diluted 1:10 in a 10 mM Tris–HCl (pH 8.0) buffer with 1 mM disodium ethylenediaminetetraacetic acid (EDTA), and then boiled for 10 minutes. The mixture was centrifuged at 12,000×g for 10 min at 4 °C, and the resulting supernatant was used as the PCR template22.
PCR primers
The forward and reverse primers are listed here after each V. cholerae target, respectively: O139-rfb: O139F2 [5′-AGCCTCTTTATTACGGGTGG-3′] and O139R2 [5′-GTCAAACCCGATCGTAAAGG-3′], O1-rfb: O1F2-1 [5′-GTTTCACTGAACAGATGGG-3′] and O1R2-2 [5′-GGTCATCTGTAAGTACAAC-3′], ctxA: VCT1 [5′- ACAGAGTGAGTACTTTGACC-3′] and VCT2 [5′- ATACCATCCATATATTTGGGAG-3′]. All primers were purchased from Integrated DNA Technologies (IDT), Coralville, IA.
PCR protocol
Simultaneous amplification with the three primer pairs was performed in 0.2 mL microcentrifuge tubes. PCR reactions were prepared using ready-to-load Solis Biodyne FIREPol Master Mix (Tartu, Estonia, 04-12-00115) to which 3 μL of sample were added. The thermal cycling conditions used were as follows: 5 min at 94 °C for initial denaturation of DNA and 35 cycles, each consisting of 1 min at 94 °C, 1 min at 55 °C, 1 min at 72 °C, with a final extension step for 7 min at 72 °C in a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Hercules, CA, 1855196). After amplification, 6 μL of each reaction mixture was separated by electrophoresis on a 3% agarose gel and the amplified gene products were visualised under UV light after staining with ethidium bromide.
Antimicrobial drug susceptibility testing
Antimicrobial drug susceptibility testing was performed by the disk diffusion method in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines. Tests were performed for 14 antimicrobial agents: piperacillin/tazobactam, cefotaxime, ceftazidime, meropenem, nalidixic acid, ciprofloxacin, levofloxacin, erythromycin, azithromycin, trimethoprim-sulfamethoxazole (cotrimoxazole), tetracycline, doxycycline, nitrofurantoin, and vibriostatic agent O/129. CLSI interpretative criteria for the antibiotic susceptibility testing of Vibrio spp. (M45 document) were used when available23 For vibriostatic agent O/129 (equivalent to trimethoprim), nitrofurantoin, nalidixic acid, and ciprofloxacin, the interpretative criteria for Enterobacteriaceae/Salmonella spp. (M100-S30 document) were used24 The minimum inhibitory concentrations (MICs) of colistin for 19 isolates—16 from the LMSE and 3 from the AUB—were determined at Institut Pasteur with the SensititreTM system (Thermo Fisher Scientific, Waltham, MA, V3000-VZ). Isolates resistant to at least one of the tested antimicrobial drugs were classified as antimicrobial drug-resistant (AMR). Isolates simultaneously resistant to azithromycin, cotrimoxazole, and third-generation cephalosporins (cefotaxime and/or ceftazidime) were classified as extensively drug-resistant (XDR).
Whole-genome sequencing
We studied 34 V. cholerae isolates (Supplementary Data 1), 16 originating from the LMSE, which were sequenced at the Institut Pasteur in Paris, and 18 isolates originating from AUB, which were sequenced in-house. At Institut Pasteur, genomic DNA was extracted with the Maxwell 16-cell purification kit (Promega, Madison, WI, AS1020). The DNA libraries were then prepared at the Institut Pasteur Mutualized Platform for Microbiology (P2M) with the Nextera XT kit (Illumina, San Diego, CA, USA, FC-131) and sequencing was performed with the NextSeq 500 system (Illumina, 20024904), generating 150 bp paired-end reads. At AUB, genomic DNA was extracted with the Quick-DNA™ Fungal/Bacterial Miniprep kit (Zymo Research, Irvine, CA, D6005) and purified with the Genomic DNA Clean & Concentrator™ kit (Zymo Research, D4003), according to the manufacturer’s protocols. DNA libraries were prepared with the Illumina DNA prep kit and subjected to 2 × 150 bp paired-end sequencing by Illumina MiSeq (Illumina, SY-410-1003).
The resulting reads were filtered with FqCleanER version 21.10 (https://gitlab.pasteur.fr/GIPhy/fqCleanER) with options -q 28 -l 70 to discard low-quality reads with phred scores <28 and length <70 bp and to remove adaptor sequences25.
Long-read sequencing and complete genome circularisation
Two isolates were selected for long-read sequencing, based on geographic origin and representativity of the two circulating strains. Long-read sequencing was performed on isolate CNRVC220127 (alternative name CMUL 009) at Institut Pasteur, with a MinION nanopore sequencer (Oxford Nanopore Technologies), the library was prepared according to the instructions of the “Native barcoding genomic DNA (with EXP-NBD104, 54 EXP-NBD114, and SQK-LSK109)” procedure provided by Oxford Nanopore Technologies. The sequencing was then completed using the MinION Mk1C apparatus (Oxford Nanopore Technologies, Oxford, UK, MIN-101C). The second isolate sequenced (VIC_202210_72, alternative name VIC11A) was cultured on MacConkey agar. DNA was extracted with the Quick DNA Fungal/Bacterial Miniprep Kit (by ZYMO Research) and cleaned with DNA Clean & Concentrator -5 (by ZYMO Research). The clean DNA was then sequenced with an Oxford Nanopore MinION at AUB. The DNA library was prepared with Rapid Barcoding Kit 96 (SQK-RBK110.96) and sequenced on R9.4.1 flow cells (FLO-MIN106) (Oxford Nanopore Technologies).
The sequences of the two isolates were assembled from both long and short reads, by two different methods. For CNRVC220127, a hybrid approach was used in UniCycler25 v.0.4.8. For VIC_202210_72, a combination of Raven26 v.1.6.0 (https://github.com/lbcb-sci/raven), Medaka v.1.4.4 (https://github.com/nanoporetech/medaka) and Polypolish27 v.0.5.0 (https://github.com/rrwick/Polypolish/) was used. The large plasmid of VIC_202210_72 was then annotated with Bakta28 v.1.5.0, corrected manually and visualised with BRIG v.0.95 (http://sourceforge.net/projects/brig)25.
Additional genomic data
For the construction of a globally representative set of isolates, we downloaded and included in this study sequences available either in raw-read format or as assembled genomes in the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena) and GenBank (https://www.ncbi.nlm.nih.gov/genbank/) databases (Supplementary Data 2).
Genomic sequence analyses
The paired-end reads and draft or assembled genomes were mapped onto the reference genome of V. cholerae O1 El Tor strain N16961, also known as A19 (GenBank accession numbers LT907989 and LT907990) with Snippy v.4.6.0/BWA v.0.7.17 (https://github.com/tseemann/snippy). The single-nucleotide variants (SNVs) were then called with Snippy v.4.6.0/Freebayes v.1.3.2, using the following parameters: a minimum read coverage of 4, a minimum base quality of 13, a mapping quality of 60, and a 75% read concordance at a locus for a variant to be reported. Finally, core-genome SNVs were then aligned in Snippy for phylogenetic inference.
Repetitive sequences (insertion sequences and the TLC-RS1-CTX region) and recombinogenic regions (VSP-II) were masked11. Putative recombinogenic regions were identified and masked with Gubbins29 v.3.2.0. A maximum likelihood (ML) phylogenetic tree was constructed from an alignment of 10,647 chromosomal SNVs, with RAxML30 v.8.2.12, under the GTR model, with 200 repetitions for bootstrapping (Supplementary Data 3). This global tree was rooted in the A6 genome—the earliest and most ancestral seventh pandemic isolates, collected in Indonesia in 195711—and visualised with iTOL31 v.6 (https://itol.embl.de/).
Short reads from Illumina were assembled de novo with SPAdes32 v.3.15.2. The presence of various genetic markers (O1 rfb gene, whole locus of VSP-II, ctxB, and wbeT) was investigated with BLAST v.2.2.26 against reference sequences. The presence and type of acquired antibiotic resistance genes (ARGs) or ARG genetic structures were investigated with ResFinder v.4.0.1 (https://cge.cbs.dtu.dk/services/ResFinder/), PlasmidFinder v.2.1.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/), and BLAST analysis against GI-15, Tn7, and SXT/R391 integrative and conjugative elements (ICE)11 The sequences assembled de novo were examined with BLAST to look for mutations of genes encoding resistance to nitrofurans (VC_0715 and VC_A0637), resistance to quinolones (gyrA and parC) or restoring susceptibility to polymyxin B (vprA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Short reads were submitted to the ENA under study project PRJEB65303 and are listed in Supplementary Data 2. Assemblies resulting from long-read sequencing were submitted to GenBank under project PRJNA1013428 (CP134060-CP134061 for CNRVC220127/CMUL009 and CP134057-CP134059 for VIC11-A).
Code availability
The fq2dna script (genome de novo assembly from raw paired-end FASTQ files) can be found at https://gitlab.pasteur.fr/GIPhy/fq2dna.
References
Clemens, J. D., Nair, G. B., Ahmed, T., Qadri, F. & Holmgren, J. Cholera. Lancet 390, 1539–1549 (2017).
Ganesan, D., Gupta, S. S. & Legros, D. Cholera surveillance and estimation of burden of cholera. Vaccine 38, A13–A17 (2020).
World Health Organization. Cholera Situation in Yemen. WHO-EM/CSR/434/E (World Health Organization, accessed 12 January 2023); https://applications.emro.who.int/docs/WHOEMCSR434E-eng.pdf?ua=1 .
World Health Organization. Disease Outbreak News; Cholera—Haiti (World Health Organization, accessed 25 January 2023); https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON427#:~:text=Between%202%20October%20through%206,ten%20departments%20in%20the%20country.
Harb, H. Compiled Literature Report on Selected Health Conditions in Lebanon (Lebanese Ministry of Public Health, Beirut, Lebanon, 2004).
Lassalle, F. et al. Genomic epidemiology reveals multidrug resistant plasmid spread between Vibrio cholerae lineages in Yemen. Nat. Microbiol. 8, 1787–1798 (2023).
Mashe, T. et al. Highly resistant cholera outbreak strain in Zimbabwe. N. Engl. J. Med. 383, 687–689 (2020).
Lebanese Ministry of Public Health. Cholera in Lebanon (Lebanese Ministry of Public Health, accessed 23 July 2023); http://www.moph.gov.lb.
Morita, D. et al. Whole-genome analysis of clinical Vibrio cholerae O1 in Kolkata, India, and Dhaka, Bangladesh, reveals two lineages of circulating strains, indicating variation in genomic attributes. mBio 11, e01227–20 (2020).
Mutreja, A. & Dougan, G. Molecular epidemiology and intercontinental spread of cholera. Vaccine 38, A46–A51 (2020).
Weill, F.-X. et al. Genomic history of the seventh pandemic of cholera in Africa. Science (1979) 358, 785–789 (2017).
Mutreja, A. et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465 (2011).
Naha, A. et al. Development and evaluation of a PCR assay for tracking the emergence and dissemination of Haitian variant ctxB in Vibrio cholerae O1 strains isolated from Kolkata, India. J. Clin. Microbiol. 50, 1733–1736 (2012).
Domman, D. et al. Integrated view of Vibrio cholerae in the Americas. Science (1979) 358, 789–793 (2017).
Benamrouche, N. et al. Outbreak of imported seventh pandemic Vibrio cholerae O1 El Tor, Algeria, 2018. Emerg. Infect. Dis. 28, 1241–1245 (2022).
Smith, A. M. et al. Imported cholera cases, South Africa, 2023. Emerg. Infect. Dis. 29, 1687–1690 (2023).
Chabuka, L. et al. Genomic epidemiology of the cholera outbreak in Malawi 2022–2023. Preprint at medRxiv https://doi.org/10.1101/2023.08.22.23294324 (2023).
Qamar, K. et al. Rise of cholera in Iraq: a rising concern. Ann. Med. Surg. 81, 104355 (2022).
Eneh, S. C. et al. Cholera outbreak in Syria amid humanitarian crisis: the epidemic threat, future health implications, and response strategy—a review. Front. Public Health 11, 1161936 (2023).
Weill, F.-X. et al. Genomic insights into the 2016–2017 cholera epidemic in Yemen. Nature 565, 230–233 (2019).
Kumar, P., Yadav, P., Nema, A., Goel, A. K. & Yadava P. K. Re-emergence of chloramphenicol resistance and associated genetic background in Vibrio cholerae O1. FASEB 31, 907.3–907.3 (2017).
Hoshino, K. et al. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol. Med. Microbiol. 20, 201–207 (1998).
CLSI M45-ED3. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria (The Clinical & Laboratory Standards Institute, 2016).
CLSI M100-S30. Performance Standards for Antimicrobial Susceptibility Testing (The Clinical & Laboratory Standards Institute, 2020).
Lefèvre, S. et al. Rapid emergence of extensively drug-resistant Shigella sonnei in France. Nat. Commun. 14, 462 (2023).
Vaser, R. & Šikić, M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 1, 332–336 (2021).
Wick, R. R. & Holt, K. E. Polypolish: short-read polishing of long-read bacterial genome assemblies. PLoS Comput. Biol. 18, e1009802 (2022).
Schwengers, O. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 7, 000685 (2021).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15–e15 (2015).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Acknowledgements
We would like to thank all the staff and personnel of the laboratories participating in this study, including the American University of Beirut and the American University of Beirut Medical Center, the LMSE, Rafik Hariri University Hospital, the Ministry of Public Health Lebanon, The Lebanese Red Cross, Institut Pasteur, the WHO, and the CDC. This work was funded by the Centers for Disease Control (CDC) award number BAA 75D301-21-C-12132, a grant awarded to the American University of Beirut (G.M.M. and A.A.F.), WHO country office Lebanon (A.A.), the Lebanese University, Institut Pasteur, and by the French Government’s Investissement d’Avenir programme, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant no. ANR-10-LABX-62-IBEID) (F.-X.W. and M.-L.Q.).
Author information
Authors and Affiliations
Contributions
A.A.F., M.H., F.-X.W. and G.M.M. conceived and designed the study. A.A.F., R.R., E.N., J.E., H.H., S.S., J.-R.G., S.B., A.S., M.A., M.B. and L.H. performed the experimental work. R.M., F.D., R.F., Z.M., A.R., N.G., F.A., A.A., A.B., N.W., M.L.Q., A.A.F., R.R., H.H., E.N., F.-X.W. and G.M.M. conducted the data analysis. A.A.F., R.R., E.N., J.E. and H.H. wrote the initial draft and all authors contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Balaji Veeraraghavan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Abou Fayad, A., Rafei, R., Njamkepo, E. et al. An unusual two-strain cholera outbreak in Lebanon, 2022-2023: a genomic epidemiology study. Nat Commun 15, 6963 (2024). https://doi.org/10.1038/s41467-024-51428-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51428-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.